Novel Procedure for Automatic Registration between Cone-Beam Computed Tomography and Intraoral Scan Data Supported with 3D Segmentation
Abstract
:1. Introduction
2. Data Preparation
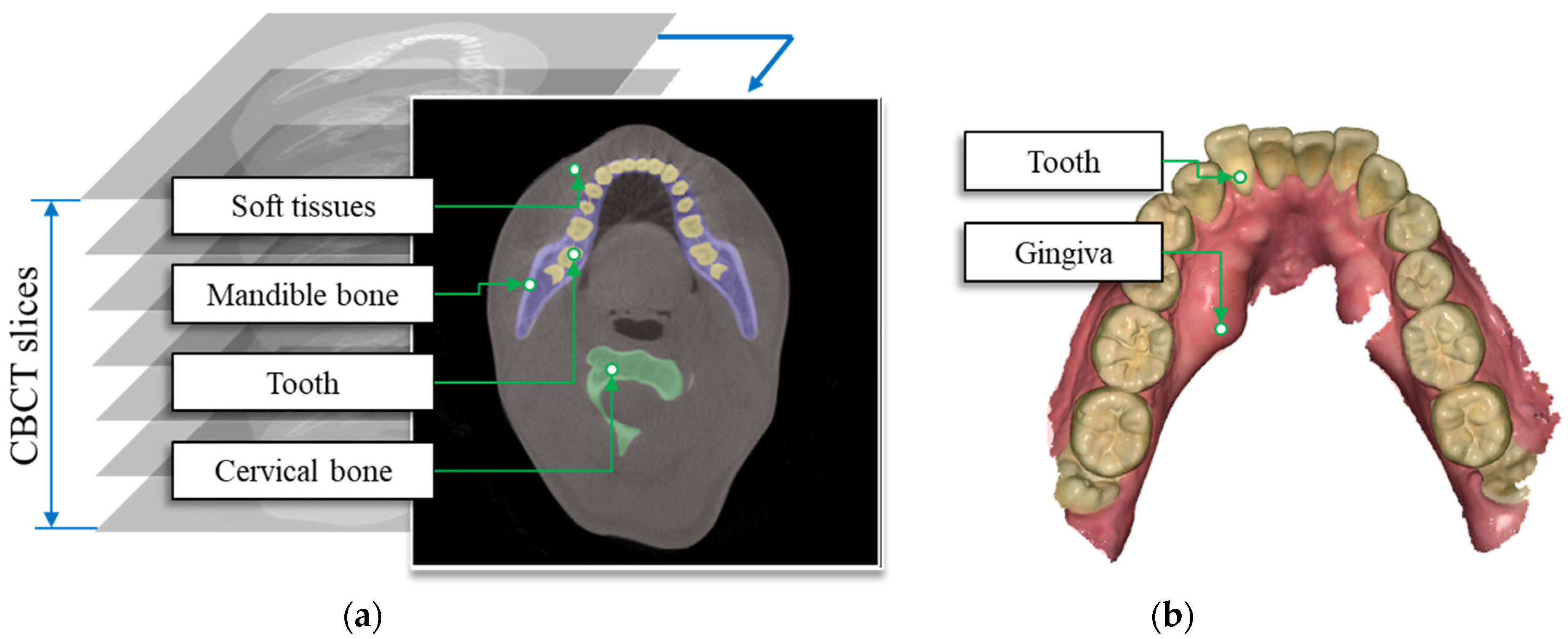
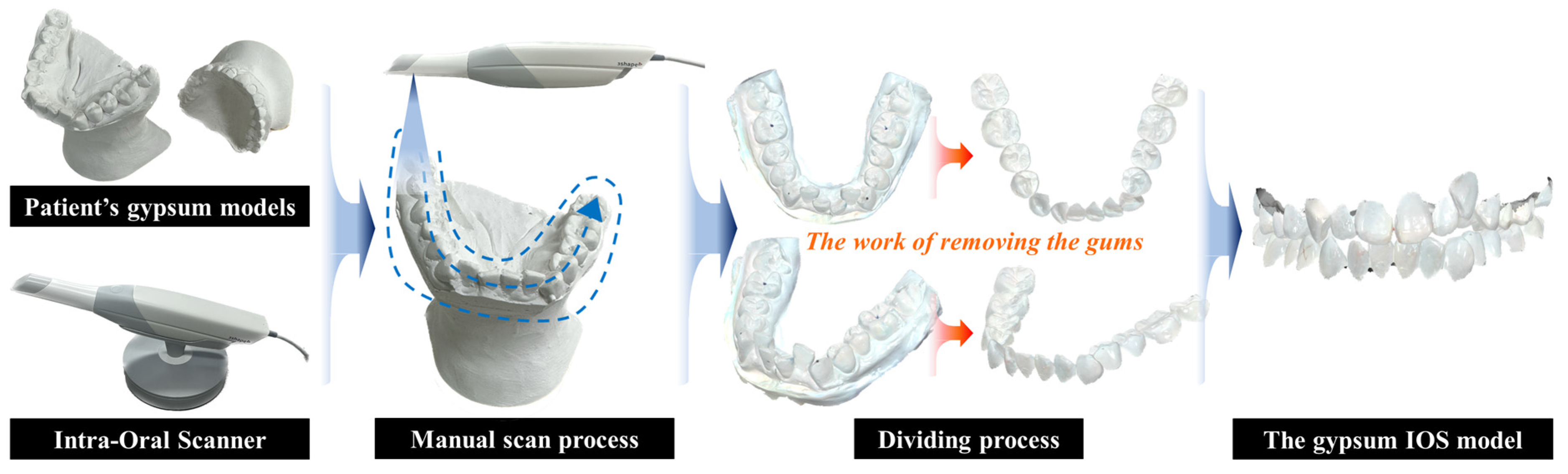
2.1. 3D Dental Data
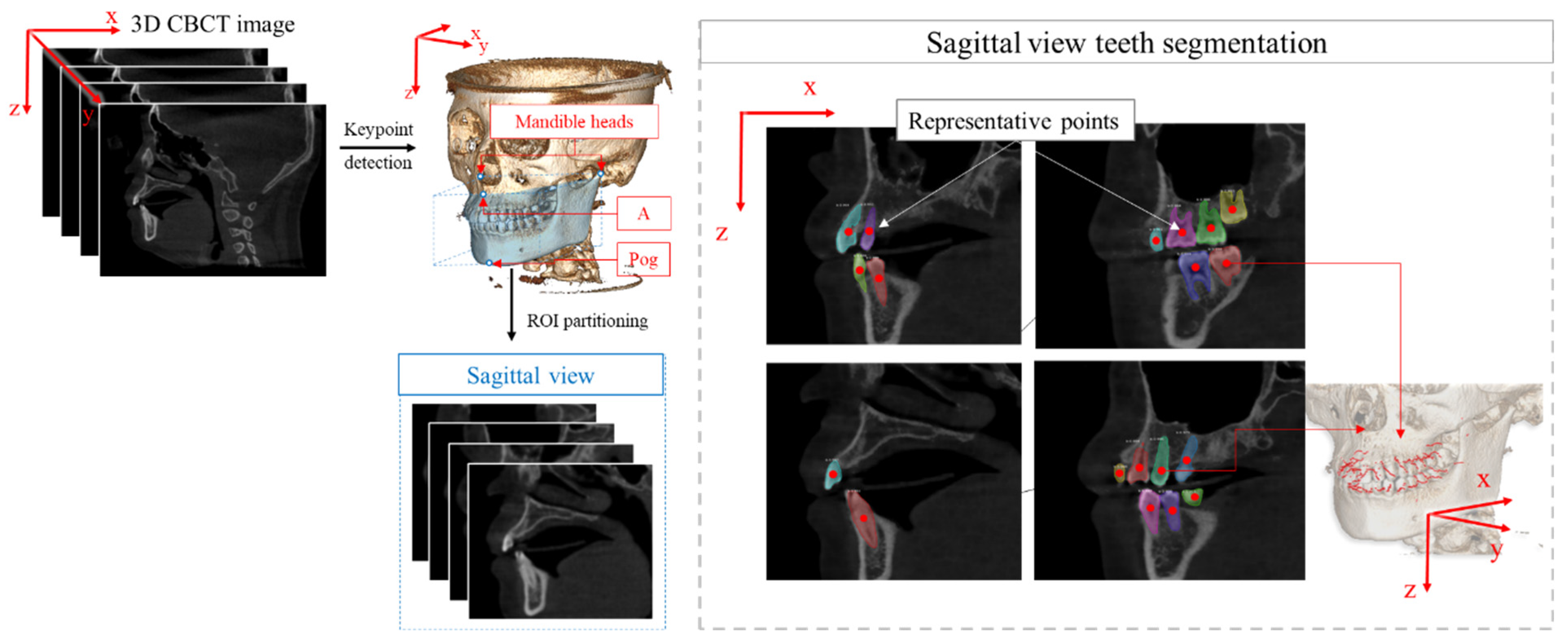
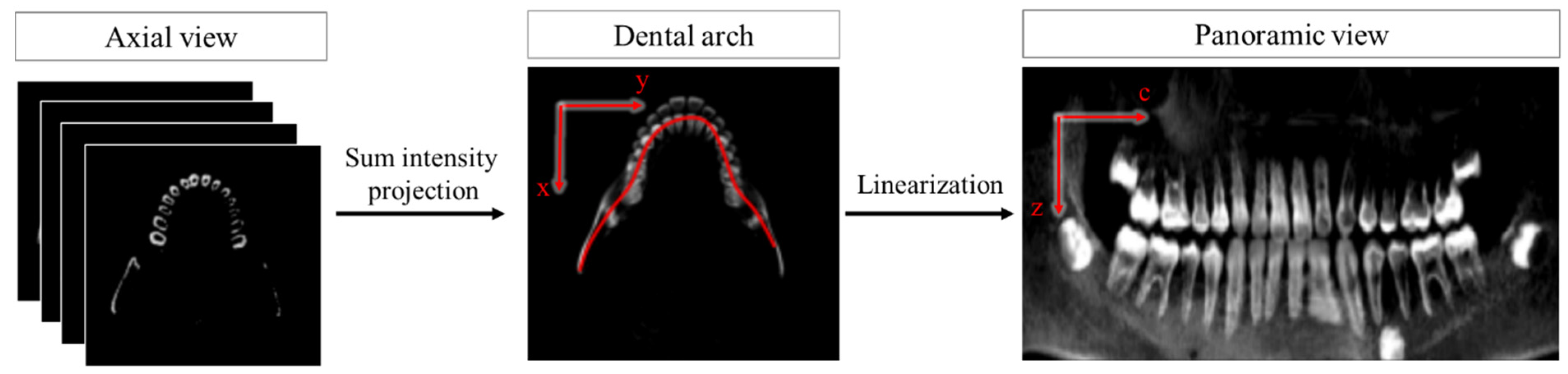
2.2. ROI Extraction
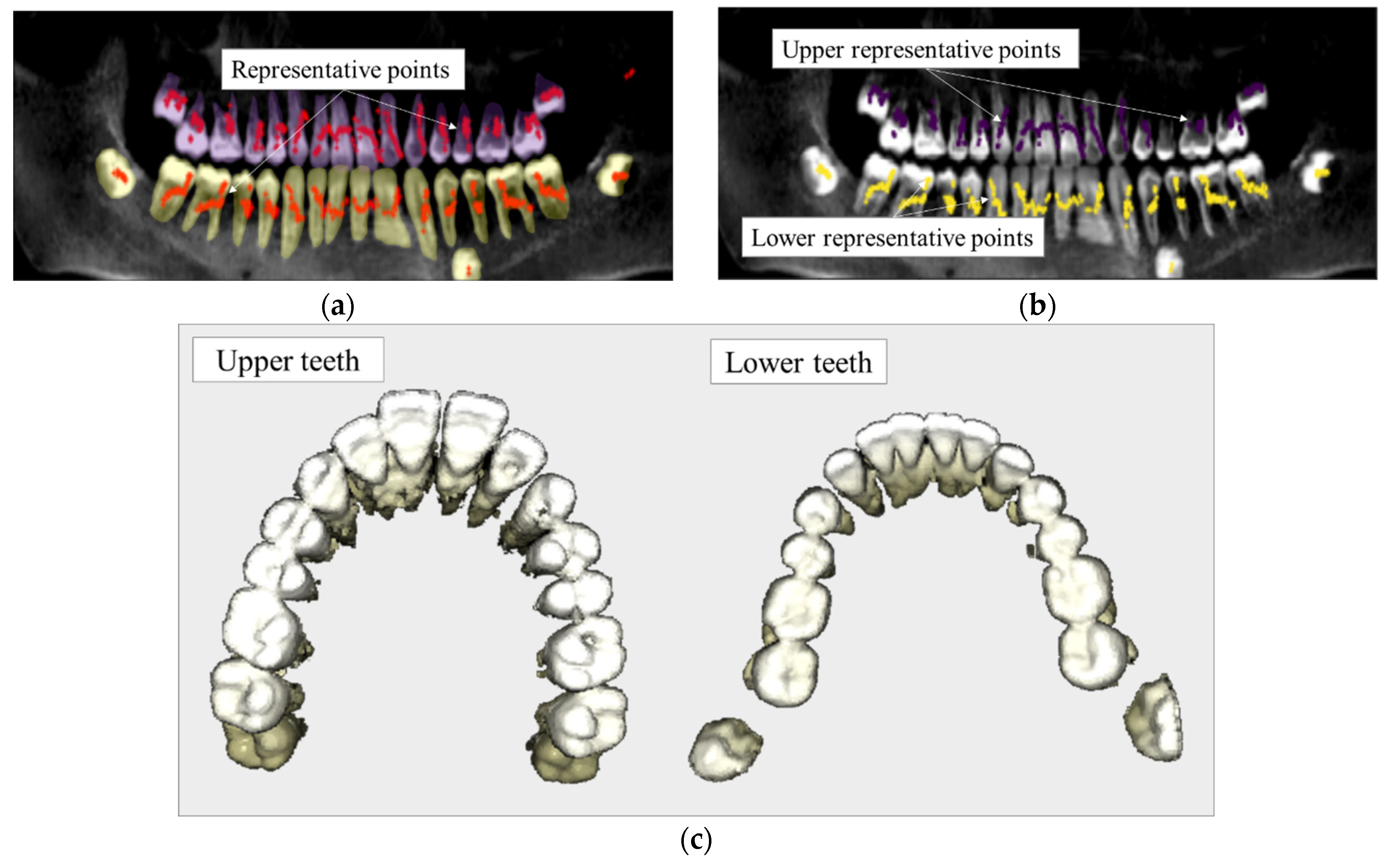
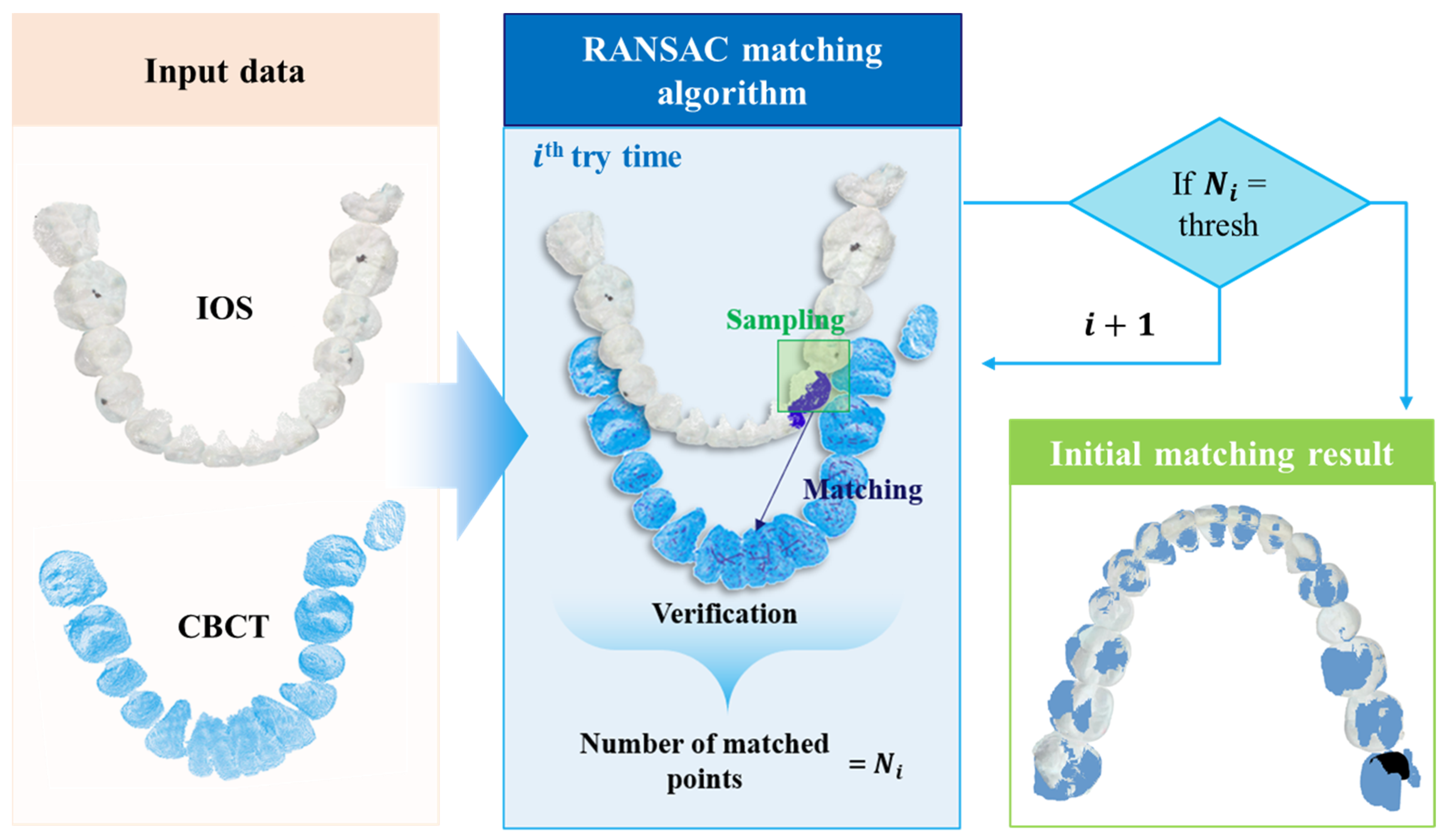
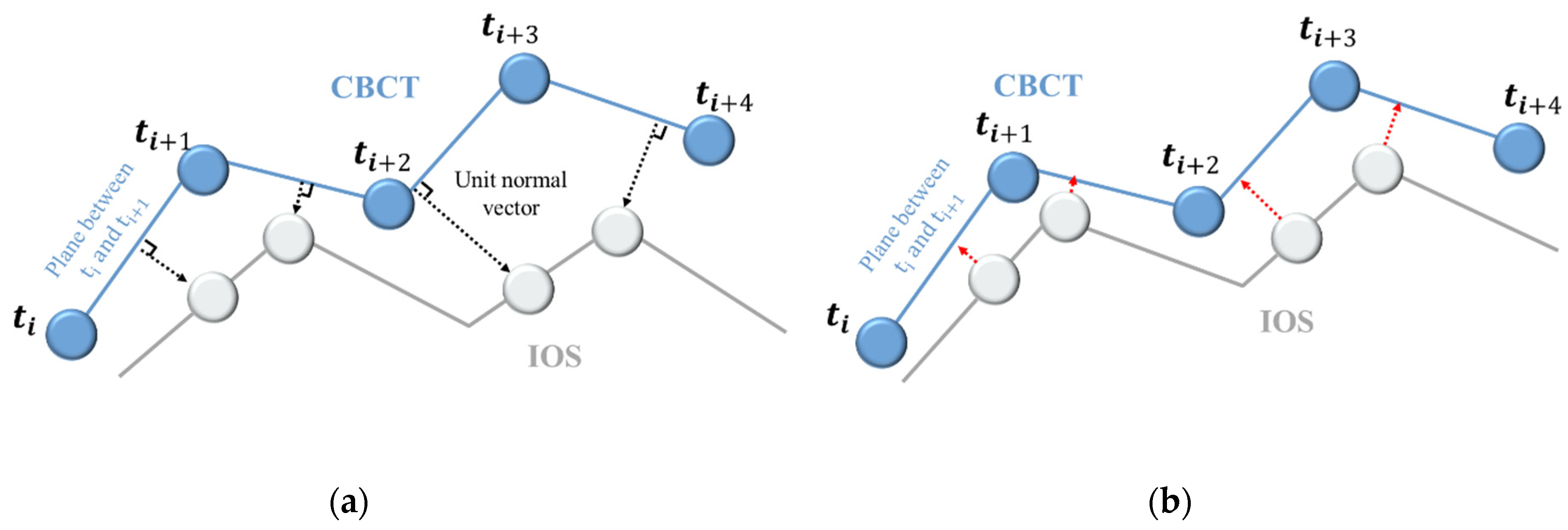
3. Registration Algorithm
4. Validation
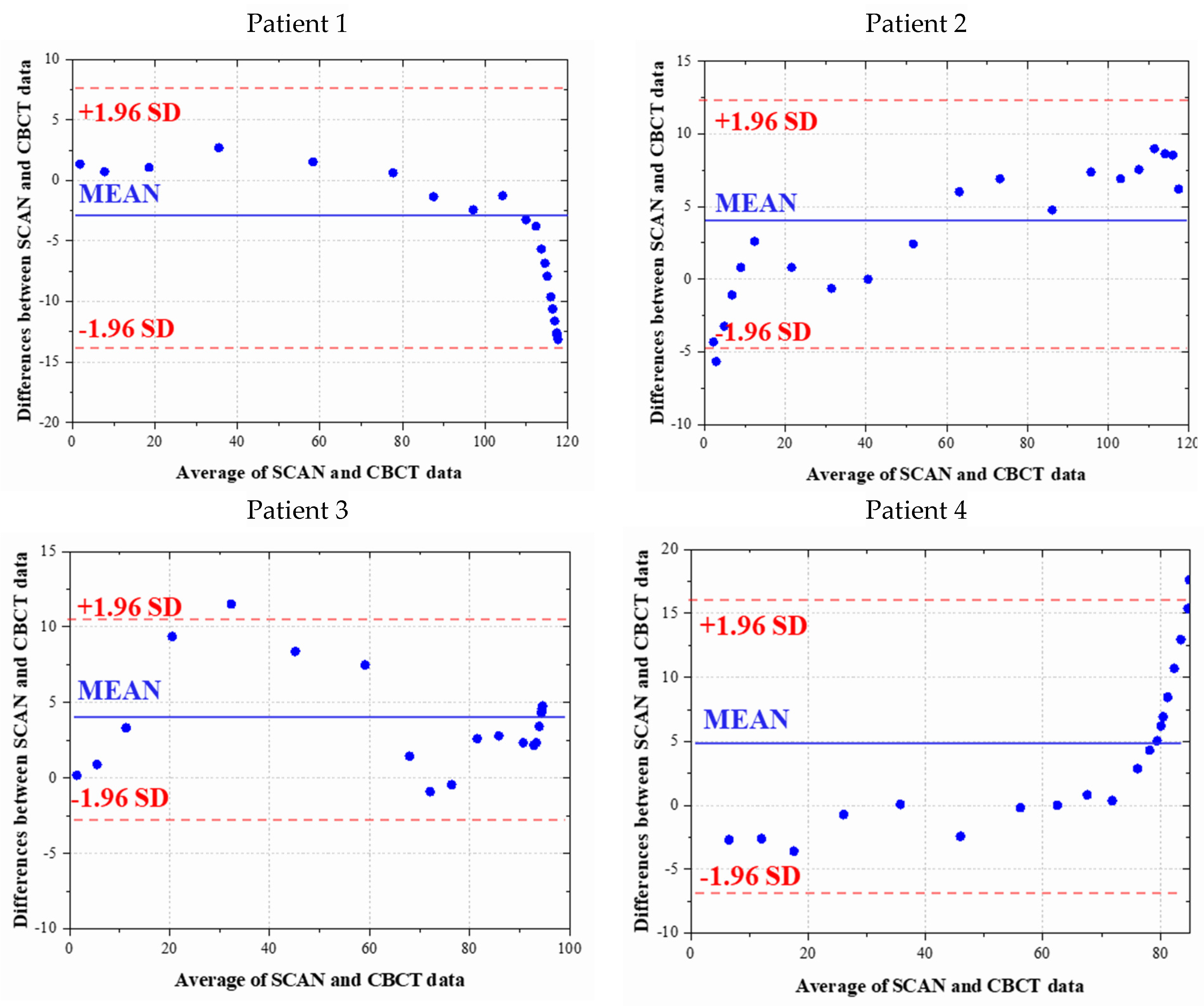
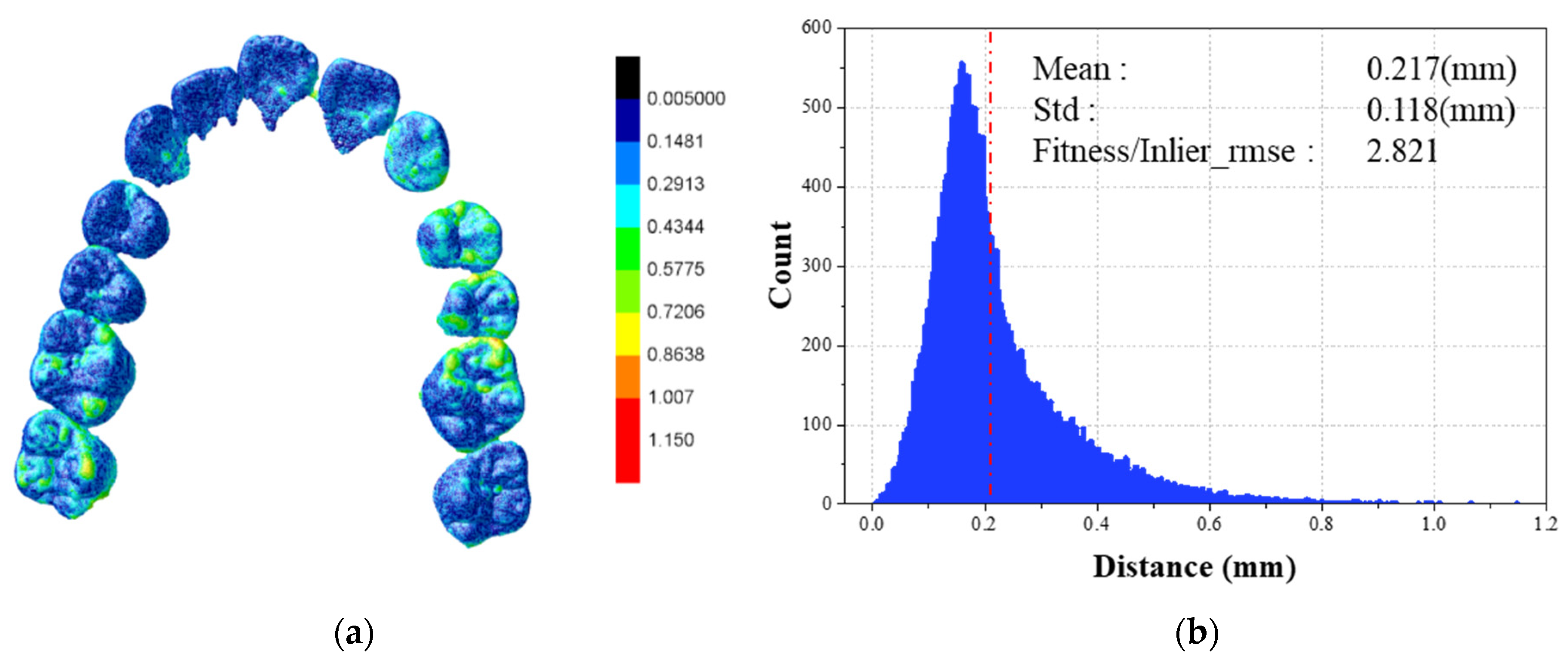
4.1. Statistical Validation
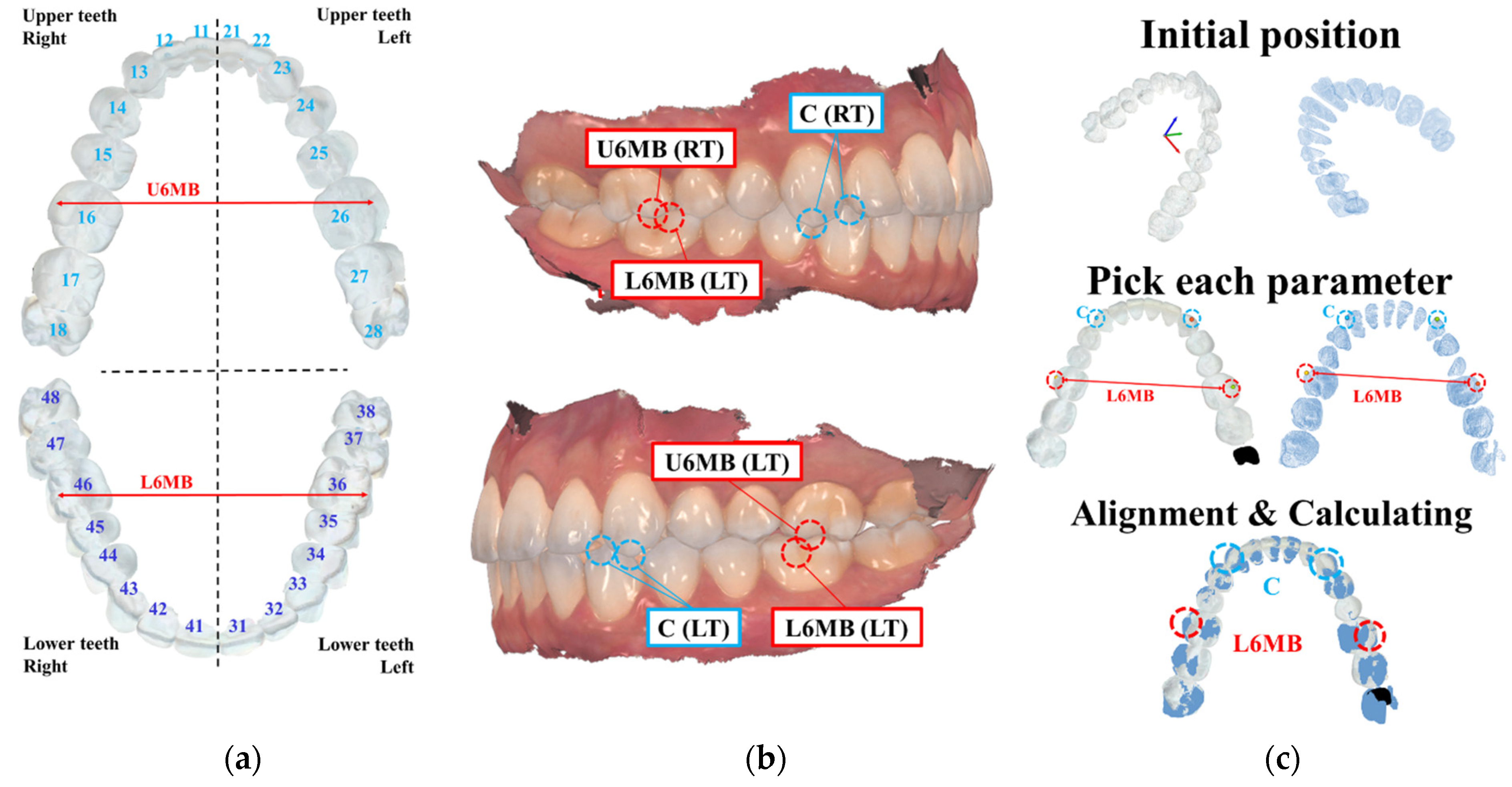
4.2. Quantitative Validation
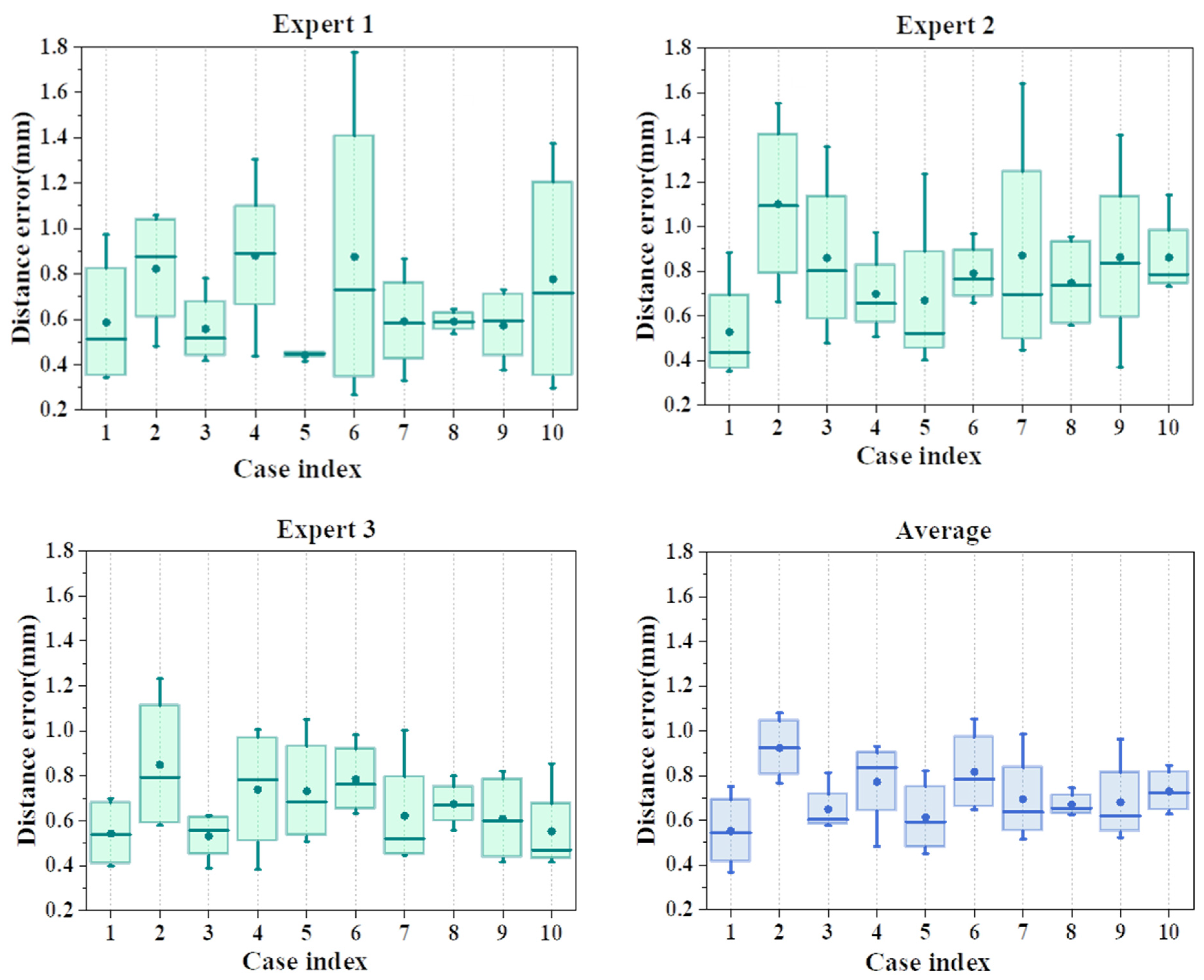
4.3. Clinical Validation
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davidowitz, G.; Kotick, P.G. The use of CAD/CAM in dentistry. Dent. Clin. 2011, 55, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Moörmann, W.H. The evolution of the CEREC system. J. Am. Dent. Assoc. 2006, 137, 7S–13S. [Google Scholar] [CrossRef] [PubMed]
- Alkhayer, A.; Piffkó, J.; Lippold, C.; Segatto, E. Accuracy of virtual planning in orthognathic surgery: A systematic review. Head Face Med. 2020, 16, 34. [Google Scholar] [CrossRef] [PubMed]
- Amorim, P.; Moraes, T.; Silva, J.; Pedrini, H. In Vesalius: An interactive rendering framework for health care support. In Proceedings of the Advances in Visual Computing: 11th International Symposium, ISVC 2015, Las Vegas, NV, USA, 14–16 December 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 45–54. [Google Scholar]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Lim, S.W.; Hwang, H.S.; Cho, I.S.; Baek, S.H.; Cho, J.H. Registration accuracy between intraoral-scanned and cone-beam computed tomography–scanned crowns in various registration methods. Am. J. Orthod. Dentofac. Orthop. 2020, 157, 348–356. [Google Scholar] [CrossRef]
- Piao, X.Y.; Park, J.M.; Kim, H.; Kim, Y.; Shim, J.S. Evaluation of different registration methods and dental restorations on the registration duration and accuracy of cone beam computed tomography data and intraoral scans: A retrospective clinical study. Clin. Oral Investig. 2022, 26, 5763–5771. [Google Scholar] [CrossRef]
- Flügge, T.; Derksen, W.; Te Poel, J.; Hassan, B.; Nelson, K.; Wismeijer, D. Registration of cone beam computed tomography data and intraoral surface scans–A prerequisite for guided implant surgery with CAD/CAM drilling guides. Clin. Oral Implant. Res. 2017, 28, 1113–1118. [Google Scholar] [CrossRef]
- Jang, T.J.; Kim, K.C.; Cho, H.C.; Seo, J.K. A fully automated method for 3D individual tooth identification and segmentation in dental CBCT. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 6562–6568. [Google Scholar] [CrossRef]
- Hyun, C.M.; Bayaraa, T.; Yun, H.S.; Jang, T.J.; Park, H.S.; Seo, J.K. Deep learning method for reducing metal artifacts in dental cone-beam CT using supplementary information from intra-oral scan. Phys. Med. Biol. 2022, 67, 175007. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis; Springer: New York, NY, USA, 1986. [Google Scholar]
- Wikipedia. Principal Component Analysis. Available online: https://en.wikipedia.org/wiki/Principal_component_analysis (accessed on 5 May 2023).
- Mitra, N.J.; Nguyen, A. Estimating surface normals in noisy point cloud data. In Proceedings of the Nineteenth Annual Symposium on Computational Geometry, San Diego, CA, USA, 8–10 June 2003; pp. 322–328. [Google Scholar]
- Bailey, S. Principal component analysis with noisy and/or missing data. Publ. Astron. Soc. Pac. 2012, 124, 1015. [Google Scholar] [CrossRef]
- Schnabel, R.; Wahl, R.; Klein, R. Efficient RANSAC for point-cloud shape detection. In Computer Graphics Forum; Blackwell Publishing Ltd.: Oxford, UK, 2007; pp. 214–226. [Google Scholar]
- Wikipedia. Random Sample Consensus. Available online: https://en.wikipedia.org/wiki/Random_sample_consensus (accessed on 5 May 2023).
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. arXiv 2017, arXiv:1703.06870v3. [Google Scholar]
- Ahn, J.; Nguyen, T.P.; Kim, Y.J.; Kim, T.; Yoon, J. Automated analysis of three-dimensional CBCT images taken in natural head position that combines facial profile processing and multiple deep-learning models. Comput. Methods Programs Biomed. 2022, 226, 107123. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, R.; Wang, Y.; Tao, W. Evaluation of the ICP algorithm in 3D point cloud registration. IEEE Access 2020, 8, 68030–68048. [Google Scholar] [CrossRef]
- Giavarina, D. Understanding bland altman analysis. Biochem. Medica 2015, 25, 141–151. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 327, 307–310. [Google Scholar] [CrossRef]
- Bunce, C. Correlation, agreement, and Bland–Altman analysis: Statistical analysis of method comparison studies. Am. J. Ophthalmol. 2009, 148, 4–6. [Google Scholar] [CrossRef]
- Liu, M.; Sheng, L.; Yang, S.; Shao, J.; Hu, S.M. Morphing and sampling network for dense point cloud completion. Proc. AAAI Conf. Artif. Intell. 2020, 34, 11596–11603. [Google Scholar] [CrossRef]
- Cignoni, P.; Rocchini, C.; Scopigno, R. Metro: Measuring error on simplified surfaces. In Computer Graphics Forum; Blackwell Publishers: Oxford, UK; Blackwell Publishers: Boston, MA, USA, 1998; pp. 167–174. [Google Scholar]
- Solow, B.; Tallgren, A. Natural head position in standing subjects. Acta Odontol. Scand. 1971, 29, 591–607. [Google Scholar] [CrossRef]
- Imandoust, S.B.; Bolandraftar, M. Application of k-nearest neighbor (knn) approach for predicting economic events: Theoretical background. Int. J. Eng. Res. Appl. 2013, 3, 605–610. [Google Scholar]
- Shaheen, E.; Leite, A.; Alqahtani, K.A.; Smolders, A.; Van Gerven, A.; Willems, H.; Jacobs, R. A novel deep learning system for multi-class tooth segmentation and classification on cone beam computed tomography. A validation study. J. Dent. 2021, 115, 103865. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Li, C.; Wang, W. ToothNet: Automatic tooth instance segmentation and identification from cone beam CT images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2019, Long Beach, CA, USA, 15–20 June 2019; pp. 6368–6377. [Google Scholar]
- Noh, H.; Nabha, W.; Cho, J.H.; Hwang, H.S. Registration accuracy in the integration of laser-scanned dental images into maxillofacial cone-beam computed tomography images. Am. J. Orthod. Dentofac. Orthop. 2011, 140, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Hwang, C.J.; Choi, Y.J.; Houschyar, K.S.; Yu, J.H.; Bae, S.Y.; Cha, J.Y. Registration of digital dental models and cone-beam computed tomography images using 3-dimensional planning software: Comparison of the accuracy according to scanning methods and software. Am. J. Orthod. Dentofac. Orthop. 2020, 157, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Deferm, J.T.; Nijsink, J.; Baan, F.; Verhamme, L.; Meijer, G.; Maal, T. Soft tissue-based registration of intraoral scan with cone beam computed tomography scan. Int. J. Oral Maxillofac. Surg. 2022, 51, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Dong, B.; Zhang, Q.; Li, J.; Yuan, Q.; Yue, L. An Indirect Digital Technique to Transfer 3D Printed Casts to a Mechanical Articulator with Individual Sagittal Condylar Inclination Settings Using CBCT and Intraoral Scans. J. Prosthodont. 2022, 31, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.; Singh, A.; Friedland, B.; Jamjoom, F.Z.; Griseto, N.; Gallucci, G.O. The impact of cone beam computer tomography field of view on the precision of digital intra-oral scan registration for static computer-assisted implant surgery: A CBCT analysis. Clin. Oral Implant. Res. 2022, 33, 1273–1281. [Google Scholar] [CrossRef]
- Lee, S.C.; Hwang, H.S.; Lee, K.C. Accuracy of deep learning-based integrated tooth models by merging intraoral scans and CBCT scans for 3D evaluation of root position during orthodontic treatment. Prog. Orthod. 2022, 23, 15. [Google Scholar] [CrossRef]


| CBCT | Intra-Oral Scanner (IOS) | |
|---|---|---|
| Device brand | Imaging sciences international | 3shape |
| Device model | Digital i-CAT FLX MV | Trios 3 |
| Accuracy | mm (voxel size) | µm |
| Measuring time | ~3 min/case | ~5 min/case |
| Measurement area | Upper part of the neck | Teeth surface, gingiva |
| Value | Case Number | Average | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Mean (mm) | 0.226 | 0.292 | 0.215 | 0.221 | 0.217 | 0.229 | 0.249 | 0.227 | 0.217 | 0.249 | 0.234 |
| Std (mm) | 0.125 | 0.202 | 0.108 | 0.114 | 0.112 | 0.118 | 0.157 | 0.112 | 0.118 | 0.155 | 0.132 |
| F/I | 2.813 | 3.167 | 2.693 | 2.880 | 2.813 | 3.025 | 2.455 | 2.989 | 2.821 | 2.746 | 2.840 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-J.; Ahn, J.-H.; Lim, H.-K.; Nguyen, T.P.; Jha, N.; Kim, A.; Yoon, J. Novel Procedure for Automatic Registration between Cone-Beam Computed Tomography and Intraoral Scan Data Supported with 3D Segmentation. Bioengineering 2023, 10, 1326. https://doi.org/10.3390/bioengineering10111326
Kim Y-J, Ahn J-H, Lim H-K, Nguyen TP, Jha N, Kim A, Yoon J. Novel Procedure for Automatic Registration between Cone-Beam Computed Tomography and Intraoral Scan Data Supported with 3D Segmentation. Bioengineering. 2023; 10(11):1326. https://doi.org/10.3390/bioengineering10111326
Chicago/Turabian StyleKim, Yoon-Ji, Jang-Hoon Ahn, Hyun-Kyo Lim, Thong Phi Nguyen, Nayansi Jha, Ami Kim, and Jonghun Yoon. 2023. "Novel Procedure for Automatic Registration between Cone-Beam Computed Tomography and Intraoral Scan Data Supported with 3D Segmentation" Bioengineering 10, no. 11: 1326. https://doi.org/10.3390/bioengineering10111326
APA StyleKim, Y.-J., Ahn, J.-H., Lim, H.-K., Nguyen, T. P., Jha, N., Kim, A., & Yoon, J. (2023). Novel Procedure for Automatic Registration between Cone-Beam Computed Tomography and Intraoral Scan Data Supported with 3D Segmentation. Bioengineering, 10(11), 1326. https://doi.org/10.3390/bioengineering10111326






