[68Ga]Ga-PSMA and [68Ga]Ga-RM2 PET/MRI vs. Histopathological Images in Prostate Cancer: A New Workflow for Spatial Co-Registration
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. In Vivo PET/MRI Acquisitions
2.3. Radical Prostatectomy
2.4. Preparation of the Ex Vivo Prostate
2.5. Ex Vivo PET/MRI Acquisitions
2.6. Specimen Cut and Digitalization
2.7. PET/MR Image Analysis
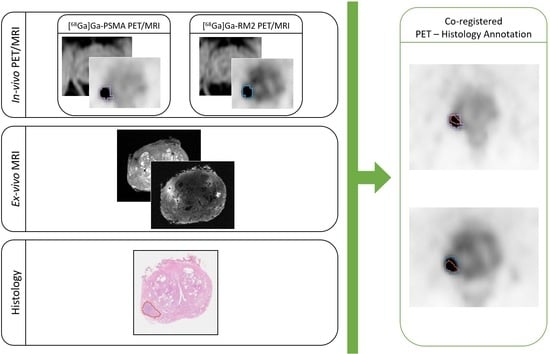
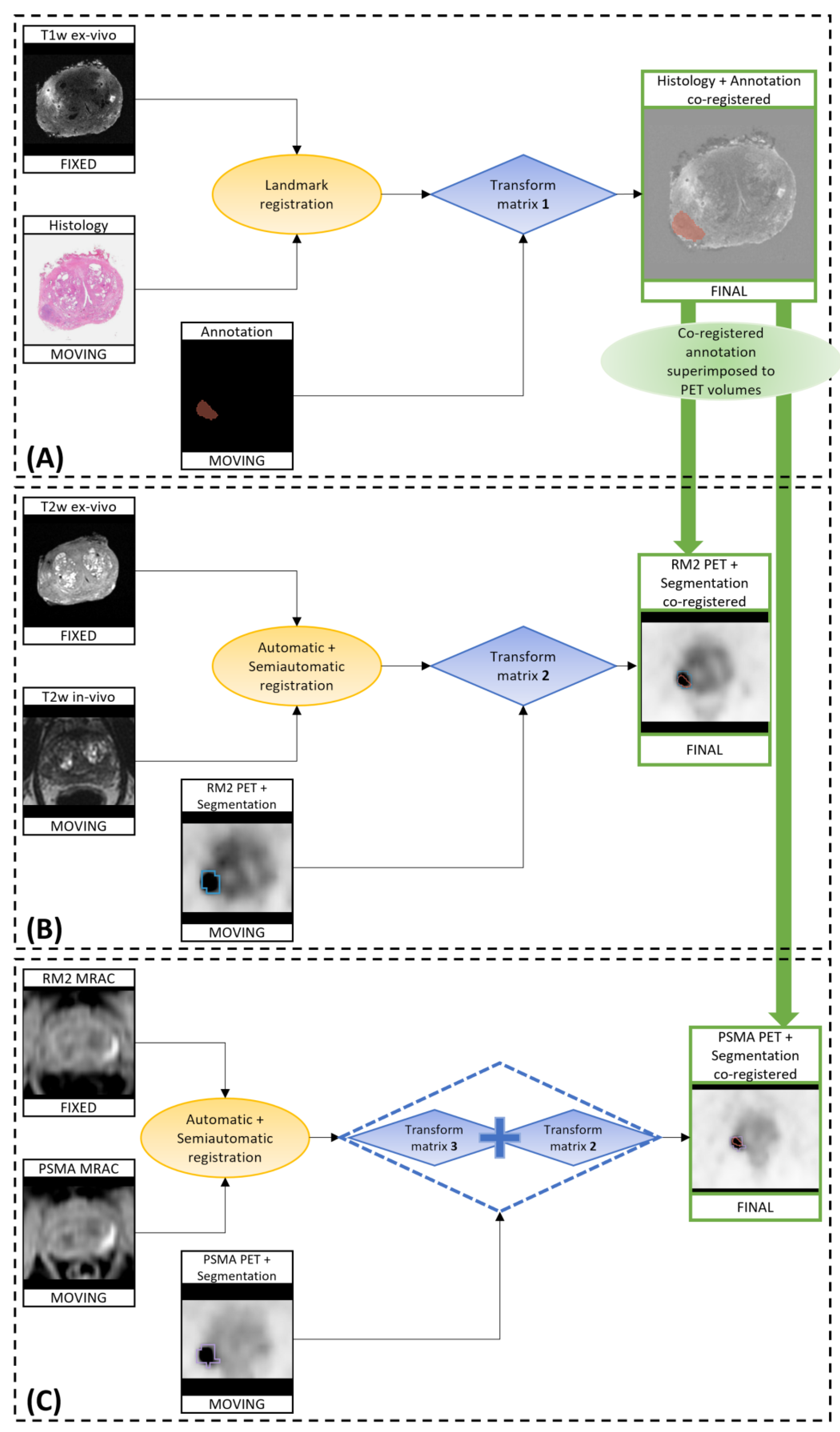
2.8. Co-Registration of In Vivo, Ex Vivo, and Digitalized Histological Images
- The co-registration of histopathological images to ex vivo 3D T1w MR images;
- The co-registration of the in vivo 3D T2w MR images to the ex vivo T2w images (originally acquired as the ex vivo T1w images, so naturally co-registered to those images);
- The application of the calculated transformations to the RM2 PET images;
- The co-registration of the Water-MRAC images of PSMA PET to the MRAC of the RM2 PET;
- The application of the calculated transformations to the PSMA PET images.
2.9. Metrics
3. Results
3.1. Patients
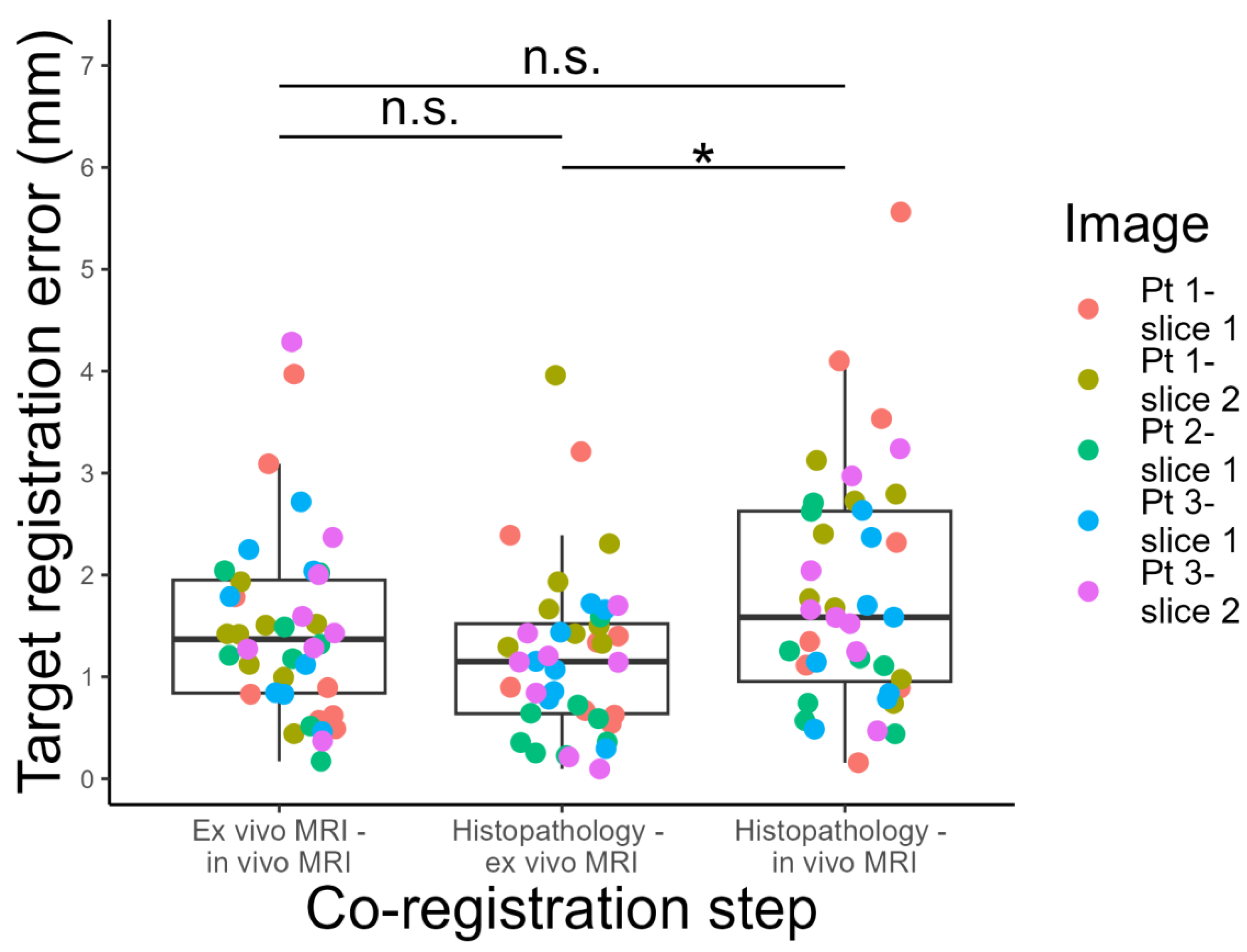
3.2. Target Registration Error Analysis
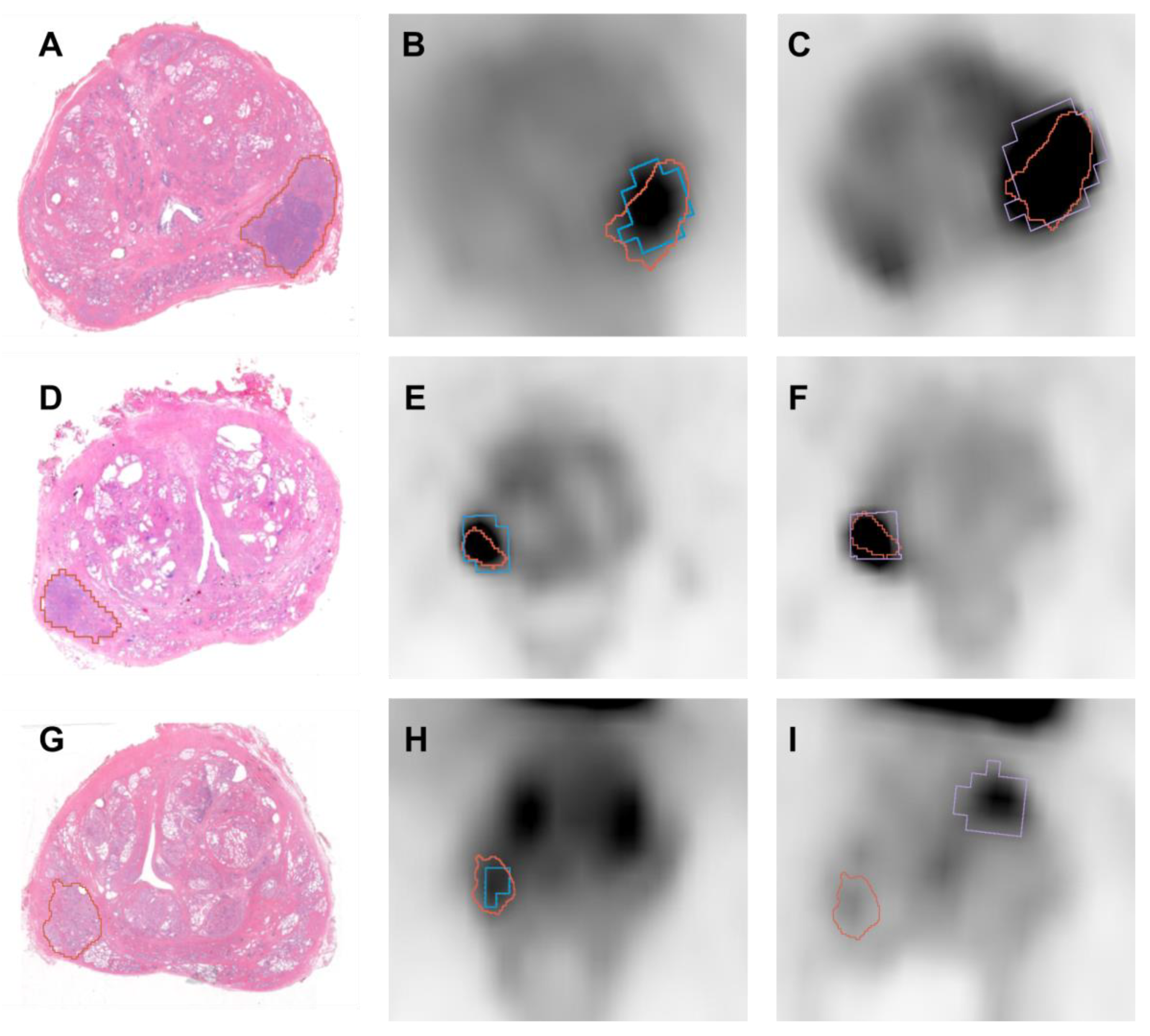
3.3. Diagnostic Accuracy (DICE)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Cancer, Research Fund International [WCRF]. Prostate Cancer Statistics. Available online: https://www.wcrf.org/cancer-trends/prostate-cancer-statistics/ (accessed on 17 July 2023).
- von Eyben, F.E.; Picchio, M.; von Eyben, R.; Rhee, H.; Bauman, G. 68 Ga-Labeled Prostate-Specific Membrane Antigen Ligand Positron Emission Tomography/Computed Tomography for Prostate Cancer: A Systematic Review and Meta-Analysis. Eur. Urol. Focus 2018, 4, 686–693. [Google Scholar] [CrossRef]
- Maurer, T.; Gschwend, J.E.; Rauscher, I.; Souvatzoglou, M.; Haller, B.; Weirich, G.; Wester, H.J.; Heck, M.; Kübler, H.; Beer, A.J.; et al. Diagnostic Efficacy of 68Gallium-PSMA Positron Emission Tomography Compared to Conventional Imaging for Lymph Node Staging of 130 Consecutive Patients with Intermediate to High Risk Prostate Cancer. J. Urol. 2016, 195, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, D.A.; Becker, A.S.; Kranzbühler, B.; Mebert, I.; Baltensperger, A.; Zeimpekis, K.G.; Grünig, H.; Messerli, M.; Rupp, N.J.; Rueschoff, J.H.; et al. Diagnostic Performance of 68Ga-PSMA-11 PET/MRI-Guided Biopsy in Patients with Suspected Prostate Cancer: A Prospective Single-Center Study. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3315–3324. [Google Scholar] [CrossRef] [PubMed]
- Hofman, M.S.; Lawrentschuk, N.; Francis, R.J.; Tang, C.; Vela, I.; Thomas, P.; Rutherford, N.; Martin, J.M.; Frydenberg, M.; Shakher, R.; et al. Prostate-Specific Membrane Antigen PET-CT in Patients with High-Risk Prostate Cancer before Curative-Intent Surgery or Radiotherapy (ProPSMA): A Prospective, Randomised, Multicentre Study. Lancet 2020, 395, 1208–1216. [Google Scholar] [CrossRef]
- Lopci, E.; Saita, A.; Lazzeri, M.; Lughezzani, G.; Colombo, P.; Buffi, N.M.; Hurle, R.; Marzo, K.; Peschechera, R.; Benetti, A.; et al. 68Ga-PSMA Positron Emission Tomography/Computerized Tomography for Primary Diagnosis of Prostate Cancer in Men with Contraindications to or Negative Multiparametric Magnetic Resonance Imaging: A Prospective Observational Study. J. Urol. 2018, 200, 95–103. [Google Scholar] [CrossRef]
- Donato, P.; Morton, A.; Yaxley, J.; Ranasinghe, S.; Teloken, P.E.; Kyle, S.; Coughlin, G.; Esler, R.; Dunglison, N.; Gardiner, R.A.; et al. 68Ga-PSMA PET/CT Better Characterises Localised Prostate Cancer after MRI and Transperineal Prostate Biopsy: Is 68Ga-PSMA PET/CT Guided Biopsy the Future? Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1843–1851. [Google Scholar] [CrossRef]
- Emmett, L.; Buteau, J.; Papa, N.; Moon, D.; Thompson, J.; Roberts, M.J.; Rasiah, K.; Pattison, D.A.; Yaxley, J.; Thomas, P.; et al. The Additive Diagnostic Value of Prostate-Specific Membrane Antigen Positron Emission Tomography Computed Tomography to Multiparametric Magnetic Resonance Imaging Triage in the Diagnosis of Prostate Cancer (PRIMARY): A Prospective Multicentre Study. Eur. Urol. 2021, 80, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Fassbender, T.F.; Schiller, F.; Mix, M.; Maecke, H.R.; Kiefer, S.; Drendel, V.; Meyer, P.T.; Jilg, C.A. Accuracy of [ 68 Ga]Ga-RM2-PET/CT for Diagnosis of Primary Prostate Cancer Compared to Histopathology. Nucl. Med. Biol. 2019, 70, 32–38. [Google Scholar] [CrossRef]
- Duan, H.; Baratto, L.; Fan, R.E.; Soerensen, S.J.C.; Liang, T.; Chung, B.I.; Thong, A.E.C.; Gill, H.; Kunder, C.; Stoyanova, T.; et al. Correlation of 68Ga-RM2 PET with Postsurgery Histopathology Findings in Patients with Newly Diagnosed Intermediate- or High-Risk Prostate Cancer. J. Nucl. Med. 2022, 63, 1829–1835. [Google Scholar] [CrossRef]
- Sandgren, K.; Nilsson, E.; Keeratijarut Lindberg, A.; Strandberg, S.; Blomqvist, L.; Bergh, A.; Friedrich, B.; Axelsson, J.; Ögren, M.; Ögren, M.; et al. Registration of Histopathology to Magnetic Resonance Imaging of Prostate Cancer. Phys. Imaging Radiat. Oncol. 2021, 18, 19–25. [Google Scholar] [CrossRef]
- Zamboglou, C.; Kramer, M.; Kiefer, S.; Bronsert, P.; Ceci, L.; Sigle, A.; Schultze-Seemann, W.; Jilg, C.A.; Sprave, T.; Fassbender, T.F.; et al. The Impact of the Co-Registration Technique and Analysis Methodology in Comparison Studies between Advanced Imaging Modalities and Whole-Mount-Histology Reference in Primary Prostate Cancer. Sci. Rep. 2021, 11, 5836. [Google Scholar] [CrossRef]
- Umutlu, L.; Kirchner, J.; Bruckmann, N.M.; Morawitz, J.; Antoch, G.; Ting, S.; Bittner, A.K.; Hoffmann, O.; Häberle, L.; Ruckhäberle, E.; et al. Multiparametric18F-FDG PET/MRI-Based Radiomics for Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancers 2022, 14, 1727. [Google Scholar] [CrossRef]
- Orczyk, C.; Rusinek, H.; Rosenkrantz, A.B.; Mikheev, A.; Deng, F.M.; Melamed, J.; Taneja, S.S. Preliminary Experience with a Novel Method of Three-Dimensional Co-Registration of Prostate Cancer Digital Histology and In Vivo Multiparametric MRI. Clin. Radiol. 2013, 68, e652–e658. [Google Scholar] [CrossRef]
- Jung, M.; Bogner, B.; Diallo, T.D.; Kim, S.; Arnold, P.; Füllgraf, H.; Kurowski, K.; Bronsert, P.; Jungmann, P.M.; Kiefer, J.; et al. Multiparametric Magnetic Resonance Imaging for Radiation Therapy Response Monitoring in Soft Tissue Sarcomas: A Histology and MRI Co-Registration Algorithm. Theranostics 2023, 13, 1594–1606. [Google Scholar] [CrossRef] [PubMed]
- Schiller, F.; Fechter, T.; Zamboglou, C.; Chirindel, A.; Salman, N.; Jilg, C.A.; Drendel, V.; Werner, M.; Meyer, P.T.; Grosu, A.L.; et al. Comparison of PET/CT and Whole-Mount Histopathology Sections of the Human Prostate: A New Strategy for Voxel-Wise Evaluation. EJNMMI Phys. 2017, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Bourne, R.M.; Bailey, C.; Johnston, E.W.; Pye, H.; Heavey, S.; Whitaker, H.; Siow, B.; Freeman, A.; Shaw, G.L.; Sridhar, A.; et al. Apparatus for Histological Validation of in Vivo and Ex Vivo Magnetic Resonance Imaging of the Human Prostate. Front. Oncol. 2017, 7, 47. [Google Scholar] [CrossRef]
- Gibson, E.; Gaed, M.; Gómez, J.A.; Moussa, M.; Romagnoli, C.; Pautler, S.; Chin, J.L.; Crukley, C.; Bauman, G.S.; Fenster, A.; et al. 3D Prostate Histology Reconstruction: An Evaluation of Image-Based and Fiducial-Based Algorithms. Med. Phys. 2013, 40, 093501. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Piert, M.R.; Khan, A.; Shah, R.; Hussain, H.; Siddiqui, J.; Chenevert, T.L.; Meyer, C.R. Registration Methodology for Histological Sections and In Vivo Imaging of Human Prostate. Acad. Radiol. 2008, 15, 1027–1039. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Le Nobin, J.; Orczyk, C.; Deng, F.M.; Melamed, J.; Rusinek, H.; Taneja, S.S.; Rosenkrantz, A.B. Prostate Tumour Volumes: Evaluation of the Agreement between Magnetic Resonance Imaging and Histology Using Novel Co-Registration Software. BJU Int. 2014, 114, E105–E112. [Google Scholar] [CrossRef]
- Rusu, M.; Shao, W.; Kunder, C.A.; Wang, J.B.; Soerensen, S.J.C.; Teslovich, N.C.; Sood, R.R.; Chen, L.C.; Fan, R.E.; Ghanouni, P.; et al. Registration of Presurgical MRI and Histopathology Images from Radical Prostatectomy via RAPSODI. Med. Phys. 2020, 47, 4177–4188. [Google Scholar] [CrossRef]
- Fassbender, T.F.; Schiller, F.; Zamboglou, C.; Drendel, V.; Kiefer, S.; Jilg, C.A.; Grosu, A.L.; Mix, M. Voxel-Based Comparison of [68Ga]Ga-RM2-PET/CT and [68Ga]Ga-PSMA-11-PET/CT with Histopathology for Diagnosis of Primary Prostate Cancer. EJNMMI Res. 2020, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, H.M.; Williams, S.; Zhang, A.; Chakravorty, R.; Rawlinson, D.; Ong, C.S.; Esteva, M.; Mitchell, C.; Parameswaran, B.; Finnegan, M.; et al. Development of a Registration Framework to Validate MRI with Histology for Prostate Focal Therapy. Med. Phys. 2015, 42, 7078–7089. [Google Scholar] [CrossRef]
- Eiber, M.; Weirich, G.; Holzapfel, K.; Souvatzoglou, M.; Haller, B.; Rauscher, I.; Beer, A.J.; Wester, H.J.; Gschwend, J.; Schwaiger, M.; et al. Simultaneous 68Ga-PSMA HBED-CC PET/MRI Improves the Localization of Primary Prostate Cancer. Eur. Urol. 2016, 70, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Zamboglou, C.; Schiller, F.; Fechter, T.; Wieser, G.; Jilg, C.A.; Chirindel, A.; Salman, N.; Drendel, V.; Werner, M.; Mix, M.; et al. 68Ga-HBED-CC-PSMA PET/CT versus Histopathology in Primary Localized Prostate Cancer: A Voxel-Wise Comparison. Theranostics 2016, 6, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Zamboglou, C.; Drendel, V.; Jilg, C.A.; Rischke, H.C.; Beck, T.I.; Schultze-Seemann, W.; Krauss, T.; Mix, M.; Schiller, F.; Wetterauer, U.; et al. Comparison of 68Ga-HBED-CC PSMA-PET/CT and Multiparametric MRI for Gross Tumour Volume Detection in Patients with Primary Prostate Cancer Based on Slice by Slice Comparison with Histopathology. Theranostics 2017, 7, 228–237. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Mapelli, P.; Ghezzo, S.; Samanes Gajate, A.M.; Preza, E.; Brembilla, G.; Cucchiara, V.; Ahmed, N.; Bezzi, C.; Presotto, L.; Bettinardi, V.; et al. Preliminary Results of an Ongoing Prospective Clinical Trial on the Use Of68ga-Psma And68ga-Dota-Rm2 Pet/Mri in Staging of High-Risk Prostate Cancer Patients. Diagnostics 2021, 11, 2068. [Google Scholar] [CrossRef]
- Gibson, E.; Crukley, C.; Gaed, M.; Gõmez, J.A.; Moussa, M.; Chin, J.L.; Bauman, G.S.; Fenster, A.; Ward, A.D. Registration of Prostate Histology Images to Ex Vivo MR Images via Strand-Shaped Fiducials. J. Magn. Reson. Imaging 2012, 36, 1402–1412. [Google Scholar] [CrossRef]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A.; Al-Hussain, T.; Algaba, F.; Aron, M.; Berman, D.; et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Pantazis, D.; Joshi, A.; Jiang, J.; Shattuck, D.W.; Bernstein, L.E.; Damasio, H.; Leahy, R.M. Comparison of Landmark-Based and Automatic Methods for Cortical Surface Registration. Neuroimage 2010, 49, 2479–2493. [Google Scholar] [CrossRef] [PubMed]
- Wildeboer, R.R.; van Sloun, R.J.G.; Postema, A.W.; Mannaerts, C.K.; Gayet, M.; Beerlage, H.P.; Wijkstra, H.; Mischi, M. Accurate Validation of Ultrasound Imaging of Prostate Cancer: A Review of Challenges in Registration of Imaging and Histopathology. J. Ultrasound 2018, 21, 197–207. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Chan, T.H.; Haworth, A.; Wang, A.; Osanlouy, M.; Williams, S.; Mitchell, C.; Hofman, M.S.; Hicks, R.J.; Murphy, D.G.; Reynolds, H.M. Detecting Localised Prostate Cancer Using Radiomic Features in PSMA PET and Multiparametric MRI for Biologically Targeted Radiation Therapy. EJNMMI Res. 2023, 13, 34. [Google Scholar] [CrossRef]
- Shao, W.; Banh, L.; Kunder, C.A.; Fan, R.E.; Soerensen, S.J.C.; Wang, J.B.; Teslovich, N.C.; Madhuripan, N.; Jawahar, A.; Ghanouni, P.; et al. ProsRegNet: A Deep Learning Framework for Registration of MRI and Histopathology Images of the Prostate. Med. Image Anal. 2021, 68, 101919. [Google Scholar] [CrossRef]
- de Vos, B.D.; Berendsen, F.F.; Viergever, M.A.; Sokooti, H.; Staring, M.; Išgum, I. A Deep Learning Framework for Unsupervised Affine and Deformable Image Registration. Med. Image Anal. 2019, 52, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Han, X.; Xu, Z.; Niethammer, M. Networks for Joint Affine and Non-Parametric Image Registration. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Fort Collins, CO, USA, 23–25 June 1999; Volume 2019, pp. 4219–4228. [Google Scholar]


| N | Age (Years) | PSA Level at Diagnosis (ng/mL) | GS at Biopsy | GS at Prostatectomy | Pathological T Stage |
|---|---|---|---|---|---|
| 1 | 74 | 6.37 | 9 (5 + 4) | 9 (4 + 5) | pT2c |
| 2 | 64 | 4.4 | 8 (4 + 4) | 9 (4 + 5) | pT3b |
| 3 | 66 | 6.37 | 9 (4 + 5) | 7 (4 + 3) | pT3a |
| Patient | [68Ga]Ga-RM2 PET-Histology | [68GA]Ga-PSMA PET-Histology |
|---|---|---|
| Patient 1–slice 1 | 0.773 | 0.555 |
| Patient 1–slice 2 | 0.814 | 0.542 |
| Patient 2 | 0.746 | 0.751 |
| Patient 3–slice 1 | 0.624 | 0 |
| Patient 3–slice 2 | 0.596 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghezzo, S.; Neri, I.; Mapelli, P.; Savi, A.; Samanes Gajate, A.M.; Brembilla, G.; Bezzi, C.; Maghini, B.; Villa, T.; Briganti, A.; et al. [68Ga]Ga-PSMA and [68Ga]Ga-RM2 PET/MRI vs. Histopathological Images in Prostate Cancer: A New Workflow for Spatial Co-Registration. Bioengineering 2023, 10, 953. https://doi.org/10.3390/bioengineering10080953
Ghezzo S, Neri I, Mapelli P, Savi A, Samanes Gajate AM, Brembilla G, Bezzi C, Maghini B, Villa T, Briganti A, et al. [68Ga]Ga-PSMA and [68Ga]Ga-RM2 PET/MRI vs. Histopathological Images in Prostate Cancer: A New Workflow for Spatial Co-Registration. Bioengineering. 2023; 10(8):953. https://doi.org/10.3390/bioengineering10080953
Chicago/Turabian StyleGhezzo, Samuele, Ilaria Neri, Paola Mapelli, Annarita Savi, Ana Maria Samanes Gajate, Giorgio Brembilla, Carolina Bezzi, Beatrice Maghini, Tommaso Villa, Alberto Briganti, and et al. 2023. "[68Ga]Ga-PSMA and [68Ga]Ga-RM2 PET/MRI vs. Histopathological Images in Prostate Cancer: A New Workflow for Spatial Co-Registration" Bioengineering 10, no. 8: 953. https://doi.org/10.3390/bioengineering10080953
APA StyleGhezzo, S., Neri, I., Mapelli, P., Savi, A., Samanes Gajate, A. M., Brembilla, G., Bezzi, C., Maghini, B., Villa, T., Briganti, A., Montorsi, F., De Cobelli, F., Freschi, M., Chiti, A., Picchio, M., & Scifo, P. (2023). [68Ga]Ga-PSMA and [68Ga]Ga-RM2 PET/MRI vs. Histopathological Images in Prostate Cancer: A New Workflow for Spatial Co-Registration. Bioengineering, 10(8), 953. https://doi.org/10.3390/bioengineering10080953