Bacterial Membrane Vesicles for In Vitro Catalysis
Abstract
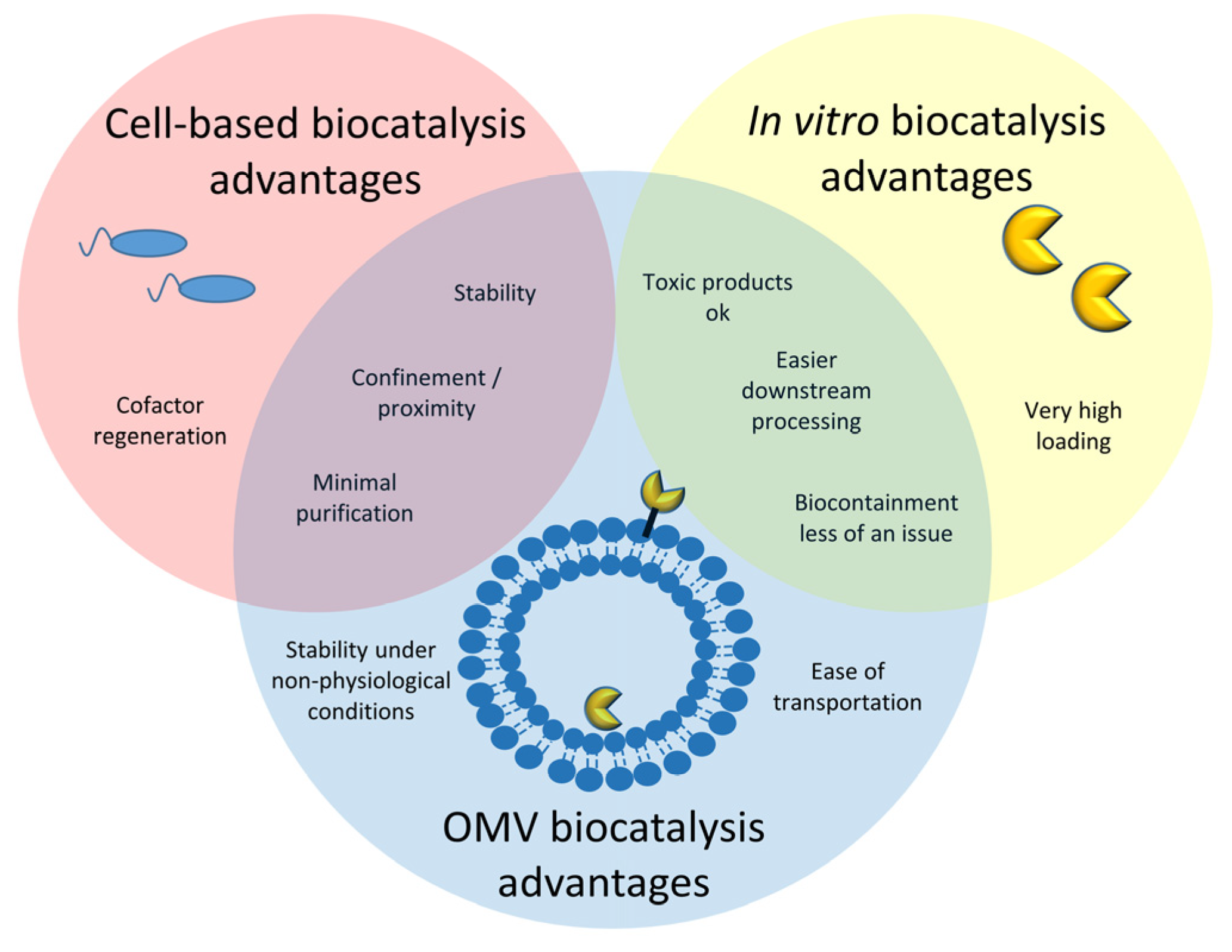
:1. Introduction
2. Bacterial Membrane Vesicles in Biocatalysis—OMVs Naturally Involved in Catalysis
3. Engineered Outer Membrane Vesicles for Biocatalysis
3.1. Cellulose Hydrolysis
3.2. Bioremediation
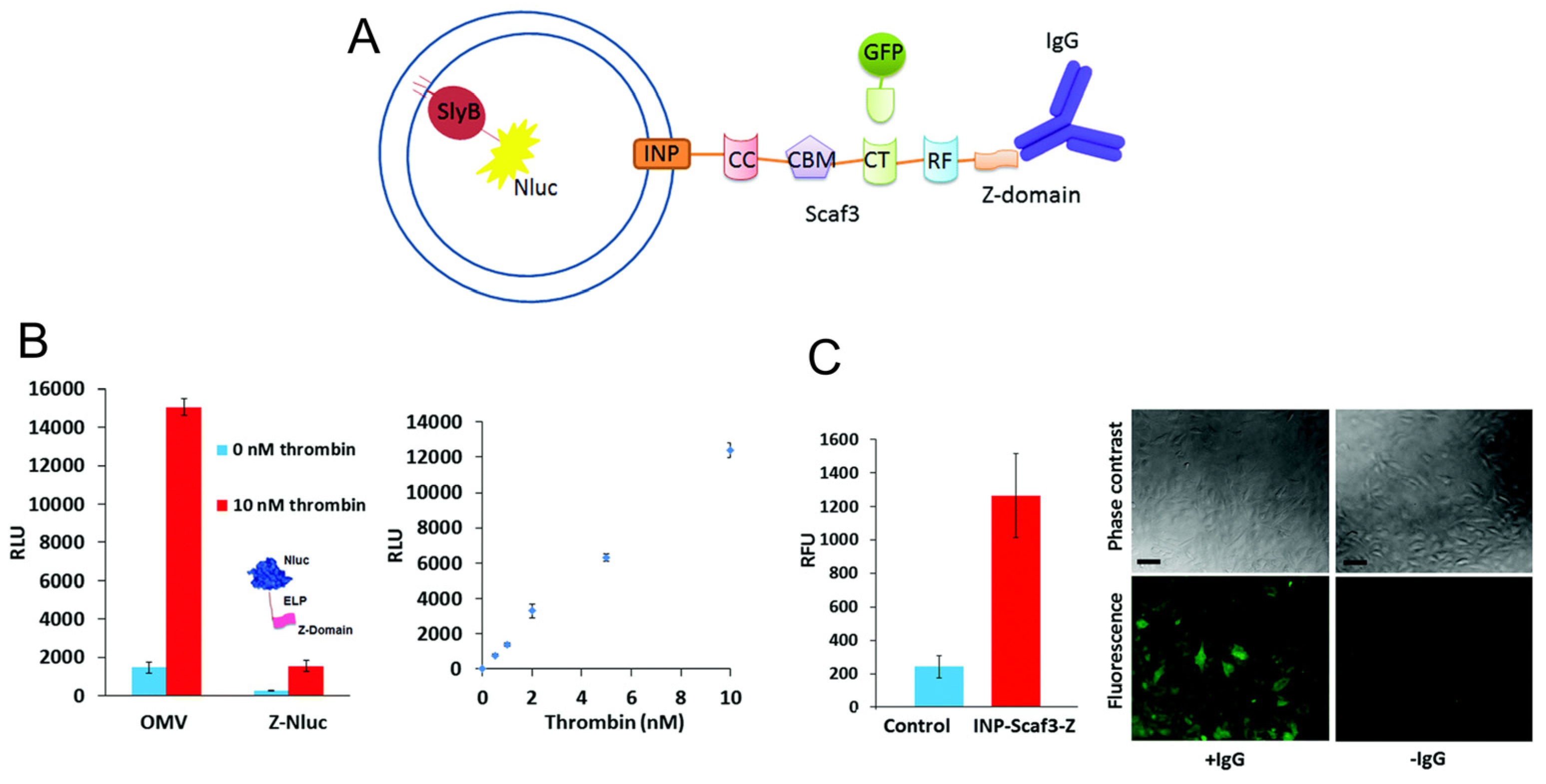
3.3. Biosensing and Bioimaging
4. Other Applications
5. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, M.R.; Khera, E.; Wen, F. Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res. 2015, 54, 4021–4032. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Factories 2017, 16, 106. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.C.; Mi, L.; Pontrelli, S.; Luo, S. Fuelling the future: Microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 2016, 14, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef]
- Clomburg, J.M.; Crumbley, A.M.; Gonzalez, R. Industrial biomanufacturing: The future of chemical production. Science 2017, 355, aag0804. [Google Scholar] [CrossRef]
- Muangphrom, P.; Seki, H.; Fukushima, E.O.; Muranaka, T. Artemisinin-based antimalarial research: Application of biotechnology to the production of artemisinin, its mode of action, and the mechanism of resistance of Plasmodium parasites. J. Nat. Med. 2016, 70, 318–334. [Google Scholar] [CrossRef]
- Chen, G.Q.; Liu, X. On the future fermentation. Microb. Biotechnol. 2021, 14, 18–21. [Google Scholar] [CrossRef]
- Gargalo, C.L.; Udugama, I.; Pontius, K.; Lopez, P.C.; Nielsen, R.F.; Hasanzadeh, A.; Mansouri, S.S.; Bayer, C.; Junicke, H.; Gernaey, K.V. Towards smart biomanufacturing: A perspective on recent developments in industrial measurement and monitoring technologies for bio-based production processes. J. Ind. Microbiol. Biotechnol. 2020, 47, 947–964. [Google Scholar] [CrossRef]
- de Carvalho, C.C.R. Whole cell biocatalysts: Essential workers from nature to the industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef]
- Selas Castiñeiras, T.; Williams, S.G.; Hitchcock, A.G.; Smith, D.C. E. coli strain engineering for the production of advanced biopharmaceutical products. FEMS Microbiol. Lett. 2018, 365, fny162. [Google Scholar] [CrossRef]
- Arbige, M.V.; Shetty, J.K.; Chotani, G.K. Industrial enzymology: The next chapter. Trends Biotechnol. 2019, 37, 1355–1366. [Google Scholar] [CrossRef]
- Bergquist, P.L.; Siddiqui, S.; Sunna, A. Cell-free biocatalysis for the production of platform chemicals. Front. Energy Res. 2020, 8, 193. [Google Scholar] [CrossRef]
- Ellis, G.A.; Klein, W.P.; Lasarte-Aragonés, G.; Thakur, M.; Walper, S.A.; Medintz, I.L. Artificial multienzyme scaffolds: Pursuing in vitro substrate channeling with an overview of current progress. ACS Catal. 2019, 9, 10812–10869. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Gan, Y.; Zhao, G.; Wang, Z.; Zhang, X.; Wu, M.X.; Lu, M. Bacterial Membrane Vesicles: Physiological Roles, Infection Immunology, and Applications. Adv. Sci. 2023, 10, 2301357. [Google Scholar] [CrossRef] [PubMed]
- Alves, N.J.; Turner, K.B.; Daniele, M.A.; Oh, E.; Medintz, I.L.; Walper, S.A. Bacterial nanobioreactors–directing enzyme packaging into bacterial outer membrane vesicles. ACS Appl. Mater. Interfaces 2015, 7, 24963–24972. [Google Scholar] [CrossRef] [PubMed]
- Alves, N.J.; Turner, K.B.; Medintz, I.L.; Walper, S.A. Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci. Rep. 2016, 6, 24866. [Google Scholar] [CrossRef]
- Park, M.; Sun, Q.; Liu, F.; DeLisa, M.P.; Chen, W. Positional assembly of enzymes on bacterial outer membrane vesicles for cascade reactions. PLoS ONE 2014, 9, e97103. [Google Scholar]
- Nieves, L.M.; Panyon, L.A.; Wang, X. Engineering sugar utilization and microbial tolerance toward lignocellulose conversion. Front. Bioeng. Biotechnol. 2015, 3, 17. [Google Scholar] [CrossRef]
- Liu, Z.L.; Blaschek, H.P. Biomass conversion inhibitors and in situ detoxification. In Biomass to Biofuels: Strategies for Global Industries; Vertès, A., Qureshi, N., Blaschek, H., Yukawa, H., Eds.; John Wiley and Sons Ltd.: West Sussex, UK, 2010; pp. 233–259. [Google Scholar]
- Arntzen, M.Ø.; Várnai, A.; Mackie, R.I.; Eijsink, V.G.; Pope, P.B. Outer membrane vesicles from Fibrobacter succinogenes S85 contain an array of carbohydrate-active enzymes with versatile polysaccharide-degrading capacity. Environ. Microbiol. 2017, 19, 2701–2714. [Google Scholar] [CrossRef]
- Ichikawa, S.; Ogawa, S.; Nishida, A.; Kobayashi, Y.; Kurosawa, T.; Karita, S. Cellulosomes localise on the surface of membrane vesicles from the cellulolytic bacterium Clostridium thermocellum. FEMS Microbiol. Lett. 2019, 366, fnz145. [Google Scholar] [CrossRef]
- Salvachúa, D.; Werner, A.Z.; Pardo, I.; Michalska, M.; Black, B.A.; Donohoe, B.S.; Haugen, S.J.; Katahira, R.; Notonier, S.; Ramirez, K.J.; et al. Outer membrane vesicles catabolize lignin-derived aromatic compounds in Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. USA 2020, 117, 9302–9310. [Google Scholar] [CrossRef] [PubMed]
- Puiggené, Ò.; Espinosa, M.J.C.; Schlosser, D.; Thies, S.; Jehmlich, N.; Kappelmeyer, U.; Schreiber, S.; Wibberg, D.; Kalinowski, J.; Harms, H.; et al. Extracellular degradation of a polyurethane oligomer involving outer membrane vesicles and further insights on the degradation of 2,4-diaminotoluene in Pseudomonas capeferrum TDA1. Sci. Rep. 2022, 12, 2666. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Coyne, M.; Comstock, L. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 2014, 24, 40–49. [Google Scholar] [CrossRef]
- Elhenawy, W.; Debelyy, M.O.; Feldman, M.F. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. MBio 2014, 5, e00909-14. [Google Scholar] [CrossRef] [PubMed]
- Naval, P.; Chandra, T. Characterization of membrane vesicles secreted by seaweed associated bacterium Alteromonas macleodii KS62. Biochem. Biophys. Res. Commun. 2019, 514, 422–427. [Google Scholar] [CrossRef]
- de Paula, R.G.; Antoniêto, A.C.C.; Nogueira, K.M.V.; Ribeiro, L.F.C.; Rocha, M.C.; Malavazi, I.; Almeida, F.; Silva, R.N. Extracellular vesicles carry cellulases in the industrial fungus Trichoderma reesei. Biotechnol. Biofuels 2019, 12, 146. [Google Scholar] [CrossRef]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Factories 2016, 15, 106. [Google Scholar] [CrossRef]
- Naas, A.E.; Pope, P.B. A mechanistic overview of ruminal fibre digestion. PeerJ Prepr. 2019, 7, e27831v1. [Google Scholar]
- Gong, J.; Egbosimba, E.E.; Forsberg, C.W. Cellulose-binding proteins of Fibrobacter succinogenes and the possible role of a 180-kDa cellulose-binding glycoprotein inadhesion to cellulose. Can. J. Microbiol. 1996, 42, 453–460. [Google Scholar] [CrossRef]
- Blatch, G.L.; Lässle, M. The tetratricopeptiderepeat: A structural motif mediating protein–protein interactions. BioEssays 1999, 21, 932–939. [Google Scholar] [CrossRef]
- Zha, J.; Yuwen, M.; Qian, W.; Wu, X. Yeast-based biosynthesis of natural products from xylose. Front. Bioeng. Biotechnol. 2021, 9, 634919. [Google Scholar] [CrossRef] [PubMed]
- Brameyer, S.; Plener, L.; Müller, A.; Klingl, A.; Wanner, G.; Jung, K. Outer membrane vesicles facilitate trafficking of the hydrophobic signaling molecule CAI-1 between Vibrio harveyi cells. J. Bacteriol. 2018, 200, e00740-17. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.B.; Alegado, R.A. Spheres of hope, packets of doom: The good and bad of outer membrane vesicles in interspecies and ecological dynamics. J. Bacteriol. 2017, 199, e00012-17. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Brown, A.C. Bacterial Outer Membrane Vesicles as Antibiotic Delivery Vehicles. Front. Immunol. 2021, 12, 733064. [Google Scholar] [CrossRef]
- Gerritzen, M.J.H.; Martens, D.E.; Wijffels, R.H.; van der Pol, L.; Stork, M. Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnol. Adv. 2017, 35, 565–574. [Google Scholar] [CrossRef]
- Huang, Y.; Nieh, M.P.; Chen, W.; Lei, Y. Outer membrane vesicles (OMVs) enabled bio-applications: A critical review. Biotechnol. Bioeng. 2022, 119, 34–47. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Doody, A.M.; Chen, D.J.; Cremona, G.H.; Shuler, M.L.; Putnam, D.; DeLisa, M.P. Engineered bacterial outer membrane vesicles with enhanced functionality. J. Mol. Biol. 2008, 380, 51–66. [Google Scholar] [CrossRef]
- Chen, D.J.; Osterrieder, N.; Metzger, S.M.; Buckles, E.; Doody, A.M.; DeLisa, M.P.; Putnam, D. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. USA 2010, 107, 3099–3104. [Google Scholar] [CrossRef]
- van den Berg van Saparoea, H.B.; Houben, D.; Kuijl, C.; Luirink, J.; Jong, W.S.P. Combining Protein Ligation Systems to Expand the Functionality of Semi-Synthetic Outer Membrane Vesicle Nanoparticles. Front. Microbiol. 2020, 11, 890. [Google Scholar] [CrossRef]
- Cheng, K.; Zhao, R.; Li, Y.; Qi, Y.; Wang, Y.; Zhang, Y.; Qin, H.; Qin, Y.; Chen, L.; Li, C.; et al. Bioengineered bacteria-derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via Plug-and-Display technology. Nat. Commun. 2021, 12, 2041. [Google Scholar] [CrossRef] [PubMed]
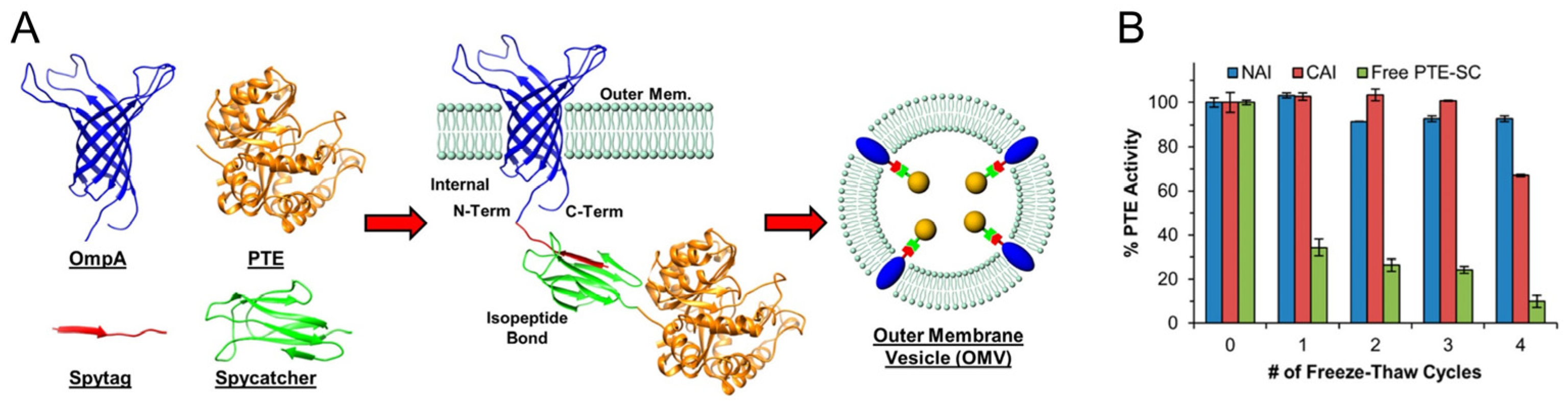
- Dean, S.N.; Thakur, M.; Spangler, J.R.; Smith, A.D.; Garin, S.P.; Walper, S.A.; Ellis, G.A. Different Strategies Affect Enzyme Packaging into Bacterial Outer Membrane Vesicles. Bioengineering 2023, 10, 583. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.M.; Kim, M.Y.; Song, J.-W.; Baeg, Y.; Jo, H.-J.; Cha, S.S.; Park, J.-B. Engineering of a bacterial outer membrane vesicle to a nano-scale reactor for the biodegradation of beta-lactam antibiotics. J. Biotechnol. 2022, 356, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Su, F.-H.; Tabañag, I.D.F.; Wu, C.-Y.; Tsai, S.-L. Decorating outer membrane vesicles with organophosphorus hydrolase and cellulose binding domain for organophosphate pesticide degradation. Chem. Eng. J. 2017, 308, 1–7. [Google Scholar] [CrossRef]
- Thakur, M.; Dean, S.N.; Moore, M.; Spangler, J.R.; Johnson, B.J.; Medintz, I.L.; Walper, S.A. Packaging of Diisopropyl Fluorophosphatase (DFPase) in Bacterial Outer Membrane Vesicles Protects Its Activity at Extreme Temperature. ACS Biomater. Sci. Eng. 2022, 8, 493–501. [Google Scholar] [CrossRef]
- Chen, Q.; Rozovsky, S.; Chen, W. Engineering multi-functional bacterial outer membrane vesicles as modular nanodevices for biosensing and bioimaging. Chem. Commun. 2017, 53, 7569–7572. [Google Scholar] [CrossRef]
- Yur, D.; Lieser, R.M.; Sullivan, M.O.; Chen, W. Engineering bionanoparticles for improved biosensing and bioimaging. Curr. Opin. Biotechnol. 2021, 71, 41–48. [Google Scholar] [CrossRef]
- Song, J.W.; Baeg, Y.; Jeong, H.; Lee, J.; Oh, D.K.; Hollmann, F.; Park, J.B. Bacterial Outer Membrane Vesicles as Nano-Scale Bioreactors: A Fatty Acid Conversion Case Study. ChemCatChem 2021, 13, 4080–4086. [Google Scholar] [CrossRef]
- Thakur, M.; Medintz, I.L.; Walper, S.A. Enzymatic Bioremediation of Organophosphate Compounds-Progress and Remaining Challenges. Front. Bioeng. Biotechnol. 2019, 7, 289. [Google Scholar] [CrossRef]
- Vyas, T.; Singh, V.; Kodgire, P.; Joshi, A. Insights in detection and analysis of organophosphates using organophosphorus acid anhydrolyases (OPAA) enzyme-based biosensors. Crit. Rev. Biotechnol. 2023, 43, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Bigley, A.N.; Raushel, F.M. The evolution of phosphotriesterase for decontamination and detoxification of organophosphorus chemical warfare agents. Chem. Biol. Interact. 2019, 308, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.M.; Mulchandani, A.; Chen, W. Bacterial cell surface display of organophosphorus hydrolase for selective screening of improved hydrolysis of organophosphate nerve agents. Appl. Environ. Microbiol. 2002, 68, 2026–2030. [Google Scholar] [CrossRef]
- Alves, N.J.; Turner, K.B.; DiVito, K.A.; Daniele, M.A.; Walper, S.A. Affinity purification of bacterial outer membrane vesicles (OMVs) utilizing a His-tag mutant. Res. Microbiol. 2017, 168, 139–146. [Google Scholar] [CrossRef]
- Turner, K.B.; Dean, S.N.; Walper, S.A. Chapter Eight-Bacterial bioreactors: Outer membrane vesicles for enzyme encapsulation. Meth. Enzymol. 2019, 617, 187–216. [Google Scholar]
- Alves, N.J.; Moore, M.; Johnson, B.J.; Dean, S.N.; Turner, K.B.; Medintz, I.L.; Walper, S.A. Environmental decontamination of a chemical warfare simulant utilizing a membrane vesicle-encapsulated phosphotriesterase. ACS Appl. Mater. Interfaces 2018, 10, 15712–15719. [Google Scholar] [CrossRef]
- Benning, M.M.; Kuo, J.M.; Raushel, F.M.; Holden, H.M. Three-Dimensional Structure of Phosphotriesterase: An Enzyme Capable of Detoxifying Organophosphate Nerve Agents. Biochemistry 1994, 33, 15001–15007. [Google Scholar] [CrossRef]
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Structural Analysis and Optimization of the Covalent Association between SpyCatcher and a Peptide Tag. J. Mol. Biol. 2014, 426, 309–317. [Google Scholar] [CrossRef]
- Cierpicki, T.; Liang, B.; Tamm, L.K.; Bushweller, J.H. Increasing the Accuracy of Solution NMR Strctures of Membrane Proteins by Application of Residual Dipolar Couplings. High-Resolution Structure of Outer Membrane Protein A. J. Am. Chem. Soc. 2006, 128, 6947–6951. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the woldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Cierpicki, T.; Liang, B.; Tamm, L.K.; Bushweller, J.H. High-Resolution Solution Structure of Outer Membrane Protein A Transmembrane Domain. 2006. Available online: https://www.rcsb.org/structure/2ge4 (accessed on 16 June 2023). [CrossRef]
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Crystal Structure of the SpyTag/SpyCatcher Complex. 2013. Available online: https://www.rcsb.org/structure/4MLI (accessed on 16 June 2023). [CrossRef]
- Kim, S.W.; Lee, J.S.; Park, S.B.; Lee, A.R.; Jung, J.W.; Chun, J.H.; Lazarte, J.M.S.; Kim, J.; Seo, J.-S.; Kim, J.-H.; et al. The Importance of Porins and beta-Lactamase in Outer Membrane Vesicles on the Hydrolysis of beta-Lactam Antibiotics. Int. J. Mol. Sci. 2020, 21, 2822. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Beringhs, A.O.R.; Chen, Q.; Song, D.; Chen, W.; Lu, X.; Fan, T.-H.; Nieh, M.-P.; Lei, Y. Genetically engineered bacterial outer membrane vesicles with expressed nanoluciferase reporter for in vivo bioluminescence kinetic modeling through noninvasive imaging. ACS Appl. Bio Mater. 2019, 2, 5608–5615. [Google Scholar] [CrossRef]
- Gujrati, V.; Prakash, J.; Malekzadeh-Najafabadi, J.; Stiel, A.; Klemm, U.; Mettenleiter, G.; Aichler, M.; Walch, A.; Ntziachristos, V. Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 2019, 10, 1114. [Google Scholar] [CrossRef] [PubMed]
- Gnopo, Y.M.; Watkins, H.C.; Stevenson, T.C.; DeLisa, M.P.; Putnam, D. Designer outer membrane vesicles as immunomodulatory systems–Reprogramming bacteria for vaccine delivery. Adv. Drug Deliv. Rev. 2017, 114, 132–142. [Google Scholar] [CrossRef] [PubMed]






| Organism | Substrate 1 | Source | Reference |
|---|---|---|---|
| Fibrobacter succinogenes | Hemicellulose and pectin | Cow microbiota | [22] |
| Clostridium thermocellum | Crystalline cellulose | Soil | [23] |
| Pseudomonas putida | Lignin | Soil | [24] |
| Pseudomonas capeferrum | Polyurethane compounds | Plastic dump site | [25] |
| Bacteroides sp. | Polysaccharides and proteins | Human microbiota | [26,27] |
| Alteromonas macleodii | κ-carrageenan and red seaweed biomass | Marine | [28] |
| Trichoderma reesei | Cellulose | Fungus/rainforest | [29,30] |
| Enzyme | OMV Anchor | Application | References |
|---|---|---|---|
| Green fluorescent protein | ClyA | Bioimaging | [40,41] |
| β-lactamase | ClyA, pectate lyase B signal sequence | Bioremediation | [40,41,45] |
| Organophosphate hydrolase | ClyA, Ice nucleation protein (INP) | Bioremediation | [40,41,46] |
| Cellulosome | INP | Biomanufacturing | [19] |
| Phosphotriesterase | SpyCatcher/SpyTag-OmpA, Lpp’-linker | Bioremediation | [17,44] |
| DFPase | Lpp’-linker | Bioremediation | [47] |
| Nanoluciferase | SlyB | Biosensing and Bioimaging | [48,49] |
| Fatty acid double-bond hydratase and fatty acid decarboxylase | pectate lyase B signal sequence | Biomanufacturing | [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, M.; Dean, S.N.; Caruana, J.C.; Walper, S.A.; Ellis, G.A. Bacterial Membrane Vesicles for In Vitro Catalysis. Bioengineering 2023, 10, 1099. https://doi.org/10.3390/bioengineering10091099
Thakur M, Dean SN, Caruana JC, Walper SA, Ellis GA. Bacterial Membrane Vesicles for In Vitro Catalysis. Bioengineering. 2023; 10(9):1099. https://doi.org/10.3390/bioengineering10091099
Chicago/Turabian StyleThakur, Meghna, Scott N. Dean, Julie C. Caruana, Scott A. Walper, and Gregory A. Ellis. 2023. "Bacterial Membrane Vesicles for In Vitro Catalysis" Bioengineering 10, no. 9: 1099. https://doi.org/10.3390/bioengineering10091099
APA StyleThakur, M., Dean, S. N., Caruana, J. C., Walper, S. A., & Ellis, G. A. (2023). Bacterial Membrane Vesicles for In Vitro Catalysis. Bioengineering, 10(9), 1099. https://doi.org/10.3390/bioengineering10091099








