Plant Cellulose as a Substrate for 3D Neural Stem Cell Culture
Abstract
:1. Introduction
2. Methods
2.1. Biomaterial Production
2.2. Scanning Electron Microscopy
2.3. Mechanical Testing
2.4. Cell Culture, Scaffold Seeding and Differentiation
2.5. Staining and Confocal Microscopy
2.6. Image Analysis
2.7. Alamar Blue Cell Proliferation Assay
2.8. Statistical Analysis
3. Results
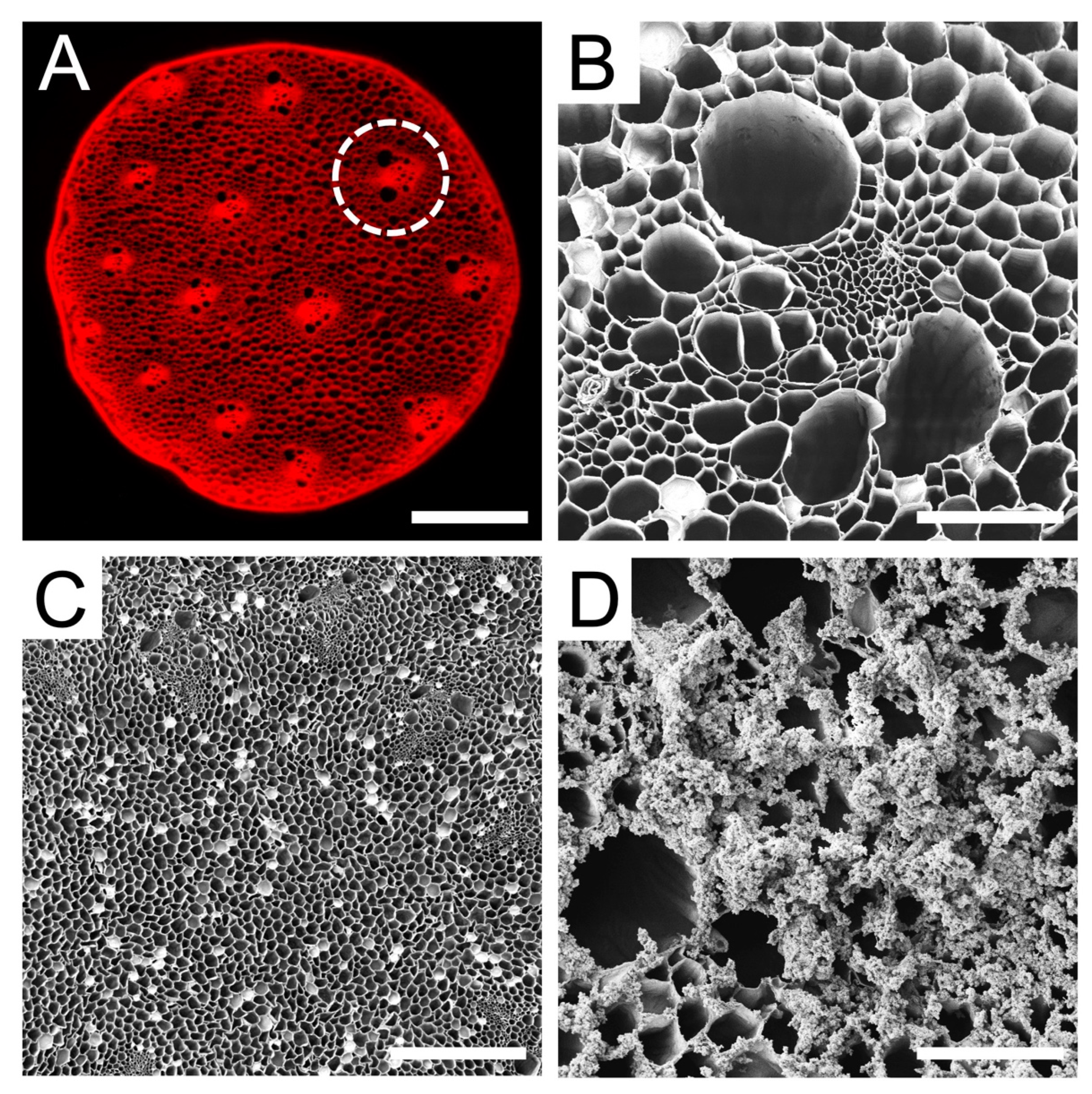
3.1. Characterization of Plant Cellulose Scaffold
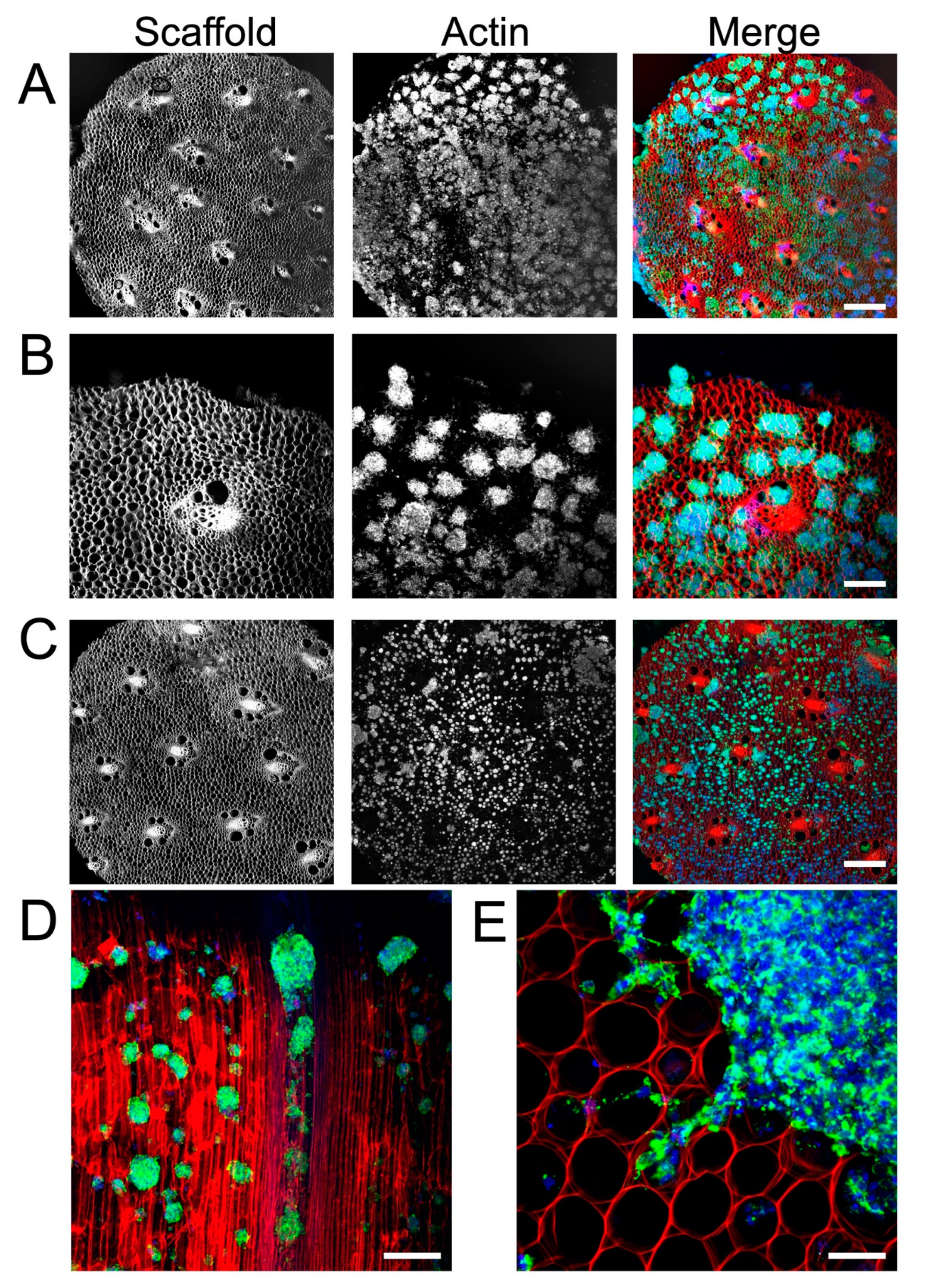
3.2. Cellulose Scaffold Supports Rat NSC Attachment and Proliferation
3.3. Plant Cellulose Scaffold Enhances Neuronal and Astrocytic Differentiation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gage, F.H. Mammalian Neural Stem Cells. Science 2000, 287, 1433–1438. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yu, P.; Cheng, L. Current progress in the derivation and therapeutic application of neural stem cells. Cell Death Dis. 2017, 8, e3108. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Benton, R.L.; Whittemore, S.R. Stem cell repair of central nervous system injury. J. Neurosci. Res. 2002, 68, 501–510. [Google Scholar] [CrossRef]
- Zhao, X.-M.; He, X.-Y.; Liu, J.; Xu, Y.; Xu, F.-F.; Tan, Y.-X.; Zhang, Z.-B.; Wang, T.-H. Neural Stem Cell Transplantation Improves Locomotor Function in Spinal Cord Transection Rats Associated with Nerve Regeneration and IGF-1 R Expression. Cell Transplant. 2019, 28, 1197–1211. [Google Scholar] [CrossRef]
- Zhu, J.; Wu, X.; Zhang, H.L. Adult Neural Stem Cell Therapy: Expansion In Vitro, Tracking In Vivo and Clinical Transplantation. Curr. Drug Targets 2005, 6, 97–110. [Google Scholar] [CrossRef]
- Tejeda, G.; Ciciriello, A.J.; Dumont, C.M. Biomaterial Strategies to Bolster Neural Stem Cell-Mediated Repair of the Central Nervous System. Cells Tissues Organs 2021, 211, 655–669. [Google Scholar] [CrossRef]
- Hwang, D.W.; Jin, Y.; Lee, D.H.; Kim, H.Y.; Na Cho, H.; Chung, H.J.; Park, Y.; Youn, H.; Lee, S.J.; Lee, H.J.; et al. In Vivo Bioluminescence Imaging for Prolonged Survival of Transplanted Human Neural Stem Cells Using 3D Biocompatible Scaffold in Corticectomized Rat Model. PLoS ONE 2014, 9, e105129. [Google Scholar] [CrossRef]
- Rammensee, S.; Kang, M.S.; Georgiou, K.; Kumar, S.; Schaffer, D.V. Dynamics of Mechanosensitive Neural Stem Cell Differentiation. Stem Cells 2016, 35, 497–506. [Google Scholar] [CrossRef]
- Chen, W.; Shao, Y.; Li, X.; Zhao, G.; Fu, J. Nanotopographical surfaces for stem cell fate control: Engineering mechanobiology from the bottom. Nano Today 2014, 9, 759–784. [Google Scholar] [CrossRef]
- Lim, S.H.; Liu, X.Y.; Song, H.; Yarema, K.J.; Mao, H.-Q. The effect of nanofiber-guided cell alignment on the preferential differentiation of neural stem cells. Biomaterials 2010, 31, 9031–9039. [Google Scholar] [CrossRef]
- Czeisler, C.; Short, A.; Nelson, T.; Gygli, P.; Ortiz, C.; Catacutan, F.P.; Stocker, B.; Cronin, J.; Lannutti, J.; Winter, J.; et al. Surface topography during neural stem cell differentiation regulates cell migration and cell morphology. J. Comp. Neurol. 2016, 524, 3485–3502. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, J.; Wu, L.; Zheng, T.; Han, Q.; Liang, Y.; Zhang, L.; Li, G.; Yang, Y. The Influence of the Surface Topographical Cues of Biomaterials on Nerve Cells in Peripheral Nerve Regeneration: A Review. Stem Cells Int. 2021, 2021, 8124444. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Keung, A.J.; Irwin, E.F.; Li, Y.; Little, L.; Schaffer, D.V.; Healy, K.E. Substrate Modulus Directs Neural Stem Cell Behavior. Biophys. J. 2008, 95, 4426–4438. [Google Scholar] [CrossRef]
- Subramony, S.D.; Dargis, B.R.; Castillo, M.; Azeloglu, E.U.; Tracey, M.S.; Su, A.; Lu, H.H. The guidance of stem cell differentiation by substrate alignment and mechanical stimulation. Biomaterials 2013, 34, 1942–1953. [Google Scholar] [CrossRef]
- Metavarayuth, K.; Sitasuwan, P.; Zhao, X.; Lin, Y.; Wang, Q. Influence of surface topographical cues on the differentiation of mesenchymal stem cells in vitro. ACS Biomater. Sci. Eng. 2016, 2, 142–151. [Google Scholar] [CrossRef]
- Li, W.; Tang, Q.Y.; Jadhav, A.D.; Narang, A.; Qian, W.X.; Shi, P.; Pang, S.W. Large-scale Topographical Screen for Investigation of Physical Neural-Guidance Cues. Sci. Rep. 2015, 5, 8644. [Google Scholar] [CrossRef]
- Wang, J.-H.; Hung, C.-H.; Young, T.-H. Proliferation and differentiation of neural stem cells on lysine–alanine sequential polymer substrates. Biomaterials 2006, 27, 3441–3450. [Google Scholar] [CrossRef]
- Flanagan, L.A.; Rebaza, L.M.; Derzic, S.; Schwartz, P.H.; Monuki, E.S. Regulation of human neural precursor cells by laminin and integrins. J. Neurosci. Res. 2006, 83, 845–856. [Google Scholar] [CrossRef]
- Ren, Y.-J.; Zhang, H.; Huang, H.; Wang, X.-M.; Zhou, Z.-Y.; Cui, F.-Z.; An, Y.-H. In vitro behavior of neural stem cells in response to different chemical functional groups. Biomaterials 2009, 30, 1036–1044. [Google Scholar] [CrossRef]
- Joseph, G.; Orme, R.P.; Kyriacou, T.; Fricker, R.A.; Roach, P. Effects of Surface Chemistry Interaction on Primary Neural Stem Cell Neurosphere Responses. ACS Omega 2021, 6, 19901–19910. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Yu, A.; Chen, J.; Yuan, J.; Yin, Y.; Duanmu, W.; Tan, L.; Yang, Y.; Lan, C.; Chen, W.; et al. Poly-L-ornithine enhances migration of neural stem/progenitor cells via promoting α-Actinin 4 binding to actin filaments. Sci. Rep. 2016, 6, 37681. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Tan, L.; Wu, P.; Yin, Y.; Liu, X.; Meng, H.; Cui, G.; Wu, N.; Lin, J.; Hu, R.; et al. Poly-L-ornithine promotes preferred differentiation of neural stem/progenitor cells via ERK signalling pathway. Sci. Rep. 2015, 5, 15535. [Google Scholar] [CrossRef] [PubMed]
- Ping, J.; Fu, H.; Xiong, Y.-J.; Soomro, S.; Huang, Z.-H.; Yu, P.-P. Poly-L-ornithine blocks the inhibitory effects of fibronectin on oligodendrocyte differentiation and promotes myelin repair. Neural Regen. Res. 2022, 18, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, M.; Williams, R.; Downes, J.; Bachhuka, A.; Vasilev, K. The Role of Controlled Surface Topography and Chemistry on Mouse Embryonic Stem Cell Attachment, Growth and Self-Renewal. Materials 2017, 10, 1081. [Google Scholar] [CrossRef] [PubMed]
- Edelbrock, A.N.; Clemons, T.D.; Chin, S.M.; Roan, J.J.W.; Bruckner, E.P.; Álvarez, Z.; Edelbrock, J.F.; Wek, K.S.; Stupp, S.I. Superstructured Biomaterials Formed by Exchange Dynamics and Host–Guest Interactions in Supramolecular Polymers. Adv. Sci. 2021, 8, 2004042. [Google Scholar] [CrossRef]
- Fontoura, J.C.; Viezzer, C.; dos Santos, F.G.; Ligabue, R.A.; Weinlich, R.; Puga, R.D.; Antonow, D.; Severino, P.; Bonorino, C. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C 2019, 107, 110264. [Google Scholar] [CrossRef]
- Belfiore, L.; Aghaei, B.; Law, A.M.; Dobrowolski, J.C.; Raftery, L.J.; Tjandra, A.D.; Yee, C.; Piloni, A.; Volkerling, A.; Ferris, C.J.; et al. Generation and analysis of 3D cell culture models for drug discovery. Eur. J. Pharm. Sci. 2021, 163, 105876. [Google Scholar] [CrossRef]
- Breslin, S.; O’driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef]
- Hong, H.; Seo, Y.B.; Kim, D.Y.; Lee, J.S.; Lee, Y.J.; Lee, H.; Ajiteru, O.; Sultan, T.; Lee, O.J.; Kim, S.H.; et al. Digital light processing 3D printed silk fibroin hydrogel for cartilage tissue engineering. Biomaterials 2019, 232, 119679. [Google Scholar] [CrossRef]
- Du, D.; Furukawa, K.S.; Ushida, T. 3D culture of osteoblast-like cells by unidirectional or oscillatory flow for bone tissue engineering. Biotechnol. Bioeng. 2009, 102, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Marei, I.; Abu Samaan, T.; Al-Quradaghi, M.A.; Farah, A.A.; Mahmud, S.H.; Ding, H.; Triggle, C.R. 3D Tissue-Engineered Vascular Drug Screening Platforms: Promise and Considerations. Front. Cardiovasc. Med. 2022, 9, 847554. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, Y.; Furukawa, T.; Waki, A.; Okuyama, H.; Inoue, M.; Itoh, M.; Zhang, M.-R.; Wakizaka, H.; Sogawa, C.; Kiyono, Y.; et al. High-throughput screening with nanoimprinting 3D culture for efficient drug development by mimicking the tumor environment. Biomaterials 2015, 51, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Xie, J.; Huck, W.T.S. Recent Advances in Engineering the Stem Cell Microniche in 3D. Adv. Sci. 2018, 5, 1800448. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. ASSAY Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Vagaska, B.; Gillham, O.; Ferretti, P. Modelling human CNS injury with human neural stem cells in 2- and 3-Dimensional cultures. Sci. Rep. 2020, 10, 6785. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, H.; Wei, H.; Cheng, H.; Cai, J.; Chen, X.; Xia, L.; Wang, H.; Chai, R. Scaffolds with anisotropic structure for neural tissue engineering. Eng. Regen. 2022, 3, 154–162. [Google Scholar] [CrossRef]
- Bilirgen, A.C.; Toker, M.; Odabas, S.; Yetisen, A.K.; Garipcan, B.; Tasoglu, S. Plant-Based Scaffolds in Tissue Engineering. ACS Biomater. Sci. Eng. 2021, 7, 926–938. [Google Scholar] [CrossRef]
- Hickey, R.J.; Modulevsky, D.J.; Cuerrier, C.M.; Pelling, A.E. Customizing the Shape and Microenvironment Biochemistry of Biocompatible Macroscopic Plant-Derived Cellulose Scaffolds. ACS Biomater. Sci. Eng. 2018, 4, 3726–3736. [Google Scholar] [CrossRef]
- Negrini, N.C.; Toffoletto, N.; Farè, S.; Altomare, L. Plant Tissues as 3D Natural Scaffolds for Adipose, Bone and Tendon Tissue Regeneration. Front. Bioeng. Biotechnol. 2020, 8, 723. [Google Scholar] [CrossRef]
- Toker, M.; Rostami, S.; Kesici, M.; Gul, O.; Kocaturk, O.; Odabas, S.; Garipcan, B. Decellularization and characterization of leek: A potential cellulose-based biomaterial. Cellulose 2020, 27, 7331–7348. [Google Scholar] [CrossRef]
- Fontana, G.; Gershlak, J.; Adamski, M.; Lee, J.S.; Matsumoto, S.; Le, H.; Binder, B.; Wirth, J.; Gaudette, G.; Murphy, W.L. Biofunctionalized Plants as Diverse Biomaterials for Human Cell Culture. Adv. Healthc. Mater. 2017, 6, 1601225. [Google Scholar] [CrossRef] [PubMed]
- Gershlak, J.R.; Hernandez, S.; Fontana, G.; Perreault, L.R.; Hansen, K.J.; Larson, S.A.; Binder, B.Y.; Dolivo, D.M.; Yang, T.; Dominko, T.; et al. Crossing kingdoms: Using decellularized plants as perfusable tissue engineering scaffolds. Biomaterials 2017, 125, 13–22. [Google Scholar] [CrossRef]
- Wang, Y.; Dominko, T.; Weathers, P.J. Using decellularized grafted leaves as tissue engineering scaffolds for mammalian cells. Vitr. Cell. Dev. Biol.-Plant 2020, 56, 765–774. [Google Scholar] [CrossRef]
- Harris, A.F.; Lacombe, J.; Liyanage, S.; Han, M.Y.; Wallace, E.; Karsunky, S.; Abidi, N.; Zenhausern, F. Supercritical carbon dioxide decellularization of plant material to generate 3D biocompatible scaffolds. Sci. Rep. 2021, 11, 3643. [Google Scholar] [CrossRef]
- Robbins, E.R.; Pins, G.D.; Laflamme, M.A.; Gaudette, G.R. Creation of a contractile biomaterial from a decellularized spinach leaf without ECM protein coating: An in vitro study. J. Biomed. Mater. Res. A 2020, 108, 2123–2132. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Shiwarski, D.J.; Ball, R.L.; Whitehead, K.A.; Feinberg, A.W.W. Engineering Aligned Skeletal Muscle Tissue Using Decellularized Plant-Derived Scaffolds. ACS Biomater. Sci. Eng. 2020, 6, 3046–3054. [Google Scholar] [CrossRef]
- Salehi, A.; Mobarhan, M.A.; Mohammadi, J.; Shahsavarani, H.; Shokrgozar, M.A.; Alipour, A. Efficient mineralization and osteogenic gene overexpression of mesenchymal stem cells on decellularized spinach leaf scaffold. Gene 2020, 757, 144852. [Google Scholar] [CrossRef]
- Dikici, S.; Claeyssens, F.; MacNeil, S. Decellularised baby spinach leaves and their potential use in tissue engineering applications: Studying and promoting neovascularisation. J. Biomater. Appl. 2019, 34, 546–559. [Google Scholar] [CrossRef]
- Lee, J.; Jung, H.; Park, N.; Park, S.-H.; Ju, J.H. Induced Osteogenesis in Plants Decellularized Scaffolds. Sci. Rep. 2019, 9, 20194. [Google Scholar] [CrossRef]
- Hickey, R.J.; Latour, M.L.; Harden, J.L.; Pelling, A.E. Designer Scaffolds for Interfacial Bioengineering. Adv. Eng. Mater. 2023, 25, 2201415. [Google Scholar] [CrossRef]
- Toker-Bayraktar, M.; Erenay, B.; Altun, B.; Odabaş, S.; Garipcan, B. Plant-derived biomaterials and scaffolds. Cellulose 2023, 30, 2731–2751. [Google Scholar] [CrossRef]
- Varhama, K.; Oda, H.; Shima, A.; Takeuchi, S. Decellularized Plant Leaves for 3D Cell Culturing. In Proceedings of the 2019 IEEE 32nd International Conference on Micro Electro Mechanical Systems (MEMS), Seoul, Republic of Korea, 27–31 January 2019; pp. 226–228. [Google Scholar]
- Chen, F.; Wu, M.; Wu, P.; Xiao, A.; Ke, M.; Huselstein, C.; Cai, L.; Tong, Z.; Chen, Y. Natural Flammulina velutipes-Based Nerve Guidance Conduit as a Potential Biomaterial for Peripheral Nerve Regeneration: In Vitro and In Vivo Studies. ACS Biomater. Sci. Eng. 2021, 7, 3821–3834. [Google Scholar] [CrossRef] [PubMed]
- Phan, N.V.; Wright, T.; Rahman, M.M.; Xu, J.; Coburn, J.M. In Vitro Biocompatibility of Decellularized Cultured Plant Cell-Derived Matrices. ACS Biomater. Sci. Eng. 2020, 6, 822–832. [Google Scholar] [CrossRef]
- Modulevsky, D.J.; Cuerrier, C.M.; Pelling, A.E. Biocompatibility of Subcutaneously Implanted Plant-Derived Cellulose Biomaterials. PLoS ONE 2016, 11, e0157894. [Google Scholar]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef]
- Modulevsky, D.J.; Lefebvre, C.; Haase, K.; Al-Rekabi, Z.; Pelling, A.E. Apple Derived Cellulose Scaffolds for 3D Mammalian Cell Culture. PLoS ONE 2014, 9, e97835. [Google Scholar] [CrossRef]
- Dai, Y.; Qiao, K.; Li, D.; Isingizwe, P.; Liu, H.; Liu, Y.; Lim, K.; Woodfield, T.; Liu, G.; Hu, J.; et al. Plant-Derived Biomaterials and Their Potential in Cardiac Tissue Repair. Adv. Healthc. Mater. 2023, 12, e2202827. [Google Scholar] [CrossRef]
- Peng, W.; Li, D.; Dai, K.; Wang, Y.; Song, P.; Li, H.; Tang, P.; Zhang, Z.; Li, Z.; Zhou, Y.; et al. Recent progress of collagen, chitosan, alginate and other hydrogels in skin repair and wound dressing applications. Int. J. Biol. Macromol. 2022, 208, 400–408. [Google Scholar] [CrossRef]
- Zhu, T.; Jiang, J.; Zhao, J.; Chen, S.; Yan, X. Regulating Preparation Of Functional Alginate-Chitosan Three-Dimensional Scaffold For Skin Tissue Engineering. Int. J. Nanomed. 2019, 14, 8891–8903. [Google Scholar] [CrossRef]
- Venkatesan, J.; Kim, S.-K. Chitosan Composites for Bone Tissue Engineering—An Overview. Mar. Drugs 2010, 8, 2252–2266. [Google Scholar] [CrossRef] [PubMed]
- Latour, M.L.; Tarar, M.; Hickey, R.J.; Cuerrier, C.M.; Catelas, I.; Pelling, A.E. Plant-derived Cellulose Scaffolds for Bone Tissue Engineering. BiorXiv 2020. [Google Scholar] [CrossRef]
- Balasundari, R.; Bishi, D.K.; Mathapati, S.; Naser, S.B.; Cherian, K.M.; Guhathakurta, S. Nanocoated Botanical Scaffold in Salvage for Human Tissue Regeneration. J. Biomater. Tissue Eng. 2012, 2, 330–335. [Google Scholar] [CrossRef]
- Loh, Q.L.; Choong, C. Three-Dimensional Scaffolds for Tissue Engineering Applications: Role of Porosity and Pore Size. Tissue Eng. Part B Rev. 2013, 19, 485–502. [Google Scholar] [CrossRef]
- Rasoulianboroujeni, M.; Kiaie, N.; Tabatabaei, F.S.; Yadegari, A.; Fahimipour, F.; Khoshroo, K.; Tayebi, L. Dual Porosity Protein-based Scaffolds with Enhanced Cell Infiltration and Proliferation. Sci. Rep. 2018, 8, 14889. [Google Scholar] [CrossRef]
- Leclech, C.; Villard, C. Cellular and Subcellular Contact Guidance on Microfabricated Substrates. Front. Bioeng. Biotechnol. 2020, 8, 551505. [Google Scholar] [CrossRef]
- Kourgiantaki, A.; Tzeranis, D.S.; Karali, K.; Georgelou, K.; Bampoula, E.; Psilodimitrakopoulos, S.; Yannas, I.V.; Stratakis, E.; Sidiropoulou, K.; Charalampopoulos, I.; et al. Neural stem cell delivery via porous collagen scaffolds promotes neuronal differentiation and locomotion recovery in spinal cord injury. NPJ Regen. Med. 2020, 5, 12. [Google Scholar] [CrossRef]
- Sheets, K.T.; Ewend, M.G.; Mohiti-Asli, M.; Tuin, S.A.; Loboa, E.G.; Aboody, K.S.; Hingtgen, S.D. Developing Implantable Scaffolds to Enhance Neural Stem Cell Therapy for Post-Operative Glioblastoma. Mol. Ther. 2020, 28, 1056–1067. [Google Scholar] [CrossRef]
- Luca, A.C.; Mersch, S.; Deenen, R.; Schmidt, S.; Messner, I.; Schäfer, K.-L.; Baldus, S.E.; Huckenbeck, W.; Piekorz, R.P.; Knoefel, W.T.; et al. Impact of the 3D Microenvironment on Phenotype, Gene Expression, and EGFR Inhibition of Colorectal Cancer Cell Lines. PLoS ONE 2013, 8, e59689. [Google Scholar] [CrossRef]
- Fallica, B.; Maffei, J.S.; Villa, S.; Makin, G.; Zaman, M. Alteration of Cellular Behavior and Response to PI3K Pathway Inhibition by Culture in 3D Collagen Gels. PLoS ONE 2012, 7, e48024. [Google Scholar] [CrossRef]
- Baek, J.; Cho, S.-Y.; Kang, H.; Ahn, H.; Jung, W.-B.; Cho, Y.; Lee, E.; Cho, S.-W.; Jung, H.-T.; Im, S.G. Distinct Mechanosensing of Human Neural Stem Cells on Extremely Limited Anisotropic Cellular Contact. ACS Appl. Mater. Interfaces 2018, 10, 33891–33900. [Google Scholar] [CrossRef] [PubMed]
- Oral, I.; Guzel, H.; Ahmetli, G. Measuring the Young’s modulus of polystyrene-based composites by tensile test and pulse-echo method. Polym. Bull. 2011, 67, 1893–1906. [Google Scholar] [CrossRef]
- Tsai, E.C.; Dalton, P.D.; Shoichet, M.S.; Tator, C.H.; Struzyna, L.A.; Wolf, J.A.; Mietus, C.J.; Adewole, D.O.; Chen, H.I.; Smith, D.H.; et al. Synthetic Hydrogel Guidance Channels Facilitate Regeneration of Adult Rat Brainstem Motor Axons after Complete Spinal Cord Transection. J. Neurotrauma 2004, 21, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.-K.; Chang, G.-L.; Lin, H.-S.; Walter, F.R.; Bunegin, L. Stress-strain relationship of the spinal cord of anesthetized cats. J. Biomech. 1981, 14, 269–276. [Google Scholar] [CrossRef]
- Koffler, J.; Zhu, W.; Qu, X.; Platoshyn, O.; Dulin, J.N.; Brock, J.; Graham, L.; Lu, P.; Sakamoto, J.; Marsala, M.; et al. Biomimetic 3D-printed scaffolds for spinal cord injury repair. Nat. Med. 2019, 25, 263–269. [Google Scholar] [CrossRef]
- Banerjee, A.; Arha, M.; Choudhary, S.; Ashton, R.S.; Bhatia, S.R.; Schaffer, D.V.; Kane, R.S. The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells. Biomaterials 2009, 30, 4695–4699. [Google Scholar] [CrossRef]
- Zhou, P.; Xu, P.; Guan, J.; Zhang, C.; Chang, J.; Yang, F.; Xiao, H.; Sun, H.; Zhang, Z.; Wang, M.; et al. Promoting 3D neuronal differentiation in hydrogel for spinal cord regeneration. Colloids Surf. B Biointerfaces 2020, 194, 111214. [Google Scholar] [CrossRef]
- Bozza, A.; Coates, E.E.; Incitti, T.; Ferlin, K.M.; Messina, A.; Menna, E.; Bozzi, Y.; Fisher, J.P.; Casarosa, S. Neural differentiation of pluripotent cells in 3D alginate-based cultures. Biomaterials 2014, 35, 4636–4645. [Google Scholar] [CrossRef]
- Lu, H.F.; Lim, S.-X.; Leong, M.F.; Narayanan, K.; Toh, R.P.; Gao, S.; Wan, A.C. Efficient neuronal differentiation and maturation of human pluripotent stem cells encapsulated in 3D microfibrous scaffolds. Biomaterials 2012, 33, 9179–9187. [Google Scholar] [CrossRef]
- Hadjiantoniou, S.V.; Sean, D.; Ignacio, M.; Godin, M.; Slater, G.W.; Pelling, A.E. Physical confinement signals regulate the organization of stem cells in three dimensions. J. R. Soc. Interface 2016, 13, 20160613. [Google Scholar] [CrossRef]
- Temple, J.; Velliou, E.; Shehata, M.; Lévy, R.; Gupta, P. Current strategies with implementation of three-dimensional cell culture: The challenge of quantification. Interface Focus 2022, 12, 20220019. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Couvrette, L.J.; Walker, K.L.A.; Bui, T.V.; Pelling, A.E. Plant Cellulose as a Substrate for 3D Neural Stem Cell Culture. Bioengineering 2023, 10, 1309. https://doi.org/10.3390/bioengineering10111309
Couvrette LJ, Walker KLA, Bui TV, Pelling AE. Plant Cellulose as a Substrate for 3D Neural Stem Cell Culture. Bioengineering. 2023; 10(11):1309. https://doi.org/10.3390/bioengineering10111309
Chicago/Turabian StyleCouvrette, Lauren J., Krystal L. A. Walker, Tuan V. Bui, and Andrew E. Pelling. 2023. "Plant Cellulose as a Substrate for 3D Neural Stem Cell Culture" Bioengineering 10, no. 11: 1309. https://doi.org/10.3390/bioengineering10111309
APA StyleCouvrette, L. J., Walker, K. L. A., Bui, T. V., & Pelling, A. E. (2023). Plant Cellulose as a Substrate for 3D Neural Stem Cell Culture. Bioengineering, 10(11), 1309. https://doi.org/10.3390/bioengineering10111309








