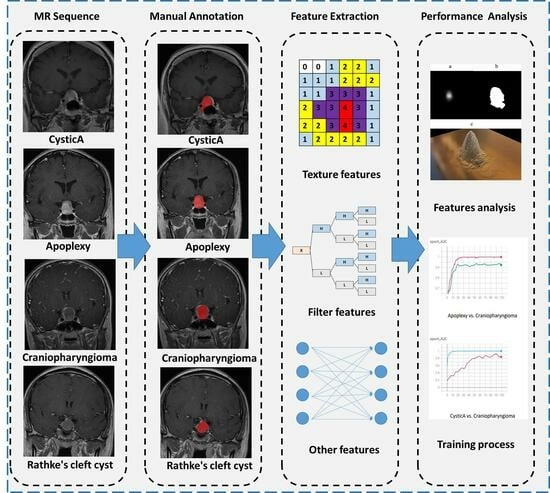
Machine Learning Approaches to Differentiate Sellar-Suprasellar Cystic Lesions on Magnetic Resonance Imaging
Abstract
:1. Introduction
- A large number of patients diagnosed with cystic sellar lesions were enrolled to fill the gap in the existing literature and provide a non-invasive method for accurately differentiating between these lesions.
- Paired imaging differentiations were performed on four subtypes, and the model achieved an average AUC value of 0.7685.
- The model achieved an average accuracy of 0.7532, which outperformed the traditional clinical knowledge-based model by approximately 8%.
2. Related Works
2.1. Basic Characteristics of Cystic Sellar Lesions
2.2. Clinical Knowledge-Based Method
2.3. Machine Learning Methods
3. Materials and Methods
3.1. Patients
3.2. MRI Data Acquisition and Preprocessing
3.3. Image Segmentation
3.4. Feature Extraction
3.5. Feature Selection, Model Construction, and Validation
3.6. Establishment of Clinical Knowledge-Based Method
3.7. Statistical Analysis
4. Results
4.1. Patient Characteristics and Pathology Types
4.2. Image Segmentation and Feature Selection
4.3. Radiomics Model Validation and Model Comparison
4.4. Comparison with Clinical Knowledge Base Methods
5. Discussion
6. Limitations
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bette, S.; Butenschön, V.M.; Wiestler, B.; von Werder, A.; Schmid, R.M.; Lehmberg, J.; Zimmer, C.; Meyer, B.; Kirschke, J.S.; Gempt, J. MRI criteria of subtypes of adenomas and epithelial cysts of the pituitary gland. Neurosurg. Rev. 2020, 43, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, A.; Hirakawa, K.; Sanno, N.; Osamura, Y. Incidental pituitary lesions in 1000 unselected autopsy specimens. Radiology 1994, 193, 161–164. [Google Scholar] [CrossRef]
- Lober, R.M.; Harsh, G.R.T. A perspective on craniopharyngioma. World Neurosurg. 2013, 79, 645–646. [Google Scholar] [CrossRef] [PubMed]
- Barkhoudarian, G.; Palejwala, S.K.; Ansari, S.; Eisenberg, A.A.; Huang, X.; Griffiths, C.F.; Kelly, D.F. Rathke’s cleft cysts: A 6-year experience of surgery vs. observation with comparative volumetric analysis. Pituitary 2019, 22, 362–371. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Vanderpump, M.; Baldeweg, S.; Drake, W.; Reddy, N.; Lanyon, M.; Markey, A.; Plant, G.; Powell, M.; Sinha, S.; et al. UK guidelines for the management of pituitary apoplexy. Clin. Endocrinol. 2011, 74, 9–20. [Google Scholar] [CrossRef]
- Muller, H.L. Craniopharyngioma. Nat. Rev. Dis. Primers 2019, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Morana, G.; Maghnie, M.; Rossi, A. Pituitary tumors: Advances in neuroimaging. Endocr. Dev. 2010, 17, 160–174. [Google Scholar]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Lu, C.-F.; Hsu, F.-T.; Hsieh, K.L.-C.; Kao, Y.-C.J.; Cheng, S.-J.; Hsu, J.B.-K.; Tsai, P.-H.; Chen, R.-J.; Huang, C.-C.; Yen, Y.; et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res. 2018, 24, 4429–4436. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Jiang, C.; Zhu, R.; Feng, S.; Wang, Y.; Li, J.; Chen, W.; Liu, P.; Zhao, D.; Ma, W.; et al. (18)F-FDG-PET-based radiomics features to distinguish primary central nervous system lymphoma from glioblastoma. Neuroimage Clin. 2019, 23, 101912. [Google Scholar] [CrossRef] [PubMed]
- Zeynalova, A.; Kocak, B.; Durmaz, E.S.; Comunoglu, N.; Ozcan, K.; Ozcan, G.; Turk, O.; Tanriover, N.; Kocer, N.; Kizilkilic, O.; et al. Preoperative evaluation of tumour consistency in pituitary macroadenomas: A machine learning-based histogram analysis on conventional T2-weighted MRI. Neuroradiology 2019, 61, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Nadezhdina, E.Y.; Rebrova, O.Y.; Grigoriev, A.Y.; Ivaschenko, O.V.; Azizyan, V.N.; Melnichenko, G.A.; Dedov, I.I. Prediction of recurrence and remission within 3 years in patients with Cushing disease after successful transnasal adenomectomy. Pituitary 2019, 22, 574–580. [Google Scholar] [CrossRef]
- Park, M.; Lee, S.K.; Choi, J.; Kim, S.H.; Kim, S.H.; Shin, N.Y.; Ahn, S.S. Differentiation between Cystic Pituitary Adenomas and Rathke Cleft Cysts: A Diagnostic Model Using MRI. Am. J. Neuroradiol. 2015, 36, 1866–1873. [Google Scholar] [CrossRef] [PubMed]
- Zada, G.; Lin, N.; Ojerholm, E.; Ramkissoon, S.; Laws, E.R. Craniopharyngioma and other cystic epithelial lesions of the sellar region: A review of clinical, imaging, and histopathological relationships. Neurosurg. Focus 2010, 28, E4. [Google Scholar] [CrossRef]
- Gadelha, M.R.; Wildemberg, L.E.; Lamback, E.B.; Barbosa, M.A.; Kasuki, L.; Ventura, N. Approach to the patient: Differential diagnosis of cystic sellar lesions. J. Clin. Endocrinol. Metab. 2022, 107, 1751–1758. [Google Scholar] [CrossRef]
- Breiman, L.J.M.l. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Schapire, R.E.; Singer, Y.J.M.L. BoosTexter: A Boosting-based System for Text Categorization. Mach. Learn. 2000, 39, 135–168. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Duchesnay, É. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Leiva, R.G.; Anta, A.F.; Mancuso, V.; Casari, P. A Novel Hyperparameter-Free Approach to Decision Tree Construction That Avoids Overfitting by Design. IEEE Access 2019, 7, 99978–99987. [Google Scholar] [CrossRef]
- Xiao, D.; Wang, S.; Huang, Y.; Zhao, L.; Wei, L.; Ding, C. Clinical analysis of infarction in pituitary adenoma. Int. J. Clin. Exp. Med. 2015, 8, 7477–7486. [Google Scholar] [PubMed]
- Semple, P.L.; Jane, J.A.; Lopes, M.B.S.; Laws, E.R. Pituitary apoplexy: Correlation between magnetic resonance imaging and histopathological results. J. Neurosurg. 2008, 108, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Ram, Z.; Hadani, M.; Berezin, M.; Findler, G.; Sahar, A.; Shacked, I. Intratumoural cyst formation in pituitary macroadenomas. Acta Neurochir. 1989, 100, 56–61. [Google Scholar] [CrossRef]
- Kurihara, N.; Takahashi, S.; Higano, S.; Ikeda, H.; Mugikura, S.; Singh, L.N.; Furuta, S.; Tamura, H.; Ishibashi, T.; Maruoka, S.; et al. Hemorrhage in pituitary adenoma: Correlation of MR imaging with operative findings. Eur. Radiol. 1998, 8, 971–976. [Google Scholar] [CrossRef]
- Andrysiak-Mamos, E.; Sagan, K.; Sagan, L.; Sowińska-Przepiera, E.; Syrenicz, A. Cystic lesions of the sellar-suprasellar region—Diagnosis and treatment. Endokrynol. Pol. 2018, 69, 212–228. [Google Scholar] [CrossRef]
- Marrero Fernandez, P.D.; Guerrero Pena, F.A.; Ren, T.; Cunha, A. FERAtt: Facial Expression Recognition with Attention Net. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic Routing Between Capsules. arXiv 2017, arXiv:abs/1710.09829. [Google Scholar]



| Type | n | Age Mean (SD) | Female (%) | Male (%) |
|---|---|---|---|---|
| Sum | 390 | 41.06 (15.14) | 249 (63.8) | 141 (36.2) |
| Apoplexy | 205 | 42.38 (14.05) | 133 (64.9) | 72 (35.1) |
| Craniopharyngioma | 37 | 35.30 (21.00) | 17 (45.9) | 20 (54.1) |
| Cystic Adenoma | 71 | 41.58 (14.54) | 50 (70.4) | 21 (29.6) |
| Rathke’s Cleft Cyst | 77 | 39.82 (14.72) | 49 (63.6) | 28 (36.4) |
| p-value | 0.094 |
| Age | n | Mean | SD | Median | p25 | p75 | Min | Max | Skew | Kurt |
|---|---|---|---|---|---|---|---|---|---|---|
| Sum | 390 | 41 | 15 | 40 | 30 | 53 | 5 | 80 | −0.03 | −0.56 |
| Apoplexy | 205 | 42 | 14 | 42 | 32 | 53 | 10 | 80 | 0.13 | −0.48 |
| Craniopharyngioma | 37 | 35 | 21 | 37 | 14 | 57 | 5 | 66 | −0.001 | −1.6 |
| Cystic Adenoma | 71 | 42 | 15 | 42 | 32 | 52 | 7 | 77 | −0.077 | −0.24 |
| Rathke’s Cleft Cyst | 77 | 40 | 15 | 36 | 28 | 53 | 10 | 75 | 0.28 | −0.85 |
| 3D | 2D | ||||
|---|---|---|---|---|---|
| T1CE | T2WI | T1CE | T2WI | ||
| Shape | 14 | 14 | 9 | 9 | |
| First-order | 18 | 18 | 18 | 18 | |
| Texture | GLCM | 24 | 24 | 24 | 24 |
| GLRLM | 16 | 16 | 16 | 16 | |
| GLSZM | 16 | 16 | 16 | 16 | |
| GLDM | 14 | 14 | 14 | 14 | |
| NGTDM | 5 | 5 | 5 | 5 | |
| LoG | 186 | 186 | 372 | 372 | |
| Wavelet | 744 | 744 | 186 | 186 | |
| Total | 1037 | 1037 | 660 | 660 | |
| Apoplexy | Apoplexy | Apoplexy | CysticA | Rathke | Rathke |
|---|---|---|---|---|---|
| Craniopharyngioma | CysticA | Rathke | Craniopharyngioma | Craniopharyngioma | CysticA |
| T1CE_wavelet-LHH_ngtdm_Complexity | T1CE_original_shape_Elongation | T2RS_original_glcm_MCC | T1CE_wavelet-LLL_ngtdm_Complexity | T2RS_log-sigma-5-0-mm-3D_glcm_Idn | T1CE_wavelet-LLH_ngtdm_Coarseness |
| T1CE_wavelet-LLL_glcm_ClusterProminence | T1CE_original_shape_Flatness | T2RS_log-sigma-3-0-mm-3D_glcm_Correlation | T2RS_original_gldm_LargeDependenceHighGrayLevelEmphasis | T2RS_wavelet-LHL_glcm_MCC | T1CE_wavelet-LLL_glcm_Correlation |
| T2RS_original_gldm_LargeDependenceHighGrayLevelEmphasis | T1CE_original_shape_LeastAxisLength | T2RS_log-sigma-3-0-mm-3D_glcm_JointAverage | T2RS_log-sigma-5-0-mm-3D_firstorder_Skewness | T2RS_wavelet-HLH_glcm_MCC | T2RS_original_firstorder_Skewness |
| Compare | Method | 3D | 3D w/o Filters | 2D | 2D w/o Filters |
|---|---|---|---|---|---|
| Apoplexy vs. Craniopharyngioma | Random Forest | 0.6591 | 0.6682 | 0.6306 | 0.5986 |
| Bagging SVM | 0.6858 | 0.7019 | 0.5855 | 0.5895 | |
| SVM | 0.6871 | 0.7011 | 0.6312 | 0.6228 | |
| AdaBoost DecisionTree | 0.6140 | 0.5666 | 0.5628 | 0.5974 | |
| AdaBoost | 0.7001 | 0.7070 | 0.6460 | 0.6110 | |
| Logistic Regression | 0.6890 | 0.7102 | 0.6660 | 0.6435 | |
| Apoplexy vs. CysticA | RandomForest | 0.6454 | 0.5556 | 0.6104 | 0.6300 |
| BaggingClassifier_SVM | 0.6108 | 0.5367 | 0.6320 | 0.6465 | |
| SVM | 0.6249 | 0.5719 | 0.6520 | 0.6524 | |
| AdaBoost_DecisionTree | 0.5911 | 0.5801 | 0.5914 | 0.5044 | |
| AdaBoost | 0.6113 | 0.6141 | 0.6641 | 0.6593 | |
| Logistic_Regression | 0.6060 | 0.5796 | 0.6406 | 0.6624 | |
| Apoplexy vs. Rathke | RandomForest | 0.7820 | 0.7944 | 0.7787 | 0.7932 |
| BaggingClassifier_SVM | 0.8046 | 0.7721 | 0.7775 | 0.7385 | |
| SVM | 0.8025 | 0.8032 | 0.7697 | 0.7728 | |
| AdaBoost_DecisionTree | 0.6647 | 0.6043 | 0.6451 | 0.6276 | |
| AdaBoost | 0.7902 | 0.7903 | 0.7827 | 0.7807 | |
| Logistic_Regression | 0.7899 | 0.7883 | 0.7712 | 0.7408 | |
| CysticA vs. Craniopharyngioma | RandomForest | 0.7387 | 0.7695 | 0.7161 | 0.7209 |
| BaggingClassifier_SVM | 0.7989 | 0.7308 | 0.6982 | 0.7769 | |
| SVM | 0.7607 | 0.7610 | 0.7770 | 0.7737 | |
| AdaBoost_DecisionTree | 0.6544 | 0.6377 | 0.7274 | 0.5924 | |
| AdaBoost | 0.8096 | 0.7452 | 0.7468 | 0.7438 | |
| Logistic_Regression | 0.7598 | 0.7202 | 0.7376 | 0.7237 | |
| Rathke vs. Craniopharyngioma | RandomForest | 0.8263 | 0.8355 | 0.8348 | 0.7842 |
| BaggingClassifier_SVM | 0.8141 | 0.8217 | 0.8451 | 0.8122 | |
| SVM | 0.8176 | 0.8165 | 0.8534 | 0.8509 | |
| AdaBoost_DecisionTree | 0.7506 | 0.6155 | 0.7358 | 0.7871 | |
| AdaBoost | 0.8224 | 0.8213 | 0.8534 | 0.8522 | |
| Logistic_Regression | 0.8085 | 0.8165 | 0.8534 | 0.8584 | |
| Rathke vs. CysticA | RandomForest | 0.6701 | 0.6716 | 0.6917 | 0.6949 |
| BaggingClassifier_SVM | 0.7660 | 0.6859 | 0.7019 | 0.6440 | |
| SVM | 0.7511 | 0.6939 | 0.7028 | 0.6570 | |
| AdaBoost_DecisionTree | 0.6418 | 0.6053 | 0.6149 | 0.5832 | |
| AdaBoost | 0.7506 | 0.6798 | 0.7361 | 0.7006 | |
| Logistic_Regression | 0.7235 | 0.6759 | 0.7254 | 0.6635 |
| Machine Learning | Clinical Knowledge-Based Method | |
|---|---|---|
| Apoplexy vs. Craniopharyngioma | 0.7708 | 0.6876 |
| Apoplexy vs. CysticA | 0.686 | 0.5404 |
| Apoplexy vs. Rathke | 0.7633 | 0.7493 |
| CysticA vs. Craniopharyngioma | 0.7675 | 0.5823 |
| Rathke vs. Craniopharyngioma | 0.8293 | 0.8135 |
| Rathke vs. CysticA | 0.7022 | 0.6758 |
| Mean Accuracy | 0.7532 | 0.6748 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, C.; Zhang, W.; Wang, H.; Jiao, Y.; Fang, Y.; Feng, F.; Feng, M.; Wang, R. Machine Learning Approaches to Differentiate Sellar-Suprasellar Cystic Lesions on Magnetic Resonance Imaging. Bioengineering 2023, 10, 1295. https://doi.org/10.3390/bioengineering10111295
Jiang C, Zhang W, Wang H, Jiao Y, Fang Y, Feng F, Feng M, Wang R. Machine Learning Approaches to Differentiate Sellar-Suprasellar Cystic Lesions on Magnetic Resonance Imaging. Bioengineering. 2023; 10(11):1295. https://doi.org/10.3390/bioengineering10111295
Chicago/Turabian StyleJiang, Chendan, Wentai Zhang, He Wang, Yixi Jiao, Yi Fang, Feng Feng, Ming Feng, and Renzhi Wang. 2023. "Machine Learning Approaches to Differentiate Sellar-Suprasellar Cystic Lesions on Magnetic Resonance Imaging" Bioengineering 10, no. 11: 1295. https://doi.org/10.3390/bioengineering10111295
APA StyleJiang, C., Zhang, W., Wang, H., Jiao, Y., Fang, Y., Feng, F., Feng, M., & Wang, R. (2023). Machine Learning Approaches to Differentiate Sellar-Suprasellar Cystic Lesions on Magnetic Resonance Imaging. Bioengineering, 10(11), 1295. https://doi.org/10.3390/bioengineering10111295