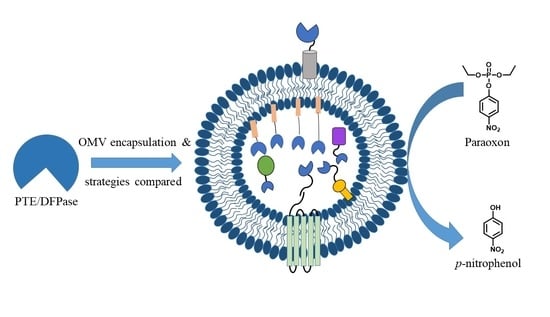
Different Strategies Affect Enzyme Packaging into Bacterial Outer Membrane Vesicles
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Growth and OMV Purification
2.2. Genetic Contstructs
2.3. NanoSight
2.4. mCherry Fluorescence
2.5. PTE and DFPase Activity Assay
2.6. SDS-PAGE and Western Blot Analysis
3. Results
3.1. PTE and DPTase OMV Packaging Strategy Design Rationale
| Name | Description/Design | Uniprot ID of Parent Protein | Membrane-Associated? | Reference |
|---|---|---|---|---|
| Lpp’ | Trunicated, 9 amino acid peptide derived from Lpp outer membrane lipoprotein from E. coli; contains modified N-terminal Cys post-cleavage | P69776 | Yes | [18] |
| L3 | A short Gly-Pro-rich, semi-rigid linker appended to Lpp’ | N/A | Yes | This work |
| L4 | (P-A-[S/G/P/T])^N linker, based on the rigid proline-rich linker (PAPAP)^N; appends 50% protein linker to Lpp’ | N/A | Yes | [21,22] |
| L34 | L4 appended to L3 (Lpp’-L3-L4) | N/A | Yes | [14] |
| OpmA-ST-SC | The two-part SpyTag-SpyCatcher system, with OmpA-fused SpyTag and PTE or DFPase fused to SpyCatcher | P0A910 | No | [11] |
| SC | PTE or DFPase fused to SpyCatcher; only the SpyCatcher protein component of the SpyTag-SpyCatcher system | N/A | No | [11,27] |
| BtuF | E. coli protein; periplasmic binding protein for the vitamin B12 transporter BtuCD | P37028 | No | [26] |
| MBP | Maltose-binding protein | P0AEX9 | No | [25] |
| SLP | Outer membrane lipoprotein; SLP may help to stabilize the outer membrane in stationary phase | P37194 | Yes | [24] |
| SlyB | Small outer membrane lipoprotein conserved in Gram-negative bacteria | P0A905 | Yes | [17] |
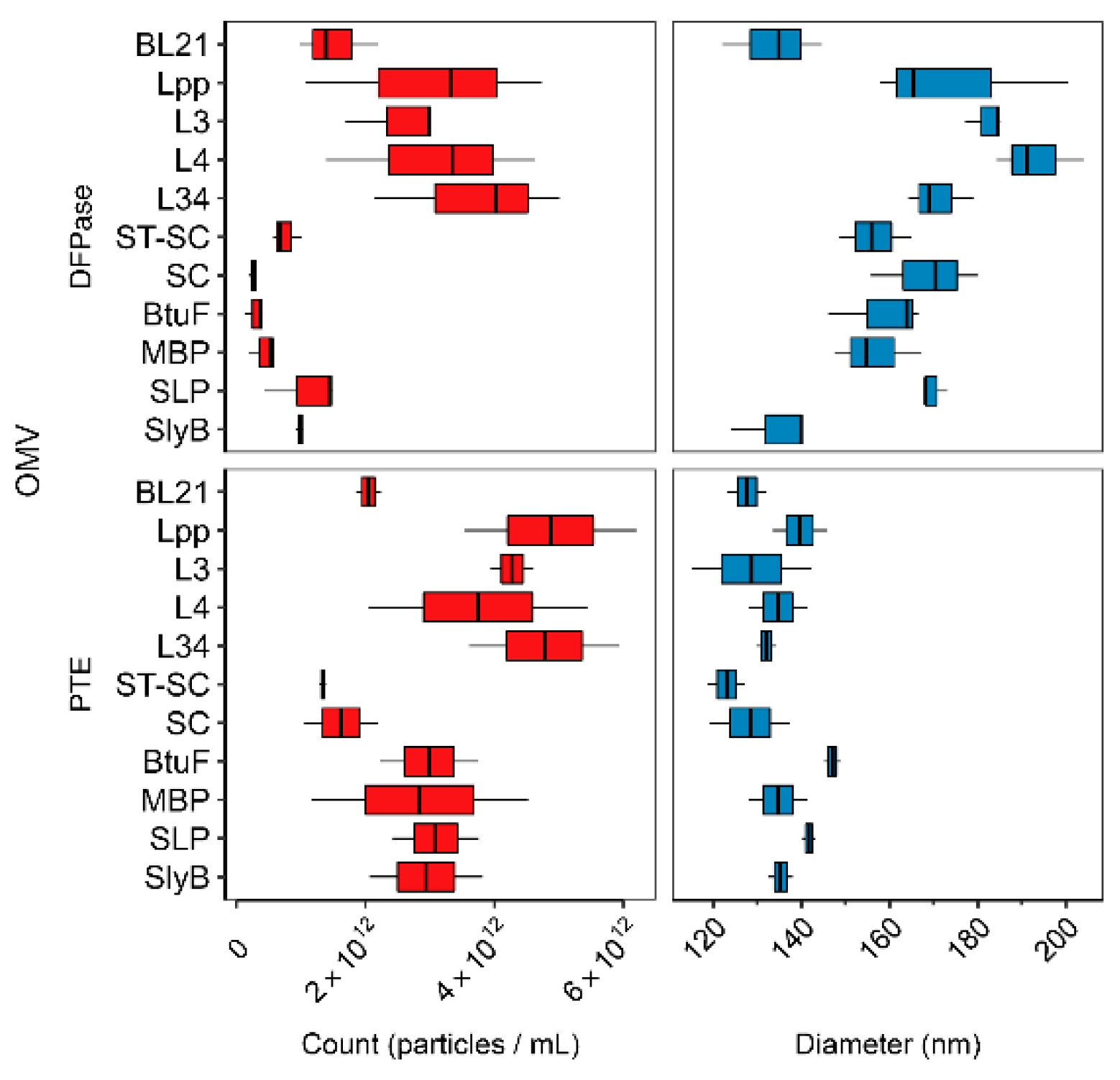
3.2. Linker Type Affects OMV Production and Size
3.3. Activity of OMV-Packaged Enzymes Was Influenced by Linkers
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Bang, J.Y.; Park, G.W.; Choi, D.S.; Kang, J.S.; Kim, H.J.; Park, K.S.; Lee, J.O.; Kim, Y.K.; Kwon, K.H.; et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 2007, 7, 3143–3153. [Google Scholar] [CrossRef] [PubMed]
- Bomberger, J.M.; Maceachran, D.P.; Coutermarsh, B.A.; Ye, S.; O’Toole, G.A.; Stanton, B.A. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009, 5, e1000382. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.G.L.; Davey, H.M.; Cookson, A.; Currinn, H.; Cooke-Fox, G.; Stanczyk, P.J.; Whitworth, D.E. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 2012, 158, 2742–2752. [Google Scholar] [CrossRef]
- Dominguez Rubio, A.P.; Martinez, J.H.; Martinez Casillas, D.C.; Coluccio Leskow, F.; Piuri, M.; Perez, O.E. Lactobacillus casei BL23 Produces Microvesicles Carrying Proteins That Have Been Associated with Its Probiotic Effect. Front. Microbiol. 2017, 8, 1783. [Google Scholar] [CrossRef]
- Liu, Y.; Alexeeva, S.; Defourny, K.A.; Smid, E.J.; Abee, T. Tiny but mighty: Bacterial membrane vesicles in food biotechnological applications. Curr. Opin. Biotechnol. 2018, 49, 179–184. [Google Scholar] [CrossRef]
- Marco, M.L.; Smid, E.J. Editorial overview: Food biotechnology: Exploration and exploitation of microbial resources to address the need for sustainable production of safe, healthy and nutritious food. Curr. Opin. Biotechnol. 2018, 49, v–vii. [Google Scholar] [CrossRef]
- Alves, N.J.; Turner, K.B.; Medintz, I.L.; Walper, S.A. Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci. Rep. 2016, 6, 24866. [Google Scholar] [CrossRef]
- Thakur, M.; Medintz, I.L.; Walper, S.A. Enzymatic Bioremediation of Organophosphate Compounds-Progress and Remaining Challenges. Front. Bioeng. Biotechnol. 2019, 7, 289. [Google Scholar] [CrossRef]
- Robb, E.L.; Baker, M.B. Organophosphate Toxicity. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Alves, N.J.; Turner, K.B.; Daniele, M.A.; Oh, E.; Medintz, I.L.; Walper, S.A. Bacterial Nanobioreactors--Directing Enzyme Packaging into Bacterial Outer Membrane Vesicles. ACS Appl. Mater. Interfaces 2015, 7, 24963–24972. [Google Scholar] [CrossRef]
- Alves, N.J.; Moore, M.; Johnson, B.J.; Dean, S.N.; Turner, K.B.; Medintz, I.L.; Walper, S.A. Environmental Decontamination of a Chemical Warfare Simulant Utilizing a Membrane Vesicle-Encapsulated Phosphotriesterase. ACS Appl. Mater. Interfaces 2018, 10, 15712–15719. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Dean, S.N.; Moore, M.; Spangler, J.R.; Johnson, B.J.; Medintz, I.L.; Walper, S.A. Packaging of Diisopropyl Fluorophosphatase (DFPase) in Bacterial Outer Membrane Vesicles Protects Its Activity at Extreme Temperature. ACS Biomater. Sci. Eng. 2022, 8, 493–501. [Google Scholar] [CrossRef]
- Melzer, M.; Heidenreich, A.; Dorandeu, F.; Gab, J.; Kehe, K.; Thiermann, H.; Letzel, T.; Blum, M.M. In vitro and in vivo efficacy of PEGylated diisopropyl fluorophosphatase (DFPase). Drug. Test. Anal. 2012, 4, 262–270. [Google Scholar] [CrossRef]
- Bigley, A.N.; Raushel, F.M. Catalytic mechanisms for phosphotriesterases. Biochim. Biophys. Acta 2013, 1834, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Rozovsky, S.; Chen, W. Engineering multi-functional bacterial outer membrane vesicles as modular nanodevices for biosensing and bioimaging. Chem. Commun. 2017, 53, 7569–7572. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Stephens, D.L.; Stathopoulos, C.; Poetschke, H.L.; Mendenhall, J.; Earhart, C.F. Display of beta-lactamase on the Escherichia coli surface: Outer membrane phenotypes conferred by Lpp’-OmpA’-beta-lactamase fusions. Protein Eng. 1996, 9, 239–247. [Google Scholar] [CrossRef]
- Irene, C.; Fantappie, L.; Caproni, E.; Zerbini, F.; Anesi, A.; Tomasi, M.; Zanella, I.; Stupia, S.; Prete, S.; Valensin, S.; et al. Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. Proc. Natl. Acad. Sci. USA 2019, 116, 21780–21788. [Google Scholar] [CrossRef]
- Huang, F.; Spangler, J.R.; Huang, A.Y. In vivo cloning of up to 16 kb plasmids in E. coli is as simple as PCR. PLoS ONE 2017, 12, e0183974. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Zhao, H.L.; Yao, X.Q.; Xue, C.; Wang, Y.; Xiong, X.H.; Liu, Z.M. Increasing the homogeneity, stability and activity of human serum albumin and interferon-alpha2b fusion protein by linker engineering. Protein Expr. Purif. 2008, 61, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Alexander, D.M.; St John, A.C. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol. Microbiol. 1994, 11, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Kapust, R.B.; Waugh, D.S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999, 8, 1668–1674. [Google Scholar] [CrossRef]
- Karpowich, N.K.; Huang, H.H.; Smith, P.C.; Hunt, J.F. Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J. Biol. Chem. 2003, 278, 8429–8434. [Google Scholar] [CrossRef] [PubMed]
- Kasaraneni, N.; Chamoun-Emanuelli, A.M.; Wright, G.; Chen, Z. Retargeting Lentiviruses via SpyCatcher-SpyTag Chemistry for Gene Delivery into Specific Cell Types. mBio 2017, 8, e01860-17. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, A.; Prevelige, P.E.; Basu, G.; Douglas, T. Coconfinement of fluorescent proteins: Spatially enforced communication of GFP and mCherry encapsulated within the P22 capsid. Biomacromolecules 2012, 13, 3902–3907. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, M.; Badieyan, S.; Marsh, E.N.G. Immobilized enzymes: Understanding enzyme—Surface interactions at the molecular level. Org. Biomol. Chem. 2017, 15, 9539–9551. [Google Scholar] [CrossRef]
- Küchler, A.; Yoshimoto, M.; Luginbuhl, S.; Mavelli, F.; Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol. 2016, 11, 409–420. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Sullivan, C.J.; Kuehn, M.J. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 2013, 52, 3031–3040. [Google Scholar] [CrossRef]
- Dean, S.N.; Turner, K.B.; Medintz, I.L.; Walper, S.A. Targeting and delivery of therapeutic enzymes. Ther. Deliv. 2017, 8, 577–595. [Google Scholar] [CrossRef] [PubMed]



| Name | PTE Initial Rate (log2) | DFPase Intial Rate (log2) | pI | Boman Index | Mol. Weight | Charge | Hp 1 | Length | Instability Index | Aliphatic Index |
|---|---|---|---|---|---|---|---|---|---|---|
| Lpp’ | 1.1 | 1.6 | 6.2 | 1.3 | 1739.7 | −0.1 | −0.06 | 21.0 | 49.9 | 23.3 |
| L3 | 1.4 | 1.5 | 6.2 | 0.9 | 2791.9 | −0.1 | −0.5 | 34.0 | 37.8 | 23.2 |
| L4 | 3.1 | 2.0 | 6.2 | 0.5 | 3285.5 | −0.1 | −0.5 | 39.0 | 108.3 | 27.9 |
| L34 | 11.6 | 2.7 | 6.2 | 0.5 | 4337.6 | −0.1 | −0.4 | 52.0 | 81.1 | 26.7 |
| BtuF | 9.6 | 1.5 | 7.3 | 1.6 | 30,093.1 | 0.0 | −0.4 | 274.0 | 39.6 | 89.7 |
| MBP | 5.5 | 1.9 | 4.8 | 1.4 | 43,378.0 | −10.6 | −0.4 | 396.0 | 20.8 | 81.4 |
| SLP | 2.8 | 1.6 | 6.5 | 1.5 | 22,299.9 | −1.7 | −0.4 | 201.0 | 34.5 | 81.9 |
| SlyB | 1.1 | 1.7 | 19.2 | 1.4 | 18,742.9 | 2.0 | −0.1 | 185.0 | 28.2 | 87.8 |
| SC | 4.9 | 1.4 | 12.2 | 2.1 | 6540.3 | 3.5 | −0.5 | 61.0 | 39.5 | 73.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dean, S.N.; Thakur, M.; Spangler, J.R.; Smith, A.D.; Garin, S.P.; Walper, S.A.; Ellis, G.A. Different Strategies Affect Enzyme Packaging into Bacterial Outer Membrane Vesicles. Bioengineering 2023, 10, 583. https://doi.org/10.3390/bioengineering10050583
Dean SN, Thakur M, Spangler JR, Smith AD, Garin SP, Walper SA, Ellis GA. Different Strategies Affect Enzyme Packaging into Bacterial Outer Membrane Vesicles. Bioengineering. 2023; 10(5):583. https://doi.org/10.3390/bioengineering10050583
Chicago/Turabian StyleDean, Scott N., Meghna Thakur, Joseph R. Spangler, Aaron D. Smith, Sean P. Garin, Scott A. Walper, and Gregory A. Ellis. 2023. "Different Strategies Affect Enzyme Packaging into Bacterial Outer Membrane Vesicles" Bioengineering 10, no. 5: 583. https://doi.org/10.3390/bioengineering10050583
APA StyleDean, S. N., Thakur, M., Spangler, J. R., Smith, A. D., Garin, S. P., Walper, S. A., & Ellis, G. A. (2023). Different Strategies Affect Enzyme Packaging into Bacterial Outer Membrane Vesicles. Bioengineering, 10(5), 583. https://doi.org/10.3390/bioengineering10050583