Reliability of Systematic and Targeted Biopsies versus Prostatectomy
Abstract
:1. Introduction
2. Materials and Methods
- Grade 1, 2, 3, 4 or 5, as per SBx in comparison to Grade 1, 2, 3, 4 or 5 as per prostatectomy (Cohen’s Kappa)
- Grade 1, 2, 3, 4 or 5, as per MRITBx in comparison to Grade 1, 2, 3, 4 or 5 as per prostatectomy (Cohen’s Kappa)
- Grade 1 vs. Grade 2, 3, 4 or 5 as per SBx in comparison to Grade 1 vs. Grade 2, 3, 4 or 5 as per prostatectomy (Logistic Regression)
- Grade 1 vs. Grade 2, 3, 4 or 5 as per MRITBx in comparison to Grade 1 vs. Grade 2, 3, 4, 5 as per prostatectomy (Logistic Regression)
- Grade 1 or 2 vs. Grade 3, 4, 5 as per SBx in comparison to Grade 1 or 2 vs. Grade 3, 4, 5 as per prostatectomy (Logistic Regression)
- Grade 1 or 2 vs. Grade 3, 4, 5 as per MRITBx in comparison to Grade 1 or 2 vs. Grade 3, 4, 5 as per prostatectomy (Logistic Regression)
- Grade 1 or 2 vs. Grade 3, 4, 5 as per MRITBx in comparison to Grade 1 or 2 vs. Grade 3, 4, 5 as per prostatectomy (Classification Tree)
3. Results
3.1. Kappa Statistic
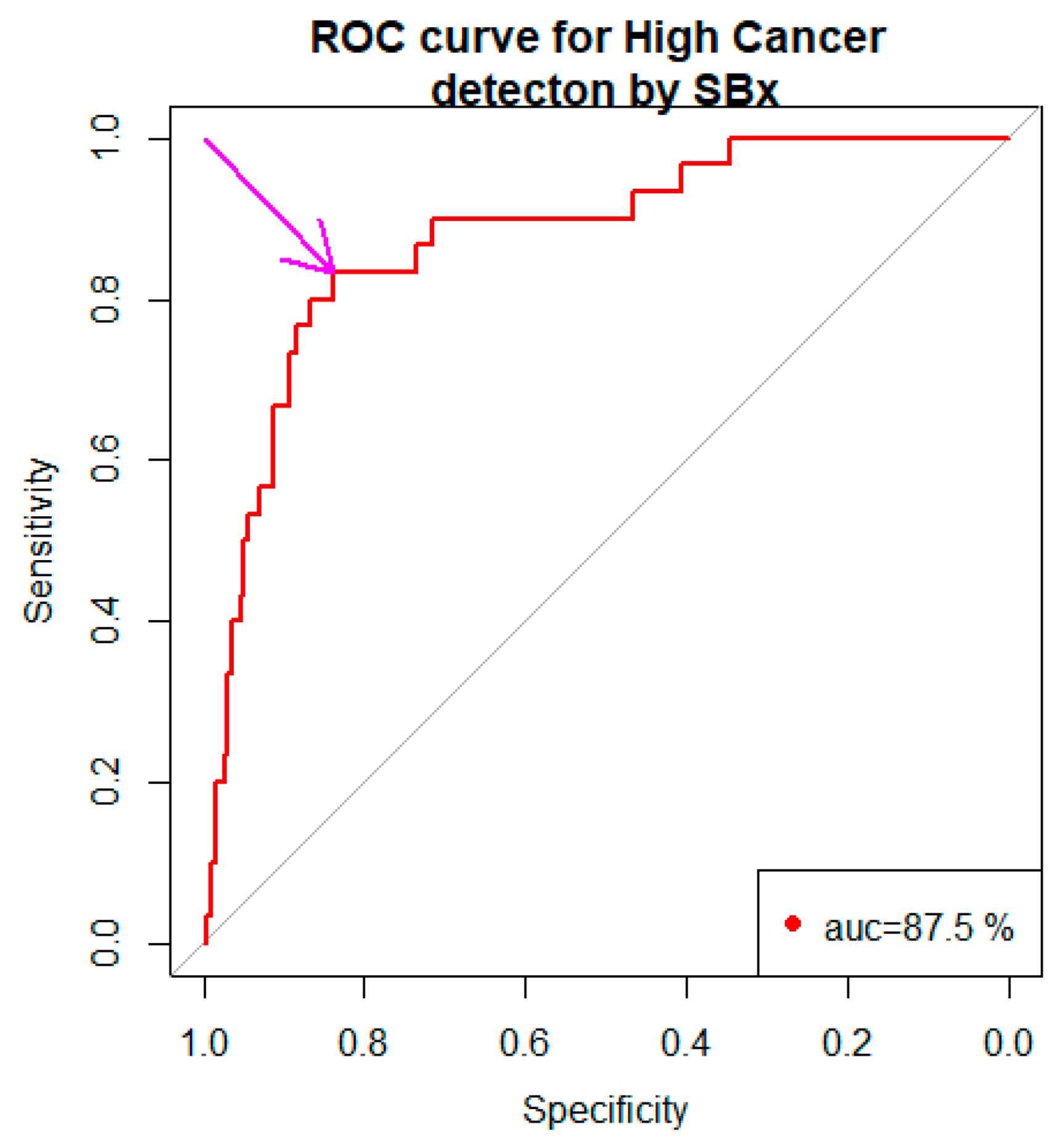
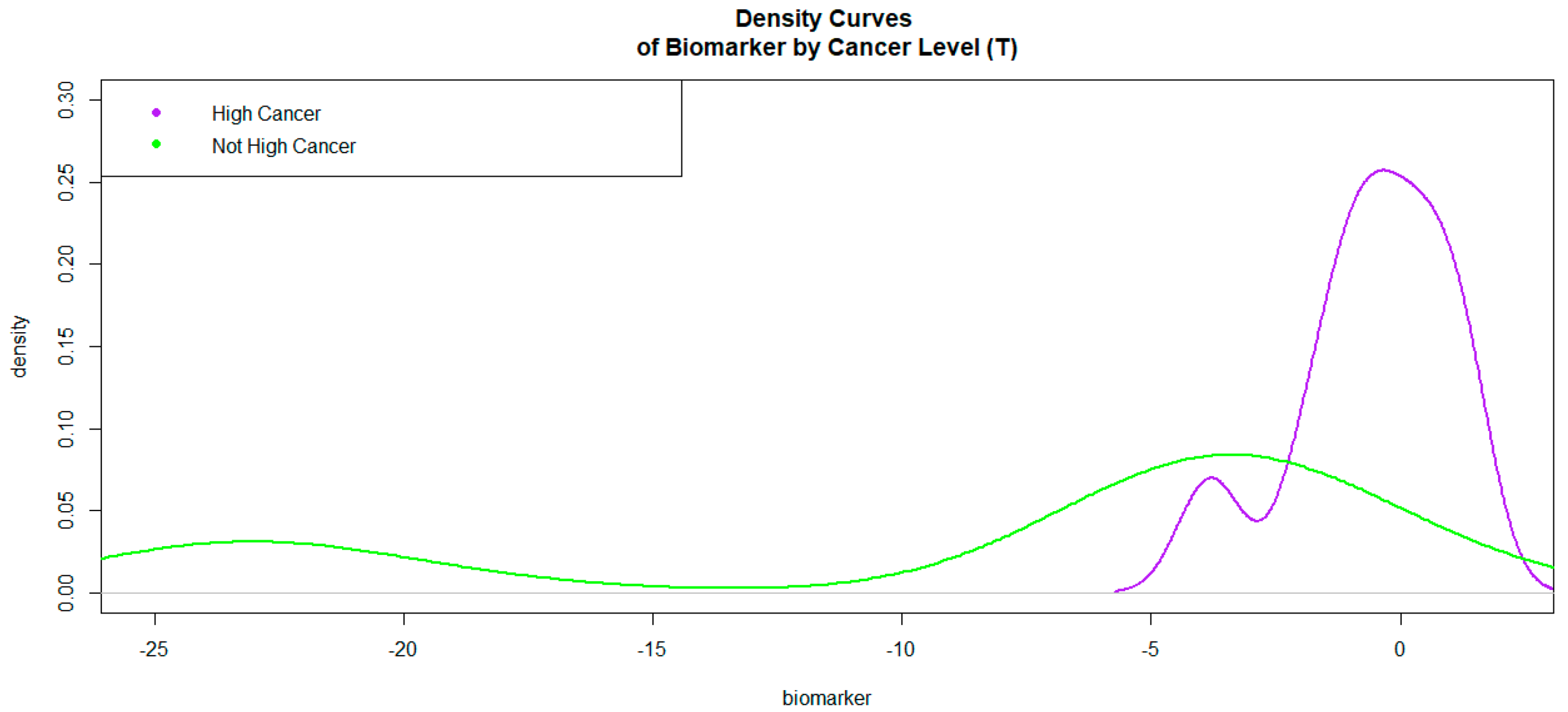
3.2. SBx Gleason Grades Binarized as a Predictor in the Model
3.3. MRITBx Gleason Grade Binarized as a Predictor in the Model
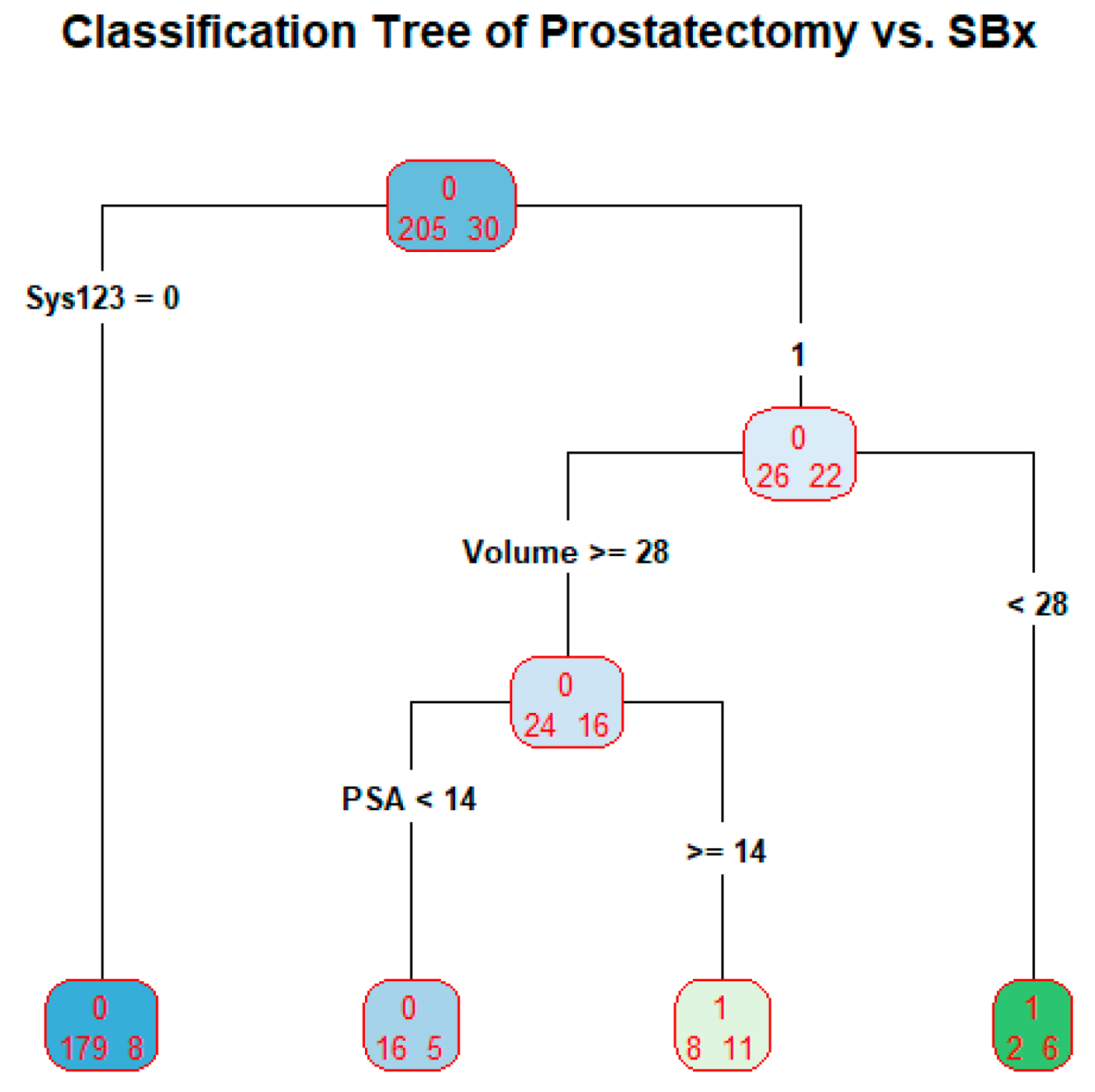
3.4. SBx as a Predictor in Classification Tree
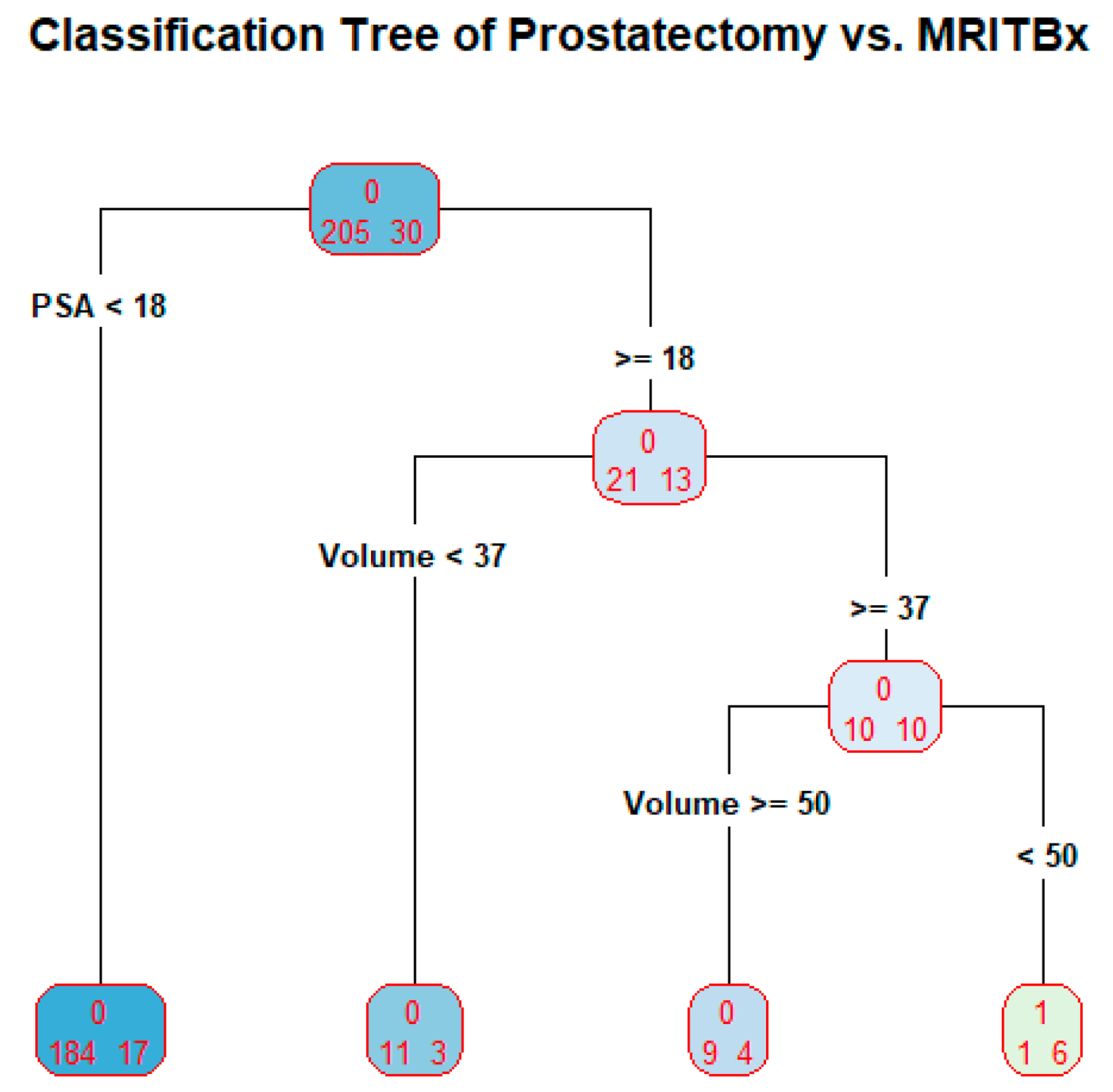
3.5. MRITBx as a Predictor in Classification Tree
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Hoge, C.; Maynor, S.; Sidana, A.; Guan Tianyuan Rao, M.B.; Naffouje, R.; Verma, S. A comparison of cancer detection rates between template systematic biopsies obtained using magnetic resonance imaging-ultrasound fusion machine and freehand transrectal ultrasound-guided systematic biopsies. J. Endourol. 2020, 3, 154–196. [Google Scholar] [CrossRef]
- Kaneko, M.; Sugano, D.; Lebastchi, A.H.; Duddalwar, V.; Nabhani, J.; Haiman, C.; Gill, I.S.; Cacciamani, G.E.; Abreu, A.L. Techniques and Outcomes of MRI-TRUS Fusion Prostate Biopsy. Curr. Urol. Rep. 2021, 22, 27. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/books/NBK556081/ (accessed on 8 May 2023).
- Cao, R.; Bajgiran, A.M.; Mirak, S.A.; Shakeri, S.; Zhong, X.; Enzmann, D.; Raman, S.; Sung, K. Joint Prostate Cancer Detection and Gleason Score Prediction in Mp-MRI via FocalNet. Med. Imaging 2019, 38, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Vente, C.d.; Vos, P.; Hosseinzadeh, M.; Pluim, J.; Veta, M. Deep Learning Regression for Prostate Cancer Detection and Grading in Bi-Parametric MRI. Biomed. Eng. 2021, 68, 374–383. [Google Scholar] [CrossRef]
- Larsen, L.K.; Jakobsen, J.S.; Abdul-Al, A.; Guldberg, P. Noninvasive Detection of High Grade Prostate Cancer by DNA Methylation Analysis of Urine Cells Captured by Microfiltration. J. Urol. 2018, 200, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Lih, T.-S.M.; Dong, M.; Mangold, L.; Partin, A.; Zhang, H. Urinary Marker Panels for Aggressive Prostate Cancer Detection. Sci. Rep. 2022, 12, 14837. [Google Scholar] [CrossRef] [PubMed]
- Sayyadi, N.; Justiniano, I.; Wang, Y.; Zheng, X.; Zhang, W.; Jiang, L.; Polikarpov, D.M.; Willows, R.D.; Gillatt, D.; Campbell, D.; et al. Detection of Rare Prostate Cancer Cells in Human Urine Offers Prospect of Non-Invasive Diagnosis. Sci. Rep. 2022, 12, 18452. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.M.; Pereira, E.M.; Salles, P.G.; Godrich, R.; Ceballos, R.; Kunz, J.D.; Casson, A.; Viret, J.; Chandarlapaty, S.; Gil Ferreira, C.; et al. Independent Real-World Application of a Clinical-Grade Automated Prostate Cancer Detection System. J. Pathol. 2021, 254, 147–158. [Google Scholar] [CrossRef]
- Yoo, S.; Gujrathi, I.; Haider, M.A.; Khalvati, F. Prostate Cancer Detection using Deep Convolutional Neural Networks. Sci. Rep. 2019, 9, 19518. [Google Scholar] [CrossRef]
- Hao, R.; Namdar, K.; Liu, L.; Haider, M.A.; Khalvati, F. A Comprehensive Study of Data Augmentation Strategies for Prostate Cancer Detection in Diffusion-Weighted MRI Using Convolutional Neural Networks. J. Digit. Imaging 2021, 34, 862–876. [Google Scholar] [CrossRef]
- Lorusso, V.; Kabre, B.; Pignot, G.; Branger, N.; Pacchetti, A.; Thomassin-Piana, J.; Brunelle, S.; Nicolai, N.; Musi, G.; Salem, N.; et al. External Validation of the Computerized Analysis of TRUS of the Prostate with the ANNA/C-TRUS System: A Potential Role of Artificial Intelligence for Improving Prostate Cancer Detection. World J. Urol. 2023, 41, 619–625. [Google Scholar] [CrossRef]
- Prostate Conditions Education Council. Available online: https://www.prostateconditions.org/about-prostate-conditions/prostate-cancer/newly-diagnosed/gleason-score (accessed on 8 May 2023).
- Rosner, B. Fundamentals of Biostatistics, 6th ed; Thomson-Brooks/Cole: Belmont, CA, USA, 2006; pp. 434–437. [Google Scholar]
- Toutenburg, H.; Fleiss, J.L. Statistical Methods for Rates and Proportions, 3rd ed.; John Wiley & Sons: New York, NY, USA, 1973; pp. 610–617. [Google Scholar]
- Reiser, B.; Faraggi, D.; Fluss, R. Estimation of the Youden Index and Its Associated Cutoff Point. Biom. J. 2005, 47, 458–472. [Google Scholar]
- Martínez-Camblor, P.; Pardo-Fernández, J.C. The Youden Index in the Generalized Receiver Operating Characteristic Curve Context. Int. J. Biostat. 2019, 15, 20180060. [Google Scholar] [CrossRef]
- Cancer Research, UK. Available online: https://www.cancerresearchuk.org/about-cancer/prostate-cancer/stages/grades (accessed on 8 May 2023).
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate Cancer Review: Genetics, Diagnosis, Treatment Options, and Alternative Approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- Nguyen-Nielsen, M.; Borre, M. Diagnostic and Therapeutic Strategies for Prostate Cancer. Semin. Nucl. Med. 2016, 46, 484–490. [Google Scholar] [CrossRef]
- Costello, A.J. Considering the Role of Radical Prostatectomy in 21st Century Prostate Cancer Care. Nat. Rev. Urol. 2020, 17, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.; Haj-Hamed, M.; Talarek, J.; Verma, S.; Sidana, A. How Does a Prebiopsy Mri Approach for Prostate Cancer Diagnosis Affect Prostatectomy Upgrade Rates? Urol. Oncol. 2021, 39, 784. [Google Scholar] [CrossRef] [PubMed]
- Autorino, R.; Porpiglia, F. Recent advances in prostate cancer: Diagnosis, patient selection and minimally invasive treatment. Minerva Urol. E Nefrol. 2015, 67, 197–200. [Google Scholar]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate Cancer. Nat. Rev. Dis. Primers 2021, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; Shoag, J.; Groß, M.; Robinson, B.; Khani, F.; Nelson, B.B.; Margolis, D.; Hu, J.C. Concordance between Biopsy and radical Prostatectomy Pathology in the era of Targeted Biopsy: A systematic review and meta-analysis. Eur. Urol. Oncol. 2020, 3, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, X.; Zhang, W.; Gao, K.; Wang, L.; Qian, L.; Mu, J.; Zheng, Z.; Cao, X. Developing a predictive model for clinically significant prostate cancer by combining age, PSA density, and mpMRI. World J. Surg. Oncol. 2023, 21, 83. [Google Scholar] [CrossRef]
- O’Connor, L.P.; Wang, A.Z.; Yerram, N.K.; Lebastchi, A.H.; Ahdoot, M.; Gurram, S.; Zeng, J.; Mehralivand, S.; Harmon, S.; Merino, M.J.; et al. Combined MRI-targeted Plus Systematic Confirmatory Biopsy Improves Risk Stratification for Patients Enrolling on Active Surveillance for Prostate Cancer. Urology 2022, 144, 164–170. [Google Scholar] [CrossRef]
- Vittinghoff, E.; McCulloch, C.E. Relaxing the Rule of Ten Events per Variable in Logistic and Cox Regression. Am. J. Epidemiol. 2007, 165, 710–718. [Google Scholar] [CrossRef]
- Van Smeden, M.; Moons, K.G.M.; Groot, J.A.H.; Eijkemans, M.J.C.; Reitsma, J.B.; Collins, G.S.; Altman, D.G. Sample size for binary logistic prediction models: Beyond events per variable criteria. Stat. Methods Med. Res. 2019, 28, 2455–2474. [Google Scholar] [CrossRef]
- Kassambara, A. Practical Guide to Cluster Analysis in R: Unsupervised Machine Learning; STHDA: Marseille, France, 2017; Chapter 2; pp. 10–23. [Google Scholar]
- Sun, Y.; Fang, J.; Shi, Y.; Li, H.; Wang, J.; Xu, J.; Zhang, B.; Liang, L. Machine Learning Based on Radiomics Features Combing B-Mode Transrectal Ultrasound and Contrast-Enhanced Ultrasound to Improve Peripheral Zone Prostate Cancer Detection. Abdom. Radiol. 2023, 1–10. [Google Scholar] [CrossRef]
- Michaely, H.J.; Aringhieri, G.; Cioni, D.; Neri, E. Current Value of Biparametric Prostate MRI with Machine-Learning or Deep-Learning in the Detection, Grading, and Characterization of Prostate Cancer: A Systematic Review. Diagnostics 2022, 12, 799. [Google Scholar] [CrossRef]


| Risk Group | Gleason Grade | Gleason Score |
|---|---|---|
| Low/Very Low | Grade 1 | Gleason Score ≤ 6 |
| Intermediate (Favorable/Unfavorable) | Grade 2 | Gleason Score 7 (3 + 4) |
| Grade 3 | Gleason Score 7 (4 + 3) | |
| High/Very High | Grade 4 | Gleason Score 8 |
| Grade 5 | Gleason Score 9–10 |
| Grades | SBx | Prostatectomy | MRITBx | Prostatectomy |
|---|---|---|---|---|
| 1 | 41 | 4 | 24 | 3 |
| 2 | 103 | 133 | 48 | 60 |
| 3 | 43 | 68 | 11 | 32 |
| 4 | 20 | 6 | 5 | 3 |
| 5 | 28 | 24 | 16 | 6 |
| Total | 235 | 235 | 104 | 104 |
| Prostatectomy (True Condition) | Diagnosed Condition by SBx | Marginal | |
|---|---|---|---|
| Grade ≥ 2 | Grade = 1 | ||
| Grade ≥ 2 | 192 | 38 | 231 |
| Grade = 1 | 2 | 2 | 4 |
| Marginal | 194 | 41 | 235 |
| Prostatectomy (True Condition) | Diagnosed Condition by MRITBx | Marginal | |
|---|---|---|---|
| Grade ≥ 2 | Grade = 1 | ||
| Grade ≥ 2 | 79 | 22 | 101 |
| Grade = 1 | 1 | 2 | 3 |
| Marginal | 80 | 24 | 104 |
| Levels of Hcpros | Min | I Quartile | Mean | Median | III Quartile | Max |
|---|---|---|---|---|---|---|
| 1 | −3.413 | −0.933 | 0.0838 | −0.693 | 0.201 | 1.079 |
| 0 | −16.776 | −3.842 | −2.934 | −3.205 | −2.789 | 1.756 |
| Levels of Hcpros | Min | I Quartile | Mean | Median | III Quartile | Max |
|---|---|---|---|---|---|---|
| 1 | −3.811 | −1.137 | 0.499 | −0.559 | 0.3428 | 1.288 |
| 0 | −24.531 | −19.515 | −4.045 | −8.649 | −3.353 | 1.463 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, T.; Sidana, A.; Rao, M.B. Reliability of Systematic and Targeted Biopsies versus Prostatectomy. Bioengineering 2023, 10, 1395. https://doi.org/10.3390/bioengineering10121395
Guan T, Sidana A, Rao MB. Reliability of Systematic and Targeted Biopsies versus Prostatectomy. Bioengineering. 2023; 10(12):1395. https://doi.org/10.3390/bioengineering10121395
Chicago/Turabian StyleGuan, Tianyuan, Abhinav Sidana, and Marepalli B. Rao. 2023. "Reliability of Systematic and Targeted Biopsies versus Prostatectomy" Bioengineering 10, no. 12: 1395. https://doi.org/10.3390/bioengineering10121395
APA StyleGuan, T., Sidana, A., & Rao, M. B. (2023). Reliability of Systematic and Targeted Biopsies versus Prostatectomy. Bioengineering, 10(12), 1395. https://doi.org/10.3390/bioengineering10121395









