Discovery Viewer (DV): Web-Based Medical AI Model Development Platform and Deployment Hub
Abstract
:1. Introduction
2. Research Projects Using DV
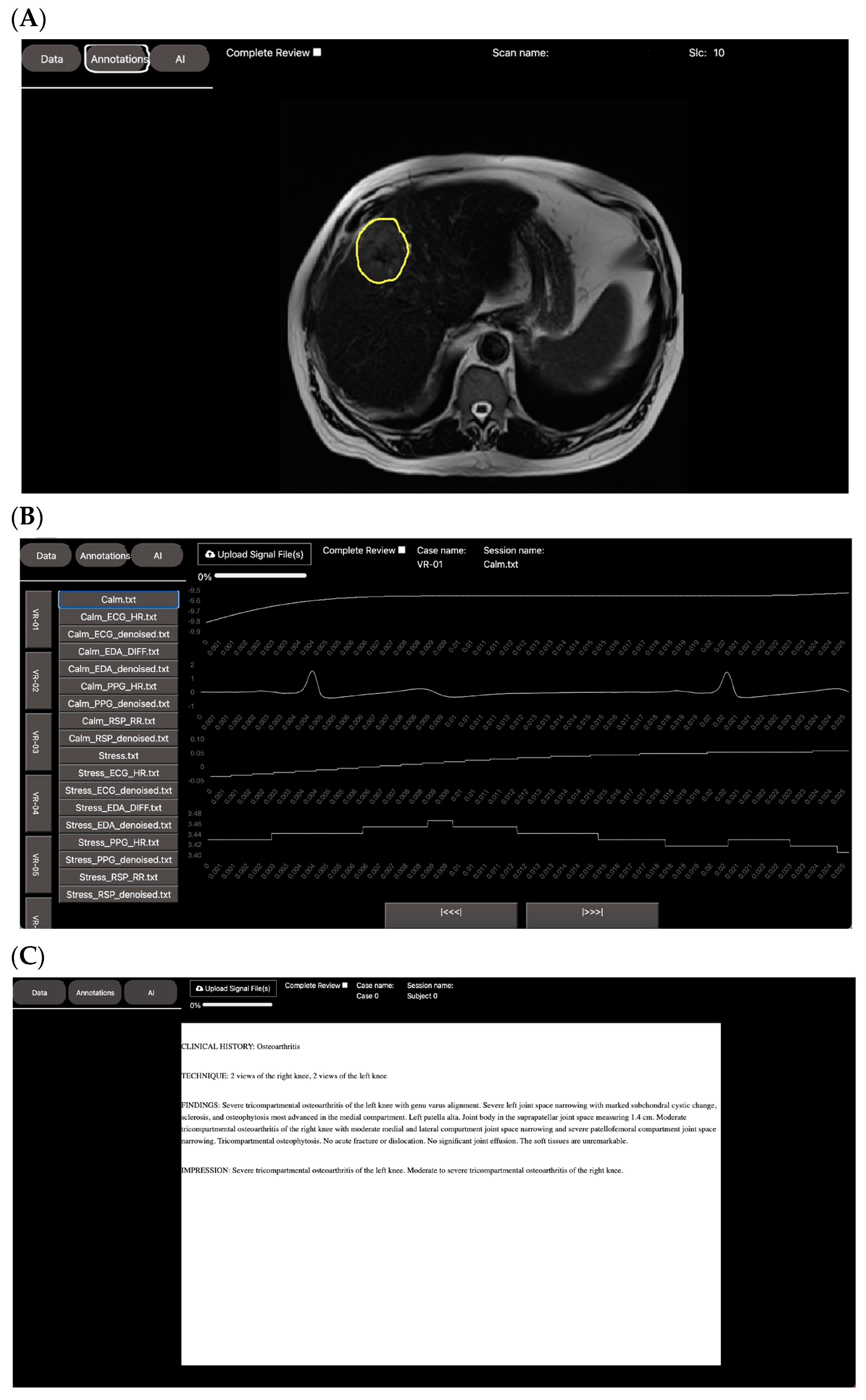
2.1. Leveraging DV for Visualization and Annotation
2.1.1. Spiral VIBE UTE-MRI for Lung Imaging in Post-COVID-19 Patients
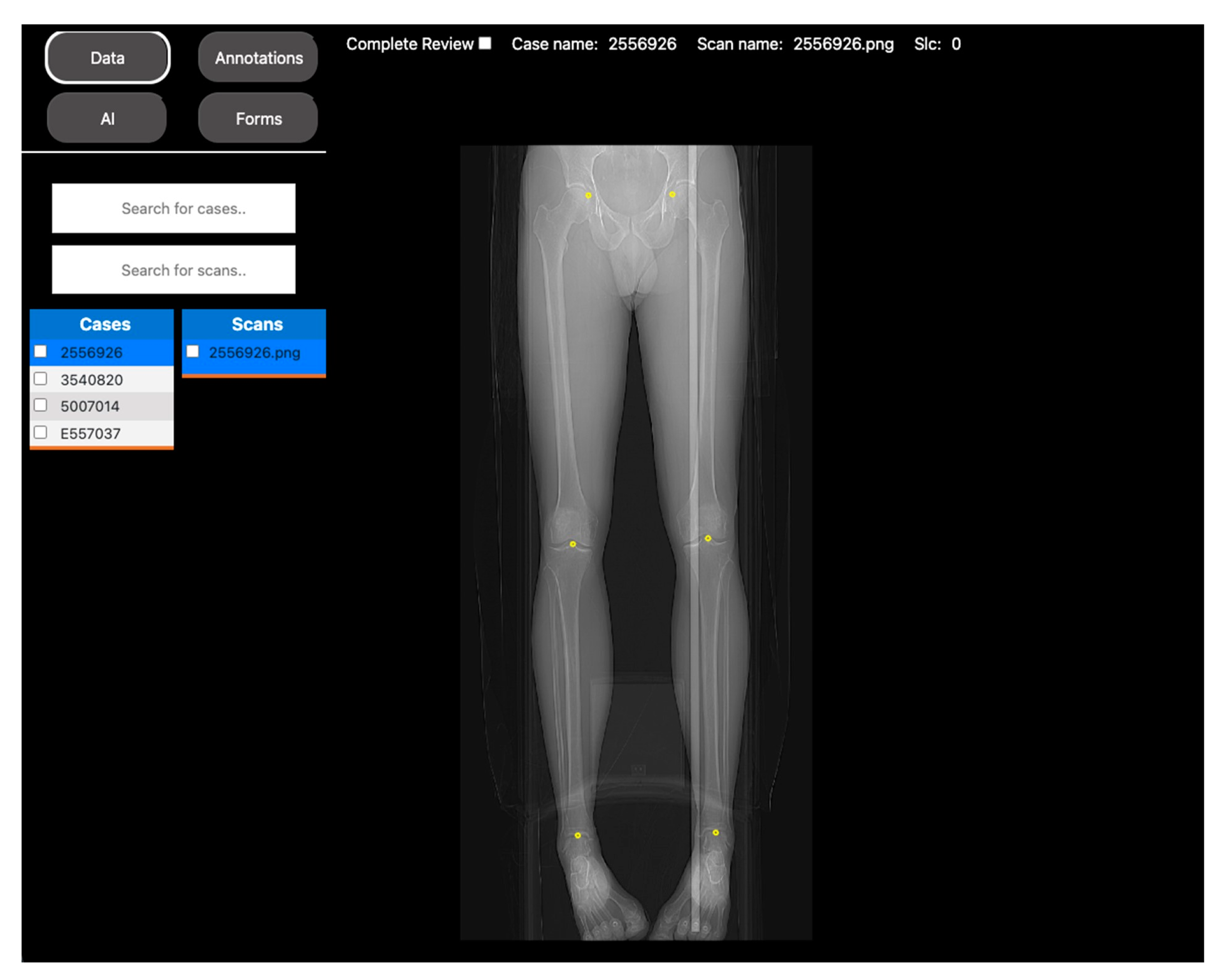
2.1.2. Automated Measurements of Leg Length on Radiographs by Deep Learning
2.2. Leveraging DV for Non-AI Expert Model Development
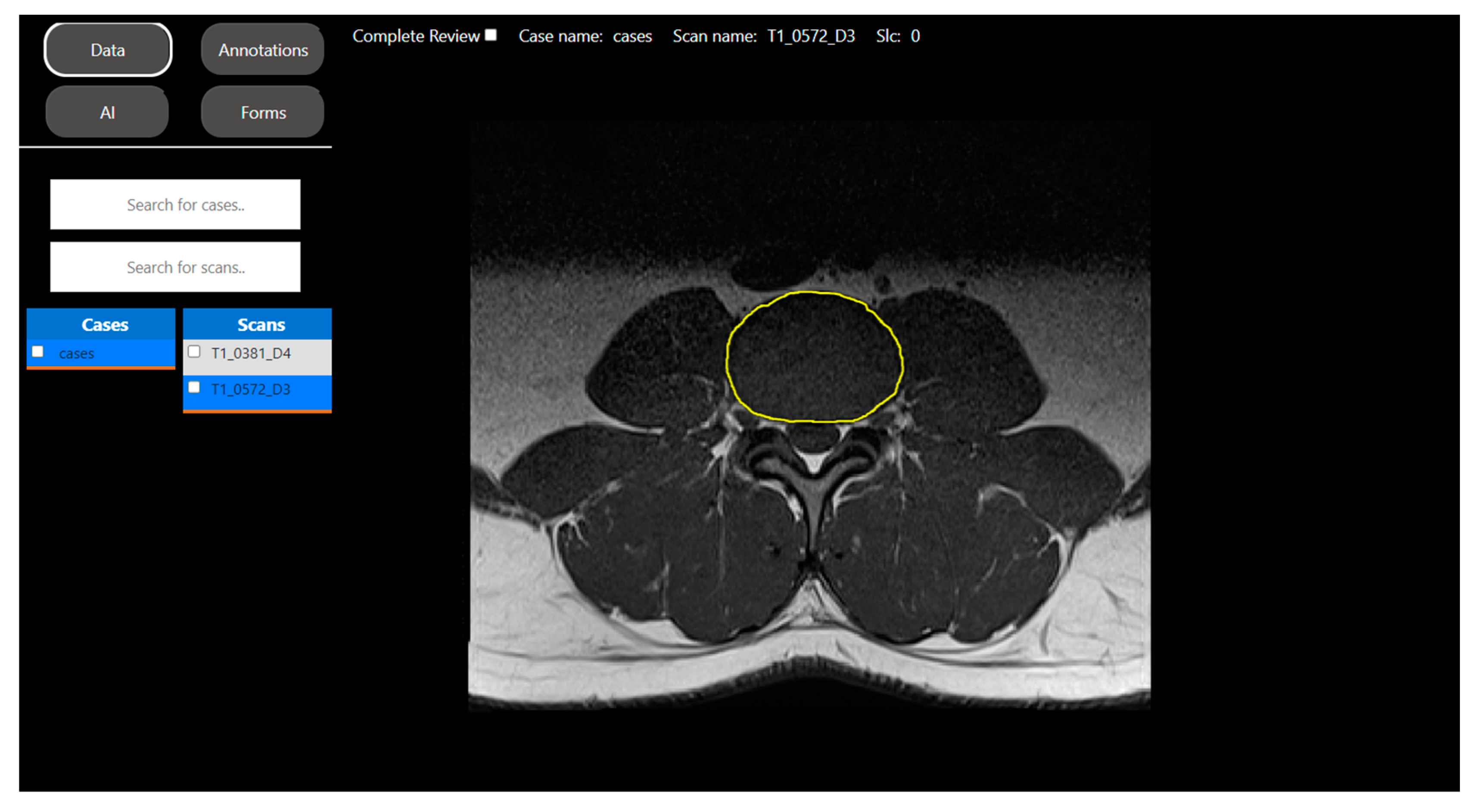
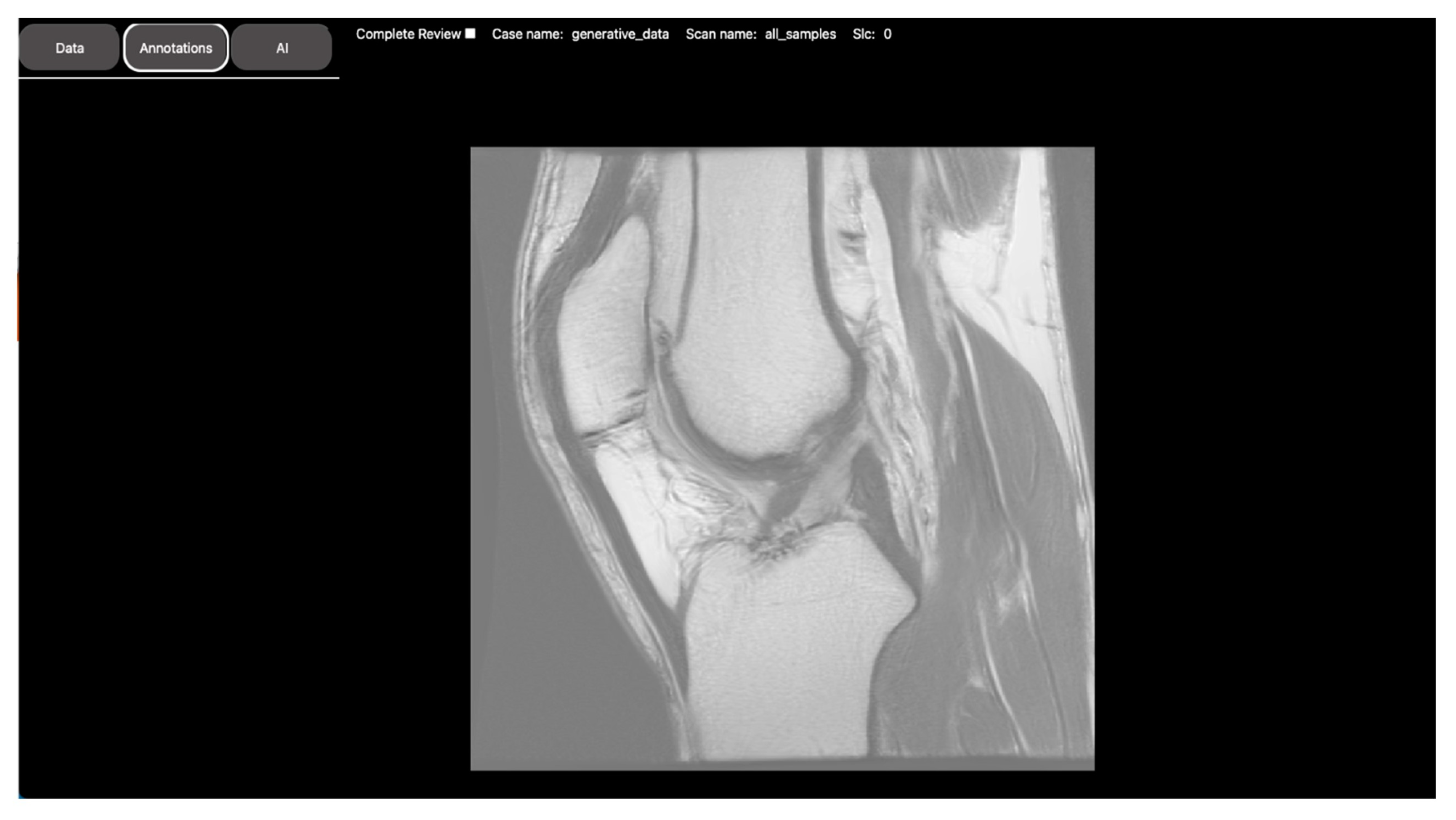
2.2.1. Segmentation Task: Intravertebral Disk Segmentation on T1-Weighted Axial-View MRI Slices
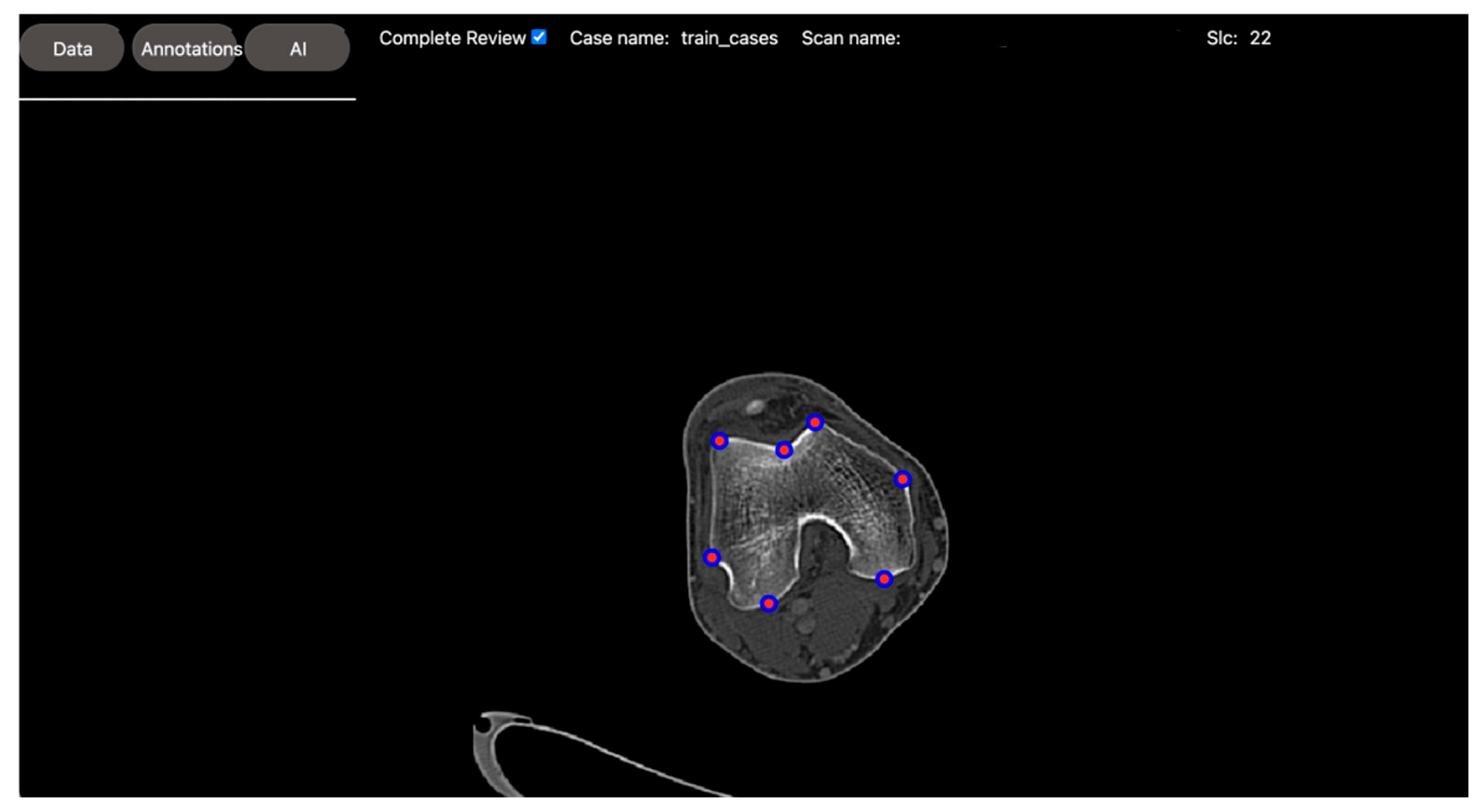
2.2.2. Regression Task: Automated Measurement of Patellofemoral Anatomic Landmarks
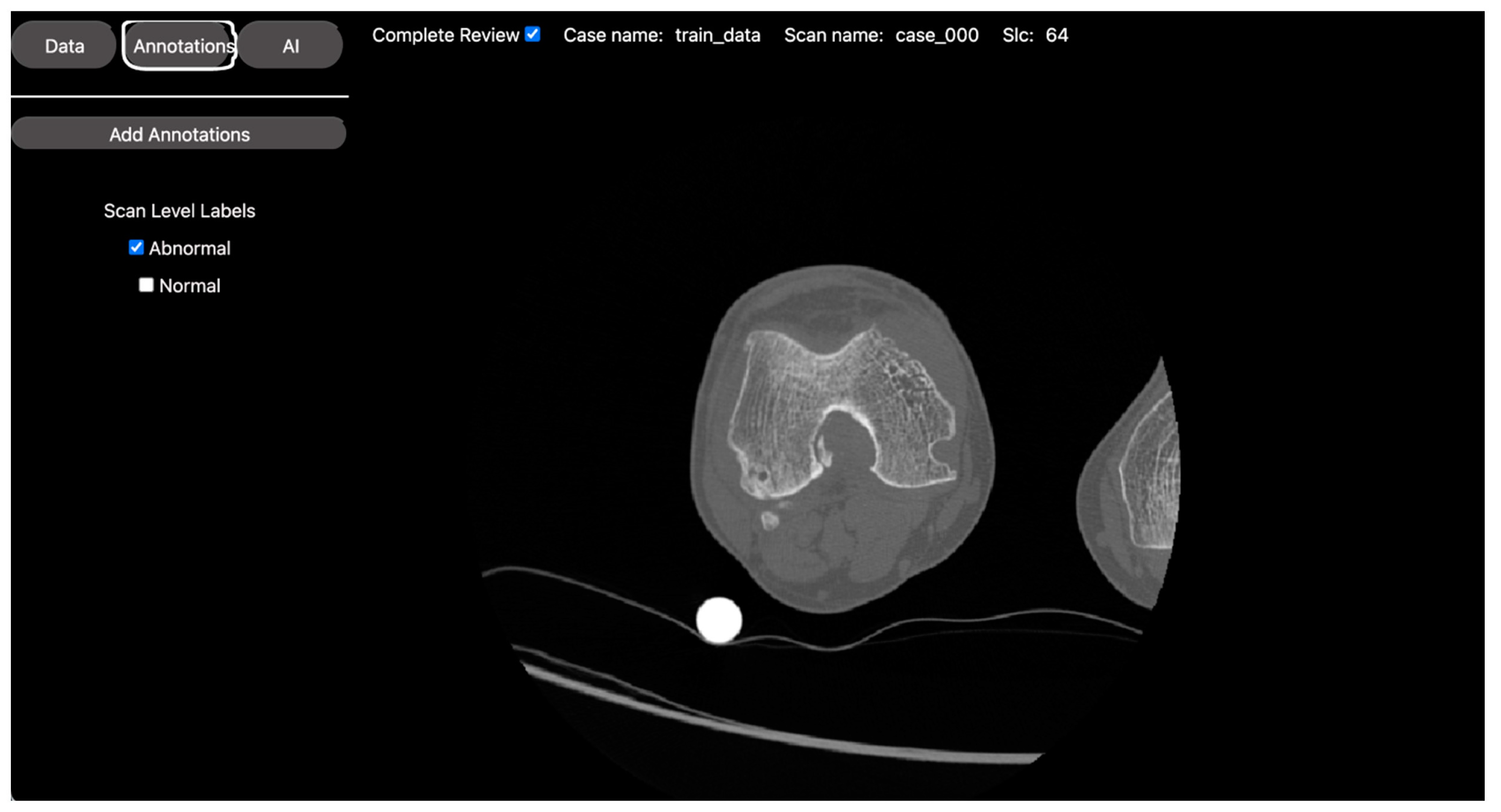
2.2.3. Classification Task: Normal vs. Osteoarthritis Knee Axial View CT Slice Classification
3. Materials and Methods
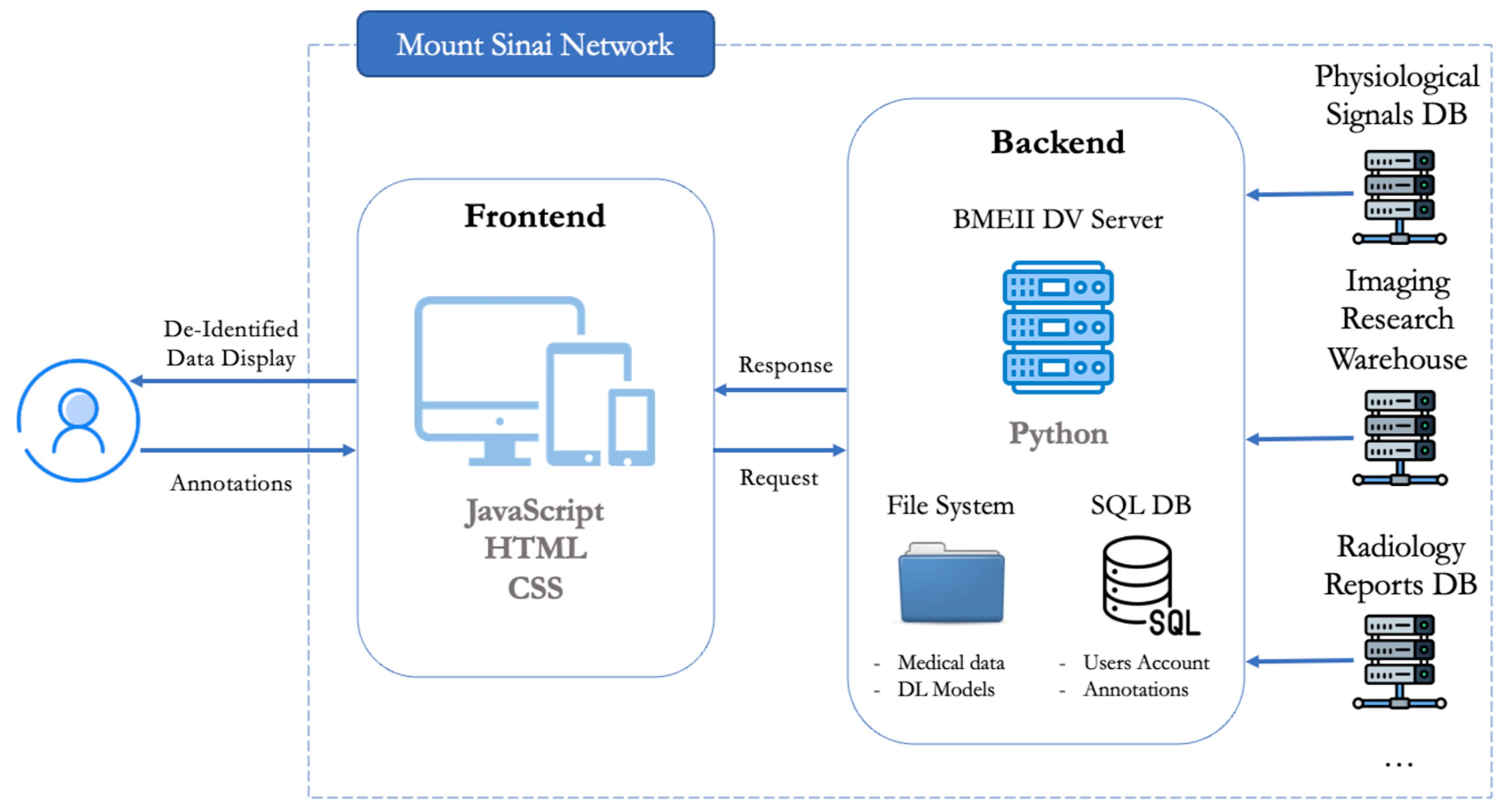
3.1. Web App Design
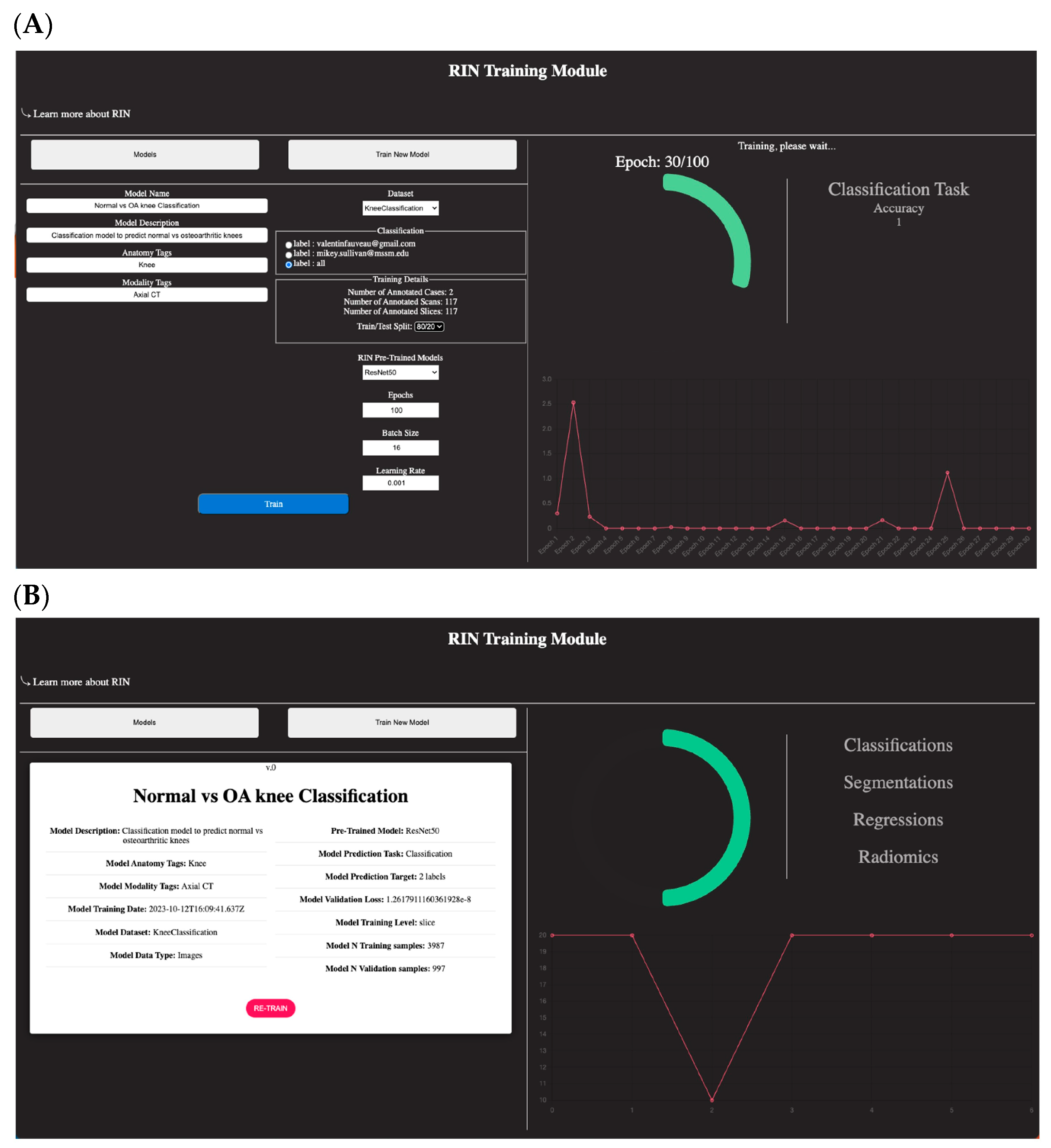
3.2. AI Project Setup
- -
- A tutorial on how to use the annotation tool (windowing, scrolling, zooming, and annotation/labeling).
- -
- A tutorial on how to use the training module to select the annotated dataset and train a new model through a transfer learning approach. The options for model training are limited to just a couple of parameters: batch size, number of epochs, learning rate, and selection of a pre-trained model. Regarding RIN pre-trained models, DV presently offers the following options. However, the plan is to expand and enhance the selection of pre-trained networks in line with the evolving architectural trends in the literature.
- ○
- For segmentation: ResNet50UNet.
- ○
- For regression: Resnet50, DenseNet121, InceptionV3, and InceptionResNetV2.
- ○
- For classification: Resnet50, DenseNet121, InceptionV3, and InceptionResNetV2.
3.3. Datasets
3.3.1. Knee CT Dataset
3.3.2. Lumbar Spine MRI Dataset
3.4. Model Building
3.4.1. Best Model for Segmentation Task
3.4.2. Best Model for Regression Task
3.4.3. Best Model for Classification Task
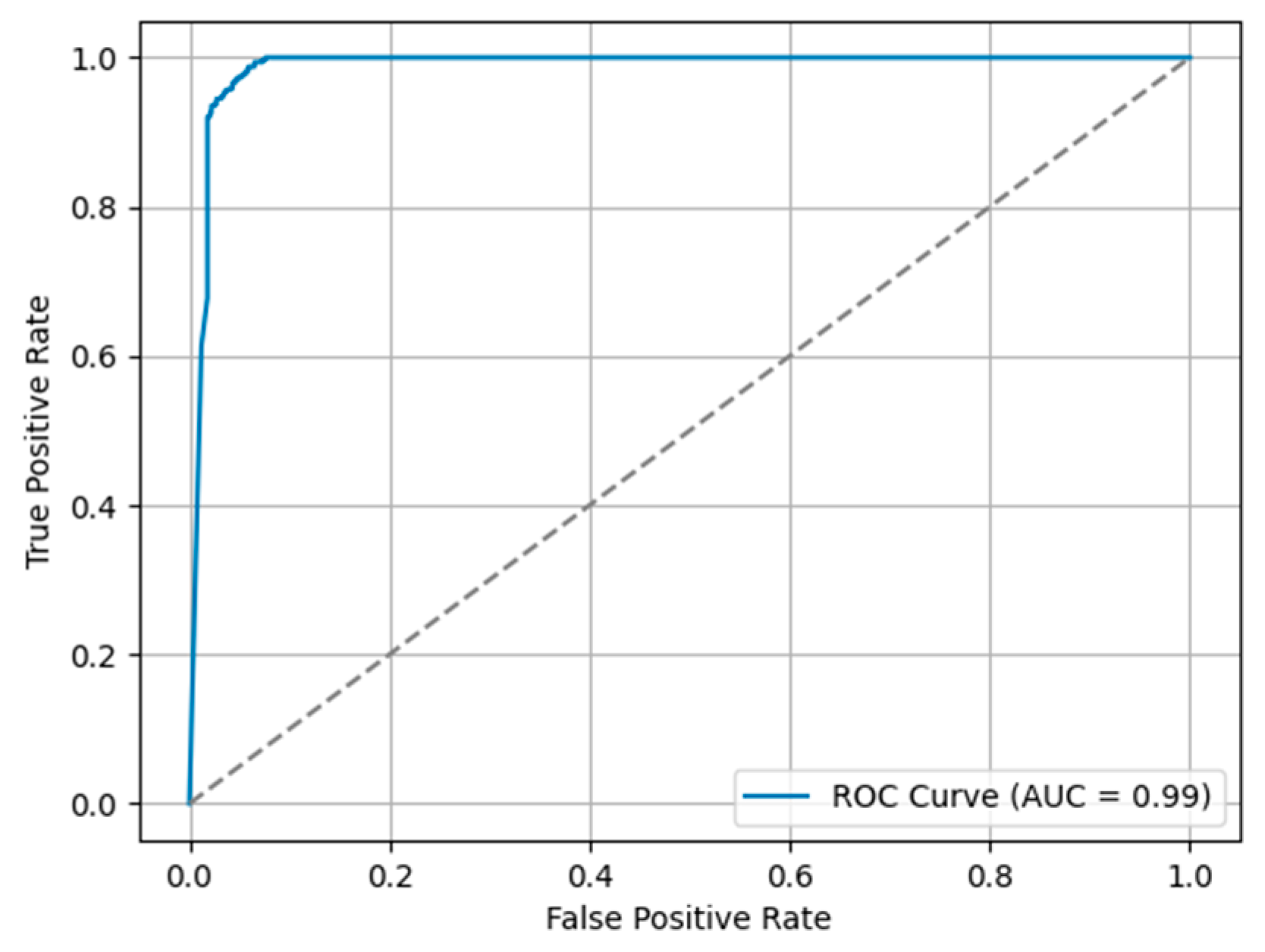
4. Results
4.1. Segmentation Model Performance
4.2. Regression Model Performance
4.3. Classification Model Performance
5. Discussion and Conclusions
6. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardoso, M.J.; Li, W.; Brown, R.; Ma, N.; Kerfoot, E.; Wang, Y.; Murrey, B.; Myronenko, A.; Zhao, C.; Yang, D.; et al. MONAI: An open-source framework for deep learning in healthcare. arXiv 2022, arXiv:arXiv:2211.02701. [Google Scholar] [CrossRef]
- Roberts, M.; Driggs, D.; Thorpe, M.; Gilbey, J.; Yeung, M.; Ursprung, S.; Aviles-Rivero, A.I.; Etmann, C.; McCague, C.; Beer, L.; et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID 19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021, 3, 199–217. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The Future of Digital Health with Federated learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Dayan, I.; Roth, H.R.; Zhong, A.; Harouni, A.; Gentili, A.; Abidin, A.Z.; Liu, A.; Costa, A.B.; Wood, B.J.; Tsai, C.-S.; et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021, 27, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Liu, Z.; Robson, P.M.; Marinelli, B.; Huang, M.; Doshi, A.; Jacobi, A.; Cao, C.; Link, K.E.; Yang, T.; et al. RadImageNet: An Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning. Radiol. Artif. Intell. 2022, 4, e210315. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Lee, H.-C.; Diao, K.-Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Liu, Z.; Singh, A.; Lange, M.; Boddu, P.; Gong, J.Q.X.; Lee, J.; DeMarco, C.; Cao, C.; Platt, S.; et al. Interstitial lung disease diagnosis and prognosis using an AI system integrating longitudinal data. Nat. Commun. 2023, 14, 2272. [Google Scholar] [CrossRef] [PubMed]
- Cahan, N.; Klang, E.; Marom, E.M.; Soffer, S.; Barash, Y.; Burshtein, E.; Konen, E.; Greenspan, H. Multimodal fusion models for pulmonary embolism mortality prediction. Sci. Rep. 2023, 13, 7544. [Google Scholar] [CrossRef]
- Fauveau, V.; Jacobi, A.; Bernheim, A.; Chung, M.; Benkert, T.; Fayad, Z.A.; Feng, L. Performance of spiral UTE-MRI of the lung in post-COVID patients. Magn. Reson. Imaging 2023, 96, 135–143. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, A.; Liu, S.; Deyer, L.; Deyer, T.; Lee, H.C.; Yang, Y.; Lee, J.; Fayad, Z.A.; Hayden, B.; et al. Automated measurements of leg length on radiographs by deep learning. In Proceedings of the 2022 IEEE 22nd International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan, 7–9 November 2022; pp. 17–22. [Google Scholar] [CrossRef]
- Jiang, H.; Qi, W.; Liao, Q.; Zhao, H.; Lei, W.; Guo, L.; Lu, H. Quantitative evaluation of lumbar disc herniation based on MRI image. In Abdominal Imaging. Computational and Clinical Applications; Springer: Berlin, Germany, 2012; pp. 91–98. [Google Scholar]
- Alomari, R.S.; Corso, J.J.; Chaudhary, V.; Dhillon, G. Lumbar spine disc herniation diagnosis with a joint shape model. In Computational Methods and Clinical Applications for Spine Imaging; Springer: Cham, Switzerland, 2014; pp. 87–98. [Google Scholar]
- Bellemans, J.; Carpentier, K.; Vandenneucker, H.; Vanlauwe, J.; Victor, J. The John Insall Award: Both Morphotype and Gender Influence the Shape of the Knee in Patients Undergoing TKA. Clin. Orthop. Relat. Res. 2010, 468, 29–36. [Google Scholar] [CrossRef]
- Altman, R.D. Early management of osteoarthritis. Am. J. Manag. Care 2010, 16, S41–S47. [Google Scholar] [PubMed]
- Liu, Z.; Zhou, A.; Fauveau, V.; Lee, J.; Marcadis, P.; Fayad, Z.A.; Chan, J.J.; Gladstone, J.; Mei, X.; Huang, M. Deep Learning for Automated Measurement of Patellofemoral Anatomic Landmarks. Bioengineering 2023, 10, 815. [Google Scholar] [CrossRef]
- Sudirman, S.; Al Kafri, A.; Natalia, F.; Meidia, H.; Afriliana, N.; Al-Rashdan, W.; Bashtawi, M.; Al-Jumaily, M. Lumbar Spine MRI Dataset. Mendeley Data, V2. 2019. Available online: https://data.mendeley.com/datasets/k57fr854j2/2 (accessed on 30 November 2023). [CrossRef]
- Yakubovskiy, P. Segmentation Models. GitHub Repository. GitHub, 2019. Available online: https://github.com/qubvel/segmentation_models (accessed on 30 November 2023).
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000, 120, 122–125. [Google Scholar]
- Icahn School of Medicine at Mount Sinai. AI Ready Mount Sinai. 2023. Available online: https://labs.icahn.mssm.edu/airms/ (accessed on 30 November 2023).


 |  |  |  | |
| Web Based | ✔ | ✔ | ⅹ | ✔ |
| Medical multi-modality data viewers | ✔ | ⅹ | ⅹ | ⅹ |
| AI model training | ✔ | ✔ (Requires programming) | ⅹ | ⅹ |
| AI model deployment | ✔ | ✔ (Requires programming) | ✔ (Requires programming) | ✔ (Requires programming) |
| Accessibility | Zero-footprint and secured web-based access | Requires Python based installation packages | Desktop software | Zero-footprint web-app |
| Project Name | Spiral VIBE UTE-MRI for Lung Imaging in Post-COVID Patients [9] | Automated Measurements of Leg Length on Radiographs by Deep Learning [10] |
|---|---|---|
| Objective | Assess the diagnostic image quality of a novel MRI sequence to image the lungs (see Figure 1). | Develop a DL model to automatically identify anatomical landmarks in full leg radiographs (see Figure 2). |
| Methodology | Three radiologists assessed the diagnostic image quality of a group of post-COVID-19 patients who underwent both an MRI and a CT scan of the lungs. | A radiologist annotated three anatomical landmarks in a cohort of full-length radiographs. |
| DV project set up | Each patient’s MRI and CT images, along with the diagnostic quality metrics for scoring, were incorporated into a DV project. Access to the project was then provided to the three radiologists to review the images. | The training set of the cohort was located into a DV project, along with the points annotation tool. Access was given to the radiologist to annotate the images. |
| Results | The scores were extracted from the platform as a CSV file for statistical analysis. | The coordinates of the annotations were extracted as a CSV file to use as the ground truth for DL model training. |
| Jaccard Score | Dice Score | Sensitivity | Specificity | |
|---|---|---|---|---|
| Mean | 0.89 | 0.93 | 0.9 | 0.99 |
| Std | 0.14 | 0.14 | 0.14 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fauveau, V.; Sun, S.; Liu, Z.; Mei, X.; Grant, J.; Sullivan, M.; Greenspan, H.; Feng, L.; Fayad, Z.A. Discovery Viewer (DV): Web-Based Medical AI Model Development Platform and Deployment Hub. Bioengineering 2023, 10, 1396. https://doi.org/10.3390/bioengineering10121396
Fauveau V, Sun S, Liu Z, Mei X, Grant J, Sullivan M, Greenspan H, Feng L, Fayad ZA. Discovery Viewer (DV): Web-Based Medical AI Model Development Platform and Deployment Hub. Bioengineering. 2023; 10(12):1396. https://doi.org/10.3390/bioengineering10121396
Chicago/Turabian StyleFauveau, Valentin, Sean Sun, Zelong Liu, Xueyan Mei, James Grant, Mikey Sullivan, Hayit Greenspan, Li Feng, and Zahi A. Fayad. 2023. "Discovery Viewer (DV): Web-Based Medical AI Model Development Platform and Deployment Hub" Bioengineering 10, no. 12: 1396. https://doi.org/10.3390/bioengineering10121396
APA StyleFauveau, V., Sun, S., Liu, Z., Mei, X., Grant, J., Sullivan, M., Greenspan, H., Feng, L., & Fayad, Z. A. (2023). Discovery Viewer (DV): Web-Based Medical AI Model Development Platform and Deployment Hub. Bioengineering, 10(12), 1396. https://doi.org/10.3390/bioengineering10121396






