Brain-Inspired Spatio-Temporal Associative Memories for Neuroimaging Data Classification: EEG and fMRI
Abstract
:1. Introduction
2. SNN, the NeuCube Framework, and the STAM on the NeuCube Concept
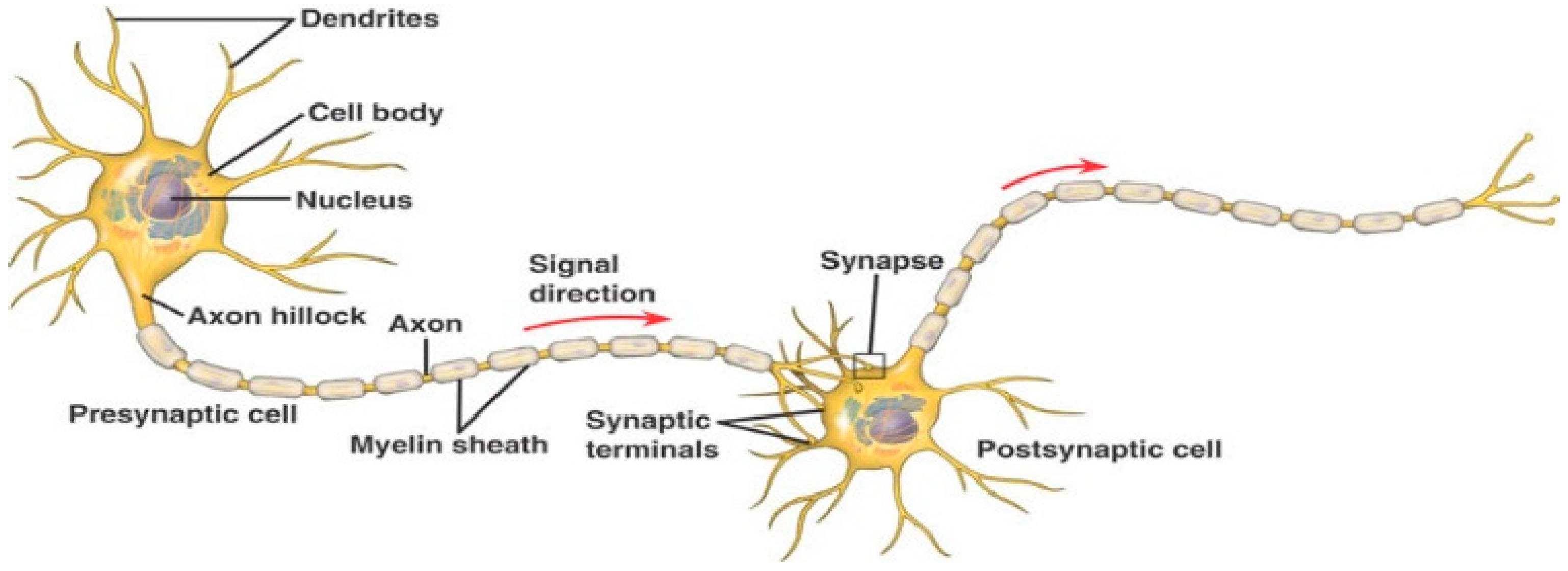
2.1. Spiking Neural Networks (SNN)
2.2. The NeuCube Framework
2.3. The STAM on NeuCube Concept
- Temporal association accuracy: validating the full model on partial temporal data of the same variables.
- Spatial association accuracy: validation of the full model on full or partial temporal data, but on a subset of variables.
- Temporal generalization accuracy: validation of the full model on partial temporal data of the same variables or a subset of them, but on a new data set.
- Spatial generalization accuracy: validation of the full model on full or partial temporal data and a subset of variables, using a new data set.
3. The Proposed STAM-NI Classification Model and Its Mathematical Description
- (1)
- (2)
- (3)
- The encoded sequences of all NI variables V were used to train a SNNcube in unsupervised mode using the STDP rule (Equation (1)), creating a connectionist structure. Before training, the SNNcube was initialized with the use of the small-world connectivity model, where neuron a was connected to other neuron b with a probability Pa,b that depended on the closeness of the two neurons. The closer they are (the smaller the distance between them Da,b), the higher the probability of connecting them (Equation (2)).λ is a parameter.
- (4)
- The trained SNNcube was recalled (activated) by all NI spatio-temporal data samples, one by one, using all variables as in step 3. For every sample Pi, a state Si of the SNNcube was defined during the propagation of the input sequence. The state Si was defined as a sequence of activated neurons Ni1, Ni2, …, Nil over time, that was used to train a deSNN classifier in a supervised mode [11], forming an l-element vector Wi of connection weights of an output neuron Oi assigned to the class of the input sequence Pi. For the supervised learning in the deSNN classifier, Equations (3) and (4) were used:Wj,i = α Modorder(j,i)ΔWj,i (t) = ej(t) Dwhere Mod is a modulation factor defining the importance of the order of the spike arriving at a synapse j of output neuron Oi, ej(t) = 1 if there is a consecutive spike at synapse j at time t during the presentation of the learned pattern by the output neuron I, and (−1) otherwise. In general, the drift parameter D can be different for ‘up’ and ‘down’ drifts, and α is a parameter.
- (5)
- When a new input sequence Pnew is presented, either as a full sequence in time and/or space (number of input variables) or as a partial one for STAM, a new SNNcube state Snew was learned as a new output neuron Onew. Its weight vector Wnew is compared with the weight vectors of the existing output neurons for classification tasks using the k-nearest neighbor method. The new sample Pnew was classified based on the pre-defined output classes of the closest, according to the Euclidean distance, output weight vectors Wi (Equation (5)).Class (Pnew) = Class (Pi), if |Wnew − Wi| < |Wnew − Wk|,for all output neurons Ok.
- (6)
- The temporal and spatial association and generalization accuracy were calculated.
4. The Proposed STAM-EEG Classification Method and Experimental Case Study
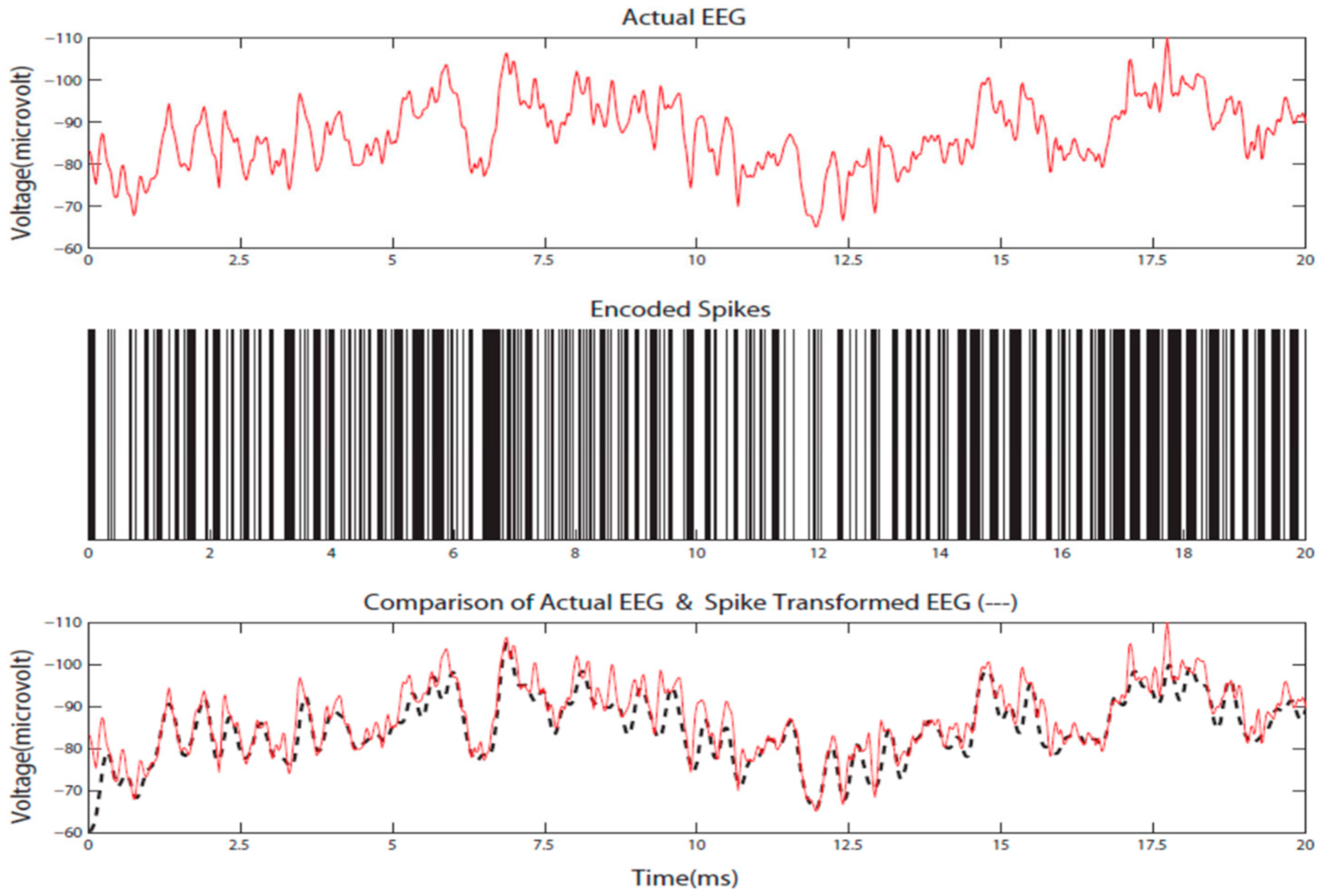
4.1. The Proposed STAM-EEG Classification Method
- i.
- Defining the spatial and temporal components of the EEG data for the classification task, e.g., EEG channels and EEG time series data.
- ii.
- Designing a SNNcube that is structured according to a brain template suitable for the EEG data (e.g., Talairach, or MNI, etc.).
- iii.
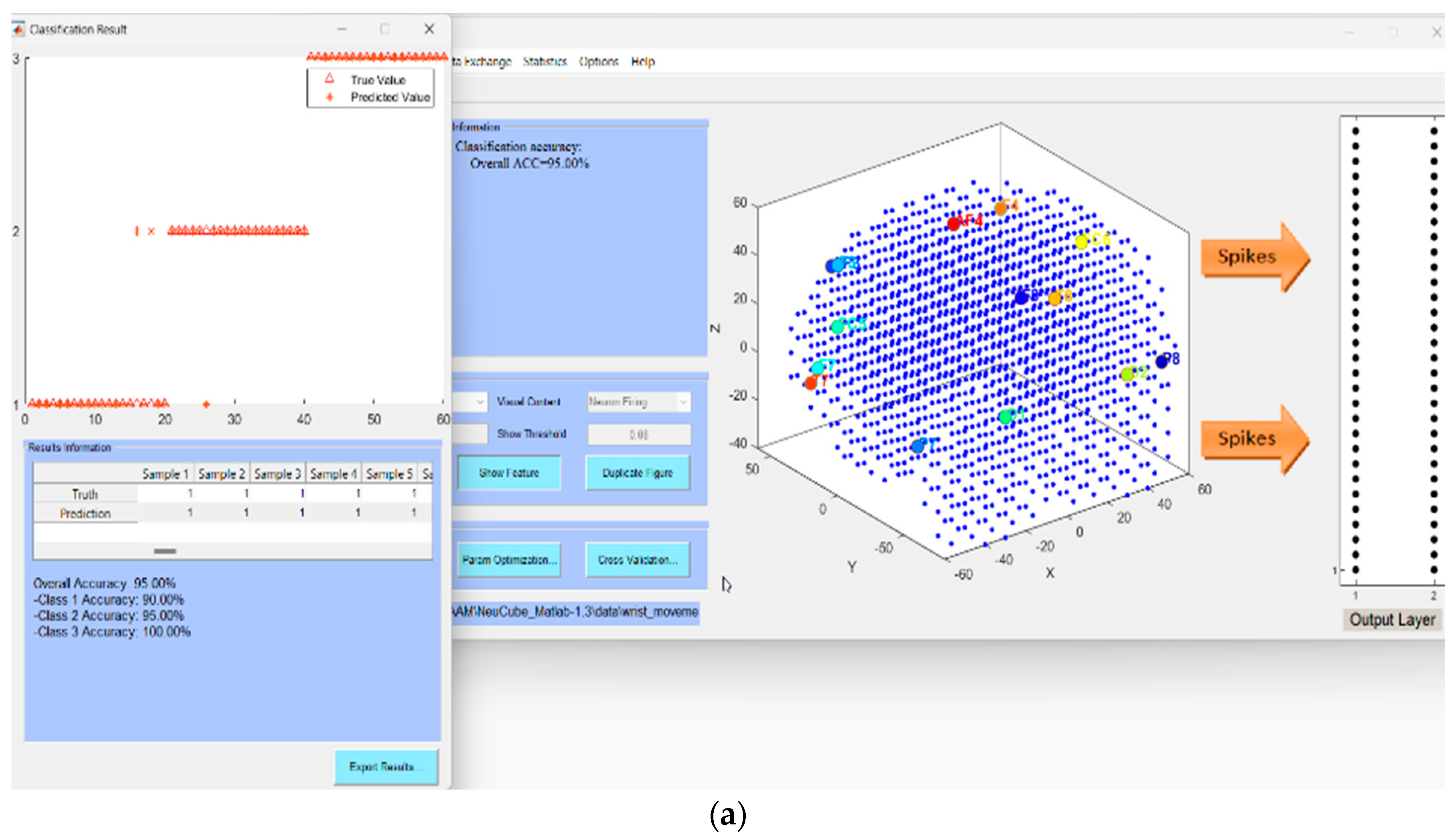
- Defining the mapping in the input EEG channels into the SNNcube 3D structure (see Figure 4a as an example of mapping 14 EEG channels in a Talairach-structured SNNcube).
- iv.
- Encoding data and training a NeuCube model to classify complete spatio-temporal EEG data, having K EEG channels measured over a full-time T.
- v.
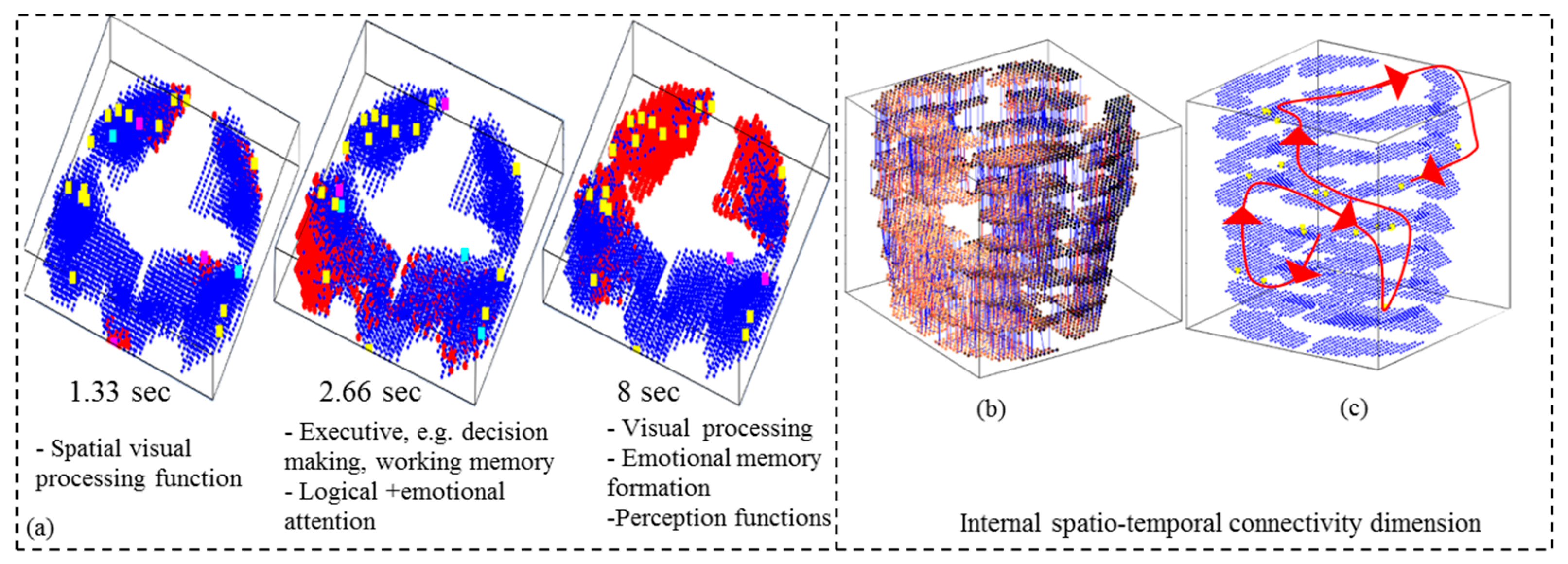
- Analyse the model through cluster analysis, spiking activity, and the EEG channel spiking proportional diagram (see for example Figure 4b,c).
- vi.
- Recall the STAM-EEG model on the same data and same variables but measured over time T1 < T to calculate the classification temporal association accuracy.
- vii.
- Recall the STAM-EEG model on K1 < K EEG channels to evaluate the classification spatial association accuracy.
- viii.
- Recall the model on the same variables, measured over time T or T1 < T on new data to calculate the classification temporal generalization accuracy.
- ix.
- Recall the NeuCube model on K1 < K EEG channels to evaluate the classification spatial generalization accuracy using a new EEG dataset.
- x.
- Evaluate the K1 EEG channels as potential classification EEG biomarkers for an early diagnosis or prognosis according to the problem at hand.
4.2. Experimental Results
5. STAM-fMRI for Classification
5.1. The Proposed STAM-fMRI Classification Method
- i.
- Defining the spatial and temporal components of the fMRI data for the classification task, e.g., fMRI voxels and the time series measurement.
- ii.
- iii.
- iv.
- Encode data and train a NeuCube model to classify a complete spatio-temporal fMRI data, having K variables as inputs measured over time T.
- v.
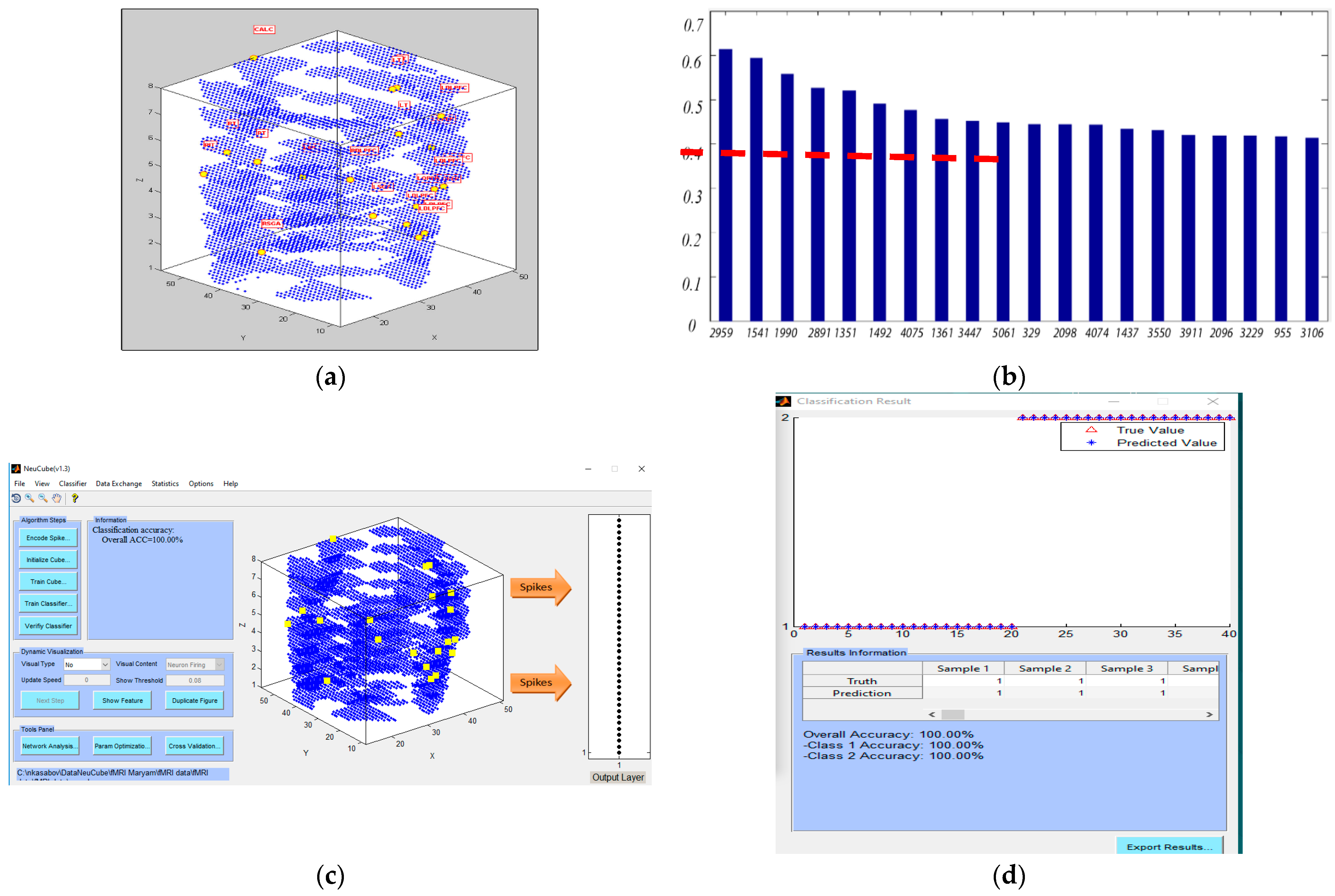
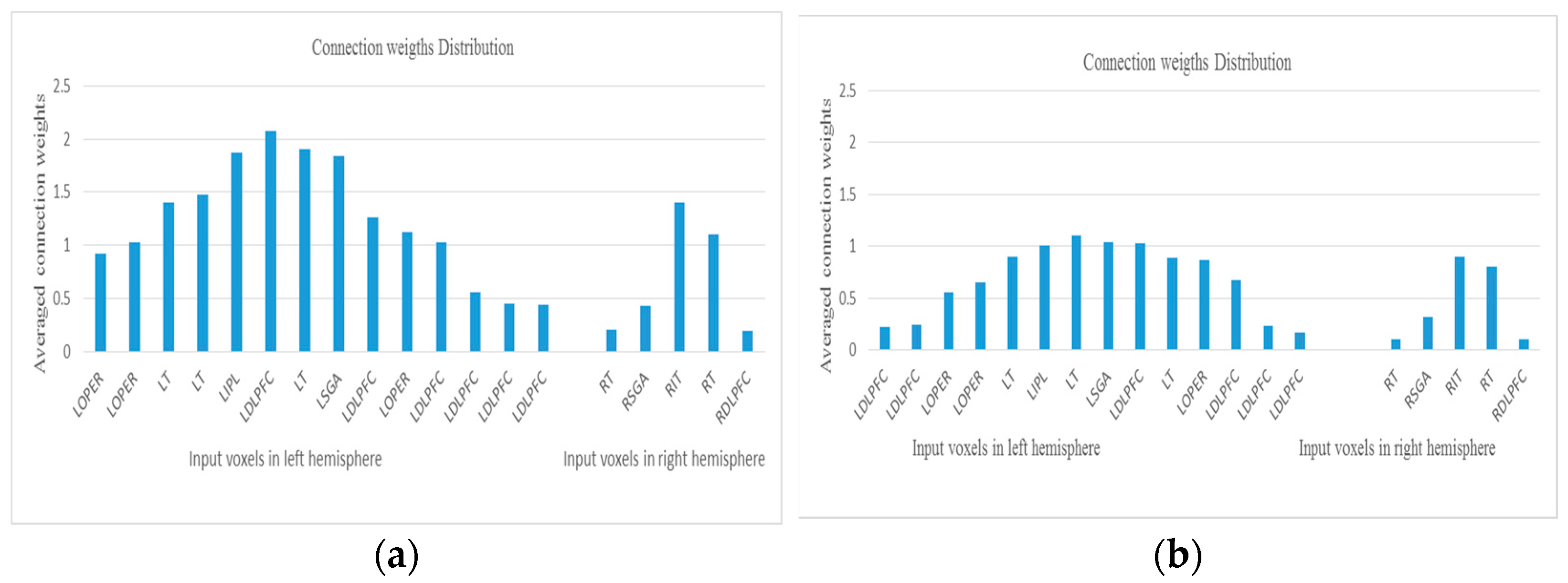
- Analyse the model through connectivity and spiking activity analysis around the input voxels (Table 3).
- vi.
- Recall the STAM-fMRI model on the same data and same variables but measured over time T1 < T to calculate the classification temporal association accuracy.
- vii.
- Recall the STAM-fMRI on K1 < K EEG channels to evaluate the classification spatial association accuracy.
- viii.
- Recall the model on the same variables, measured over time T or T1 < T on new data to calculate the classification temporal generalization accuracy.
- ix.
- Recall the NeuCube model on K1 < K variables to evaluate the classification spatial generalization accuracy using a new fMRI dataset.
- x.
- wRank and evaluate the K1 fMRI features/variables as potential classification biomarkers (Section 5.5).
5.2. STAM-fMRI for Classification of Experimental fMRI Data
5.3. The Full STAM-fMRI Classification Model Is Recalled on Partial Temporal fMRI Data
5.4. Testing the Full STAM-fMRI Model on a Smaller Portion of the Spatial Information (a Smaller Number of fMRI Variables/Features)
5.5. Potential Bio-Marker Discovery from the STAM-fMRI
5.6. STAM for Longitudinal MRI Neuroimaging
6. Discussions, Conclusions, and Directions for Further Research
6.1. Potential Applications of the Proposed STAM-NI Classification Methods
6.2. Future Development and Challenges of the STAM-NI Methods
- -
- -
- Normalizing or/and harmonizing NI data across various data sources [48]. Establishing an effective “mapping” between training variables and synchronized time units will be crucial.
- -
- -
- STAM-NI, which works under different temporal conditions, e.g., with data collected at varying intervals. At present the same time unit is used for training and for (e.g., milliseconds, seconds, etc.). If the recall data is measured in different time intervals, we can apply interpolation between the data points so that they will match the training temporal units. Such data interpolation has been successfully used in brain data analysis using the NeuCube SNN [22].
- -
- STAM-NI, for different spatial settings. At this stage, we have explored the model when data for training and recall are in the same spatial setting and same context. We can explore further the ability of the model for incremental learning of new variables, that can be mapped spatially. In this case, the network of connections in the 3D SNN will form new clusters that connect spatially the new variables and may also develop links with the “old” variables.
- -
- STAM-NI, which accounts for the variability of the variables themselves. In real-world scenarios, variables may have different characteristics, and their relationships may evolve. We will consider how a STAM framework performs with diverse types of variables, including those with different temporal dynamics and spatial distributions.
- -
- In conclusion, the proposed STAM-NI classification framework and its specific models STAM-EEG and STAM-fMRI are not aimed to substitute existing methods and systems for NI data analyses. Rather, they are extending their functionality for better NI data modeling, data understanding, and early event diagnosis and prognosis.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Squire, L.R. Memory and Brain Systems: 1969–2009. J. Neurosci. 2009, 29, 12711–12716. [Google Scholar] [CrossRef]
- Squire, L.R. Memory systems of the brain: A brief history and current perspective. Neurobiol. Learn. Mem. 2004, 82, 171–177. [Google Scholar] [CrossRef]
- Hopfield, J.J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. USA 1982, 79, 2554–2558. [Google Scholar] [CrossRef]
- Kosko, B. Bidirectional Associative Memories. IEEE Trans. Syst. Man Cybern. 1988, 18, 49–60. [Google Scholar] [CrossRef]
- Haga, T.; Fukai, T. Extended Temporal Association Memory by Modulations of Inhibitory Circuits. Phys. Rev. Lett. 2019, 123, 078101. [Google Scholar] [CrossRef]
- Kasabov, N.K. NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Netw. 2014, 52, 62–76. [Google Scholar] [CrossRef]
- Kasabov, N. Spatio-Temporal Associative Memories in Brain-inspired Spiking Neural Networks: Concepts and Perspectives. TechRxiv 2023. preprint. [Google Scholar] [CrossRef]
- Song, S.; Miller, K.D.; Abbott, L.F. Competitive hebbian learning through spike-timing-dependent synaptic plasticity. Nat. Neurosci. 2000, 3, 919–926. [Google Scholar] [CrossRef]
- Kasabov, N. Time-Space, Spiking Neural Networks and Brain-Inspired Artificial Intelligence; Springer Nature: Berlin/Heidelberg, Germany, 2019; 750p, Available online: https://www.springer.com/gp/book/9783662577134 (accessed on 12 November 2023).
- Kasabov, N. NeuroCube EvoSpike Architecture for Spatio-Temporal Modelling and Pattern Recognition of Brain Signals. In ANNPR; Mana, N., Schwenker, F., Trentin, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7477, pp. 225–243. [Google Scholar]
- Kasabov, N.; Dhoble, K.; Nuntalid, N.; Indiveri, G. Dynamic evolving spiking neural networks for on-line spatio- and spectro-temporal pattern recognition. Neural Netw. 2013, 41, 188–201. [Google Scholar] [CrossRef]
- Talairach, J.; Tournoux, P. Co-Planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System—An Approach to Cerebral Imaging; Thieme Medical Publishers: New York, NY, USA, 1988. [Google Scholar]
- Zilles, K.; Amunts, K. Centenary of Brodmann’s map—Conception and fate. Nat. Rev. Neurosci. 2010, 11, 139–145. [Google Scholar] [CrossRef]
- Mazziotta, J.C.; Toga, A.W.; Evans, A.; Fox, P.; Lancaster, J. A Probablistic Atlas of the Human Brain: Theory and Rationale for Its Development. NeuroImage 1995, 2, 89–101. [Google Scholar] [CrossRef]
- Abeles, M. Corticonics; Cambridge University Press: New York, NY, USA, 1991. [Google Scholar]
- Izhikevich, E. Polychronization: Computation with Spikes. Neural Comput. 2006, 18, 245–282. [Google Scholar] [CrossRef]
- Neftci, E.; Chicca, E.; Indiveri, G.; Douglas, R. A systematic method for configuring vlsi networks of spiking neurons. Neural Comput. 2011, 23, 2457–2497. [Google Scholar] [CrossRef]
- Szatmáry, B.; Izhikevich, E.M. Spike-Timing Theory of Working memory. PLoS Comput. Biol. 2010, 6, e1000879. [Google Scholar] [CrossRef]
- Humble, J.; Denham, S.; Wennekers, T. Spatio-temporal pattern recognizers using spiking neurons and spike-timing-dependent plasticity. Front. Comput. Neurosci. 2012, 6, 84. [Google Scholar] [CrossRef]
- Kasabov, N.K.; Doborjeh, M.G.; Doborjeh, Z.G. Mapping, learning, visualisation, classification and understanding of fMRI data in the NeuCube Evolving Spatio Temporal Data Machine of Spiking Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 887–899. [Google Scholar] [CrossRef]
- Mitchell, T.M.; Hutchinson, R.; Niculescu, R.S.; Pereira, F.; Wang, X.; Just, M.; Newman, S. Learning to Decode Cognitive States from Brain Images. Mach. Learn. 2004, 57, 145–175. [Google Scholar] [CrossRef]
- Doborjeh, M.; Doborjeh, Z.; Merkin, A.; Bahrami, H.; Sumich, A.; Krishnamurthi, R.; Medvedev, O.N.; Crook-Rumsey, M.; Morgan, C.; Kirk, I.; et al. Personalised Predictive Modelling with Spiking Neural Networks of Longitudinal MRI Neuroimaging Cohort and the Case Study of Dementia. Neural Netw. 2021, 144, 522–539. [Google Scholar] [CrossRef]
- Chong, B.; Wang, A.; Borges, V.; Byblow, W.D.; Alan Barber, P.; Stinear, C. Investigating the structure-function relationship of the corticomotor system early after stroke using machine learning. NeuroImage Clin. 2022, 33, 102935. [Google Scholar] [CrossRef]
- Karim, M.; Chakraborty, S.; Samadiani, N. Stroke Lesion Segmentation using Deep Learning Models: A Survey. IEEE Access 2021, 9, 44155–44177. [Google Scholar]
- Li, H.; Shen, D.; Wang, L. A Hybrid Deep Learning Framework for Alzheimer’s Disease Classification Based on Multimodal Brain Imaging Data. Front. Neurosci. 2021, 15, 625534. [Google Scholar]
- Niazi, F.; Bourouis, S.; Prasad, P.W.C. Deep Learning for Diagnosis of Mild Cognitive Impairment: A Systematic Review and Meta-Analysis. Front. Aging Neurosci. 2020, 12, 244. [Google Scholar]
- Sona, C.; Siddiqui, S.A.; Mehmood, R. Classification of Depression Patients and Healthy Controls Using Machine Learning Techniques. IEEE Access 2021, 9, 26804–26816. [Google Scholar]
- Fanaei, M.; Davari, A.; Shamsollahi, M.B. Autism Spectrum Disorder Diagnosis Based on fMRI Data Using Deep Learning and 3D Convolutional Neural Networks. Sensors 2020, 20, 4600. [Google Scholar]
- Zhang, X.; Liu, T.; Qian, Z. A Comprehensive Review on Parkinson’s Disease Using Deep Learning Techniques. Front. Aging Neurosci. 2021, 13, 702474. [Google Scholar]
- Hjelm, R.D.; Calhoun-Sauls, A.; Shiffrin, R.M. Deep Learning and the Audio-Visual World: Challenges and Frontiers. Front. Neurosci. 2020, 14, 219. [Google Scholar]
- Poline, J.-B.; Poldrack, R.A. Frontiers in brain imaging methods grand challenge. Front. Neurosci. 2012, 6, 96. [Google Scholar] [CrossRef]
- Pascual-Leone, A.; Hamilton, R. The metamodal organization of the brain. Prog. Brain Res. 2001, 134, 427–445. [Google Scholar]
- Honey, C.J.; Kötter, R.; Breakspear, M.; Sporns, O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl Acad. Sci. USA 2007, 104, 10240–10245. [Google Scholar] [CrossRef]
- Nicolelis, M. Mind in Motion. Sci. Am. 2012, 307, 44–49. [Google Scholar] [CrossRef]
- Paulun, V.C.; Beer, A.L.; Thompson-Schill, S.L. Distinct contributions of functional and deep neural network features to representational similarity of scenes in human brain and behavior. eLife 2019, 8, e42848. [Google Scholar]
- Liu, H.; Lu, G.; Wang, Y.; Kasabov, N. Evolving spiking neural network model for PM2.5 hourly concentration prediction based on seasonal differences: A case study on data from Beijing and Shanghai. Aerosol Air Qual. Res. 2021, 21, 200247. [Google Scholar] [CrossRef]
- Furber, S. To Build a Brain. IEEE Spectr. 2012, 49, 39–41. [Google Scholar] [CrossRef]
- Indiveri, G.; Stefanini, F.; Chicca, E. Spike-based learning with a generalized integrate and fire silicon neuron. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems (ISCAS 2010) 2010, Paris, France, 30 May–2 June 2010; pp. 1951–1954. [Google Scholar]
- Indiveri, G.; Chicca, E.; Douglas, R.J. Artificial cognitive systems: From VLSI networks of spiking neurons to neuromorphic cognition. Cogn. Comput. 2009, 1, 119–127. [Google Scholar] [CrossRef]
- Delbruck, T. jAER Open-Source Project. 2007. Available online: https://sourceforge.net/p/jaer/wiki/Home/ (accessed on 12 November 2023).
- Benuskova, L.; Kasabov, N. Computational Neuro-Genetic Modelling; Springer: New York, NY, USA, 2007. [Google Scholar]
- BrainwaveR Toolbox. Available online: http://www.nitrc.org/projects/brainwaver/ (accessed on 12 November 2023).
- Buonomano, D.; Maass, W. State-dependent computations: Spatio-temporal processing in cortical networks. Nat. Rev. Neurosci. 2009, 10, 113–125. [Google Scholar] [CrossRef]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Kasabov, N.; Tan, Y.; Doborjeh, M.; Tu, E.; Yang, J.; Goh, W.; Lee, J. Transfer Learning of Fuzzy Spatio-Temporal Rules in the NeuCube Brain-Inspired Spiking Neural Network: A Case Study on EEG Spatio-temporal Data. IEEE Trans. Fuzzy Syst. 2023, 1–12. [Google Scholar] [CrossRef]
- Kumarasinghe, K.; Kasabov, N.; Taylor, D. Brain-inspired spiking neural networks for decoding and understanding muscle activity and kinematics from electroencephalography signals during hand movements. Sci. Rep. 2021, 11, 2486. Available online: https://www.nature.com/articles/s41598-021-81805-4 (accessed on 12 November 2023). [CrossRef]
- Wu, X.; Feng, Y.; Lou, S.; Zheng, H.; Hu, B.; Hong, Z.; Tan, J. Improving NeuCube spiking neural network for EEG-based pattern recognition using transfer learning. Neurocomputing 2023, 529, 222–235. [Google Scholar] [CrossRef]
- Wen, G.; Shim, V.; Holdsworth, S.J.; Fernandez, J.; Qiao, M.; Kasabov, N.; Wang, A. Artificial Intelligence for Brain MRI Data Harmonization: A Systematic Review. Bioengineering 2023, 10, 397. [Google Scholar] [CrossRef]
- Dey, S.; Dimitrov, A. Mapping and Validating a Point Neuron Model on Intel’s Neuromorphic Hardware Loihi. Front. Neuroinform. 2022, 16, 883360. [Google Scholar] [CrossRef]


| Dataset Information | Encoding Method and Parameters | NeuCube Model | STDP Parameters | deSNNs Classifier Parameters |
|---|---|---|---|---|
| sample number: 60, feature number: 14 channels, time length: 128, class number: 3. | encoding method: Step Forward (SF) spike threshold: 0.5, window size: 5, filter type: SS. | number of neurons: 1471, brain template: Talairach, neuron model: LIF. | potential leak rate: 0.002, STDP rate: 0.01, firing threshold: 0.5, training iteration: 1, refractory time: 6, LDC probability: 0. | mod: 0.8, drift: 0.005, K: 1, sigma: 1. |
| Time T1 (% of the Full Time for Training) | Number of Input Variables | Training/Validation % of Data Samples | Temporal Association Accuracy | RMA |
|---|---|---|---|---|
| 100% (full) | 14 (100%) | 100/100 | 100% | 1 |
| 95% | 14 (100%) | 100/100 | 100% | 1 |
| 90% | 14 (100%) | 100/100 | 98% | 0.98 |
| 80% | 14 (100%) | 100/100 | 95% | 0.95 |
| Time T1 (% of the Full Time) | Number of Input Variables Used | Training/Validation % of Data Samples | Temporal Generalization Accuracy | RMA |
|---|---|---|---|---|
| 100% (full) | 14 (100%) | 50/50 | 80% | 1 |
| 95% | 14 (100%) | 50/50 | 80% | 1 |
| 90% | 14 (100%) | 50/50 | 76% | 0.95 |
| Time T1 (% of the Full Time) | Number of Input Variables | Training/Validation % of Data Samples | Spatial Association Accuracy | RMA |
|---|---|---|---|---|
| 100% (full) | 14 (100%) | 100/100 | 100% | 1 |
| 100% (full) | 13 (93%) | 100/100 | 100% | 1 |
| 95% | 13 (93%) | 100/100 | 100% | 1 |
| 90% | 13 (93%) | 100/100 | 86% | 0.86 |
| Time T1 (% of the Full Time) | Number of Input Variables | Training/Validation % of Data Samples | Temporal Association Accuracy | RMA |
|---|---|---|---|---|
| 100% (full) | 14 (100%) | 50/50 | 80% | 1 |
| 100% (full) | 13 (93%) | 50/50 | 100% | 1 |
| 95% | 13 (93%) | 50/50 | 80% | 1 |
| 90% | 13 (93%) | 50/50 | 76% | 0.95 |
| Area | LT | LOPER | LIPL | LOPER | LDLPFC | LOPER | LT | LDLPFC | RT | CALC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg | 1.4 | 0.92 | 1.87 | 1.03 | 2.08 | 1.12 | 1.48 | 0.44 | 0.2 | 0.89 | |
| Aff | 0.9 | 0.56 | 1.01 | 0.87 | 1.03 | 0.65 | 0.89 | 0.23 | 0.1 | 0.43 | |
| area | LSGA | LDLPFC | LT | LDLPFC | RT | LDLPFC | LDLPFC | RDLPFC | RSGA | RIT | Avg |
| Neg | 1.84 | 1.03 | 1.9 | 0.45 | 1.1 | 1.26 | 0.56 | 0.19 | 0.43 | 1.4 | 1.7 |
| Aff | 1.04 | 0.68 | 1.1 | 0.17 | 0.8 | 0.24 | 0.22 | 0.11 | 0.32 | 0.9 | 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasabov, N.K.; Bahrami, H.; Doborjeh, M.; Wang, A. Brain-Inspired Spatio-Temporal Associative Memories for Neuroimaging Data Classification: EEG and fMRI. Bioengineering 2023, 10, 1341. https://doi.org/10.3390/bioengineering10121341
Kasabov NK, Bahrami H, Doborjeh M, Wang A. Brain-Inspired Spatio-Temporal Associative Memories for Neuroimaging Data Classification: EEG and fMRI. Bioengineering. 2023; 10(12):1341. https://doi.org/10.3390/bioengineering10121341
Chicago/Turabian StyleKasabov, Nikola K., Helena Bahrami, Maryam Doborjeh, and Alan Wang. 2023. "Brain-Inspired Spatio-Temporal Associative Memories for Neuroimaging Data Classification: EEG and fMRI" Bioengineering 10, no. 12: 1341. https://doi.org/10.3390/bioengineering10121341
APA StyleKasabov, N. K., Bahrami, H., Doborjeh, M., & Wang, A. (2023). Brain-Inspired Spatio-Temporal Associative Memories for Neuroimaging Data Classification: EEG and fMRI. Bioengineering, 10(12), 1341. https://doi.org/10.3390/bioengineering10121341










