Ammonia Production Using Bacteria and Yeast toward a Sustainable Society
Abstract
1. Introduction
1.1. Industrial Uses and the Need for Ammonia
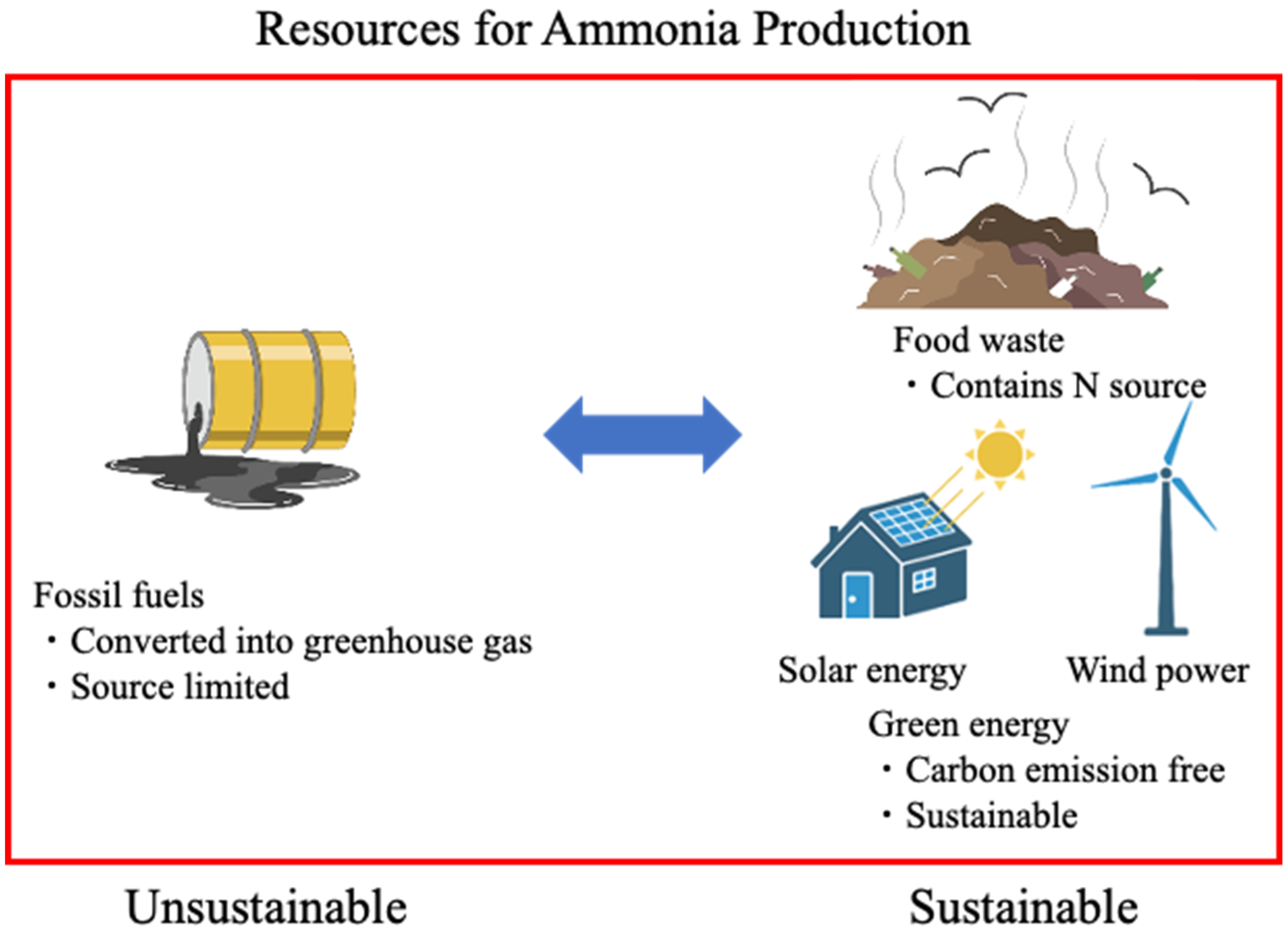
1.2. Chemical Method for Ammonia Production
2. Engineered Bacterial Method for Ammonia Production
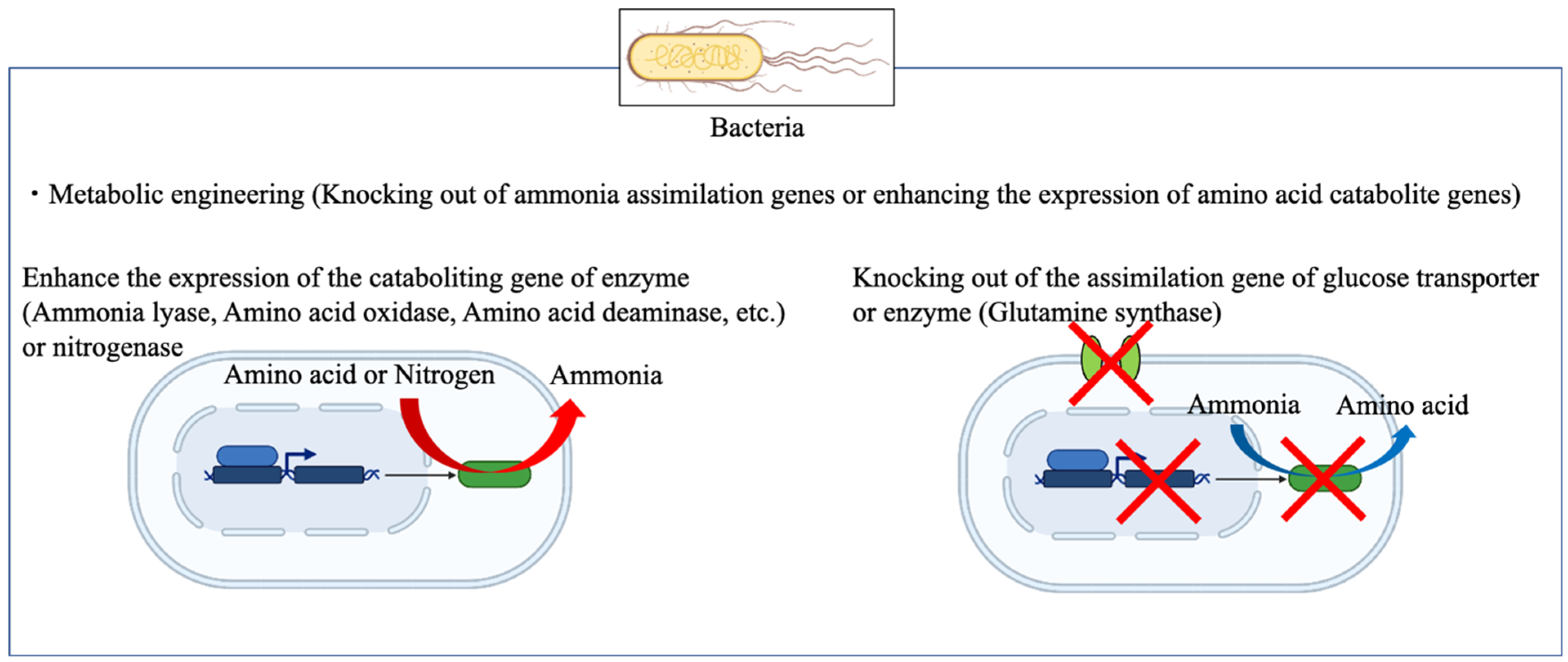
2.1. Bioengineering of Nitrogen Fixation and Metabolic Engineering for Ammonia Production
2.2. Ammonia Production from Food Waste by Using Bioengineering Methods
3. Engineered Yeast Method for Ammonia Production
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Fawzy, S.; Osman, A.I.; Doran, W.J.; Rooney, D.W. Strategies for mitigation of climate change: A review. Environ. Chem. Lett. 2020, 18, 2069–2094. [Google Scholar] [CrossRef]
- Tortell, P.D. Earth 2020: Science, society, and sustainability in the Anthropocene. Proc. Natl. Acad. Sci. USA 2020, 117, 8683–8691. [Google Scholar] [CrossRef] [PubMed]
- Capua, I.; Giovannini, E. Coding system to track research progress towards SDGs. Nature 2019, 572, 178. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Majumdar, A. Opportunities and challenges for a sustainable energy future. Nature 2012, 488, 294–303. [Google Scholar] [CrossRef]
- Chu, S.; Cui, Y.; Liu, N. The path towards sustainable energy. Nat. Mater. 2016, 16, 16–22. [Google Scholar] [CrossRef]
- Hussain, A.; Arif, S.M.; Aslam, M. Emerging renewable and sustainable energy technologies: State of the art. Renew. Sustain. Energy Rev. 2017, 71, 12–28. [Google Scholar] [CrossRef]
- Creutzig, F.; Ravindranath, N.H.; Berndes, G.; Bolwig, S.; Bright, R.; Cherubini, F.; Chum, H.; Corbera, E.; Delucchi, M.; Faaij, A.; et al. Bioenergy and climate change mitigation: An assessment. Glob. Chang. Biol. Bioenergy 2015, 7, 916–944. [Google Scholar] [CrossRef]
- Elishav, O.; Lis, B.M.; Miller, E.M.; Arent, D.J.; Valera-Medina, A.; Dana, A.G.; Shter, G.E.; Grader, G.S. Progress and Prospective of Nitrogen-Based Alternative Fuels. Chem. Rev. 2020, 120, 5352–5436. [Google Scholar] [CrossRef]
- Baliban, R.C.; Elia, J.A.; Floudas, C.A. Biomass and Natural Gas to Liquid Transportation Fuels: Process Synthesis, Global Optimization, and Topology Analysis. Ind. Eng. Chem. Res. 2013, 52, 3381–3406. [Google Scholar] [CrossRef]
- Lynd, L.R.; Larson, E.; Greene, N.; Laser, M.; Sheehan, J.; Dale, B.E.; McLaughlin, S.; Wang, M. The role of biomass in America’s energy future: Framing the analysis. Biofuels Bioprod. Biorefin. 2009, 3, 113–123. [Google Scholar] [CrossRef]
- Siddiki, S.Y.A.; Mofijur, M.; Kumar, P.S.; Ahmed, S.F.; Inayat, A.; Kusumo, F.; Badruddin, I.A.; Yunus Khan, T.M.; Nghiem, L.D.; Ong, H.C.; et al. Microalgae biomass as a sustainable source for biofuel, biochemical and biobased value-added products: An integrated biorefinery concept. Fuel 2022, 307, 121782. [Google Scholar] [CrossRef]
- Govarthanan, M.; Manikandan, S.; Subbaiya, R.; Krishnan, R.Y.; Srinivasan, S.; Karmegam, N.; Kim, W. Emerging trends and nanotechnology advances for sustainable biogas production from lignocellulosic waste biomass: A critical review. Fuel 2022, 312, 122928. [Google Scholar] [CrossRef]
- Paritosh, K.; Kushwaha, S.K.; Yadav, M.; Pareek, N.; Chawade, A.; Vivekanand, V. Food Waste to Energy: An Overview of Sustainable Approaches for Food Waste Management and Nutrient Recycling. BioMed Res. Int. 2017, 2017, 2370927. [Google Scholar] [CrossRef]
- Rosenboom, J.-G.; Langer, R.; Traverso, G. Bioplastics for a circular economy. Nat. Rev. Mater. 2022, 7, 117–137. [Google Scholar] [CrossRef]
- Erisman, J.W.; Sutton, M.A.; Galloway, J.; Klimont, Z.; Winiwarter, W. How a century of ammonia synthesis changed the world. Nat. Geosci. 2008, 1, 636–639. [Google Scholar] [CrossRef]
- Smil, V. Population Growth and Nitrogen: An Exploration of a Critical Existential Link. Popul. Dev. Rev. 1991, 17, 569. [Google Scholar] [CrossRef]
- Fedoruk, M.J.; Bronstein, R.; Kerger, B.D. Ammonia exposure and hazard assessment for selected household cleaning product uses. J. Expo. Sci. Environ. Epidemiol. 2005, 15, 534–544. [Google Scholar] [CrossRef]
- Park, S.; Jeong, J.; Fujita, K.-I.; Yamamoto, A.; Yoshida, H. Anti-Markovnikov Hydroamination of Alkenes with Aqueous Ammonia by Metal-Loaded Titanium Oxide Photocatalyst. J. Am. Chem. Soc. 2020, 142, 12708–12714. [Google Scholar] [CrossRef]
- Zumdahl, S.S. Ammonia. Encyclopedia Britannica. 2022. Available online: https://www.britannica.com/science/ammonia (accessed on 1 January 2022).
- Hu, B.; Huang, S.; Shao, Y.; Chen, J. Thermodynamic analysis of a new ammonia-water power cycle. Energy Rep. 2020, 6, 567–573. [Google Scholar] [CrossRef]
- Wakida, T.; Tokuyama, T.; Doi, C.; Lee, M.; Jeong, D.S.; Ishida, S. Mechanical Properties of Polyester/Cotton and Polyester/Rayon Fabrics Treated with Ammonia-Gas. J. Soc. Fiber Sci. Technol. 2004, 60, 34–37. [Google Scholar] [CrossRef]
- Müller, T.E.; Beller, M. Metal-Initiated Amination of Alkenes and Alkynes. Chem. Rev. 1998, 98, 675–704. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, M.; Muroyama, H.; Suzuki, S.; Saito, M.; Koide, T.; Takahashi, Y.; Horiuchi, T.; Yamasaki, H.; Matsumoto, S.; Kuboet, H.; et al. Development of 1 kW-class ammonia-fueled solid oxide fuel cell stack. Fuel Cells 2020, 20, 80–88. [Google Scholar] [CrossRef]
- Zhao, Y.; Setzler, B.P.; Wang, J.; Nash, J.; Wang, T.; Xu, B.; Yan, Y. An Efficient Direct Ammonia Fuel Cell for Affordable Carbon-Neutral Transportation. Joule 2019, 3, 2472–2484. [Google Scholar] [CrossRef]
- Valera-Medina, A.; Amer-Hatem, F.; Azad, A.K.; Dedoussi, I.C.; de Joannon, M.; Fernandes, R.X.; Glarborg, P.; Hashemi, H.; He, X.; Mashruk, S.; et al. Review on Ammonia as a Potential Fuel: From Synthesis to Economics. Energy Fuels 2021, 35, 6964–7029. [Google Scholar] [CrossRef]
- Erdemir, D.; Dincer, I. A perspective on the use of ammonia as a clean fuel: Challenges and solutions. Int. J. Energy Res. 2021, 45, 4827–4834. [Google Scholar] [CrossRef]
- Herbinet, O.; Bartocci, P.; Dana, A.G. On the use of ammonia as a fuel—A perspective. Fuel Commun. 2022, 11, 100064. [Google Scholar] [CrossRef]
- Miura, D.; Tezuka, T. A comparative study of ammonia energy systems as a future energy carrier, with particular reference to vehicle use in Japan. Energy 2014, 68, 428–436. [Google Scholar] [CrossRef]
- Valera-Medina, A.; Xiao, H.; Owen-Jones, M.; David, W.I.F.; Bowen, P.J. Ammonia for power. Prog. Energy Combust. Sci. 2018, 69, 63–102. [Google Scholar] [CrossRef]
- Wijayanta, A.T.; Oda, T.; Purnomo, C.W.; Kashiwagi, T.; Aziz, M. Liquid hydrogen, methylcyclohexane, and ammonia as potential hydrogen storage: Comparison review. Int. J. Hydrogen Energy 2019, 44, 15026–15044. [Google Scholar] [CrossRef]
- Moseley, P.T.; Garche, J. Electrochemical Energy Storage for Renewable Sources and Grid Balancing; Newnes: Oxford, UK; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Lan, R.; Irvine, J.T.S.; Tao, S. Ammonia and related chemicals as potential indirect hydrogen storage materials. Int. J. Hydrog. Energy 2012, 37, 1482–1494. [Google Scholar] [CrossRef]
- Jeerh, G.; Zhang, M.; Tao, S. Recent progress in ammonia fuel cells and their potential applications. J. Mater. Chem. A Mater. Energy Sustain. 2021, 9, 727–752. [Google Scholar] [CrossRef]
- Wang, W.; Herreros, J.M.; Tsolakis, A.; York, A.P. Ammonia as hydrogen carrier for transportation; investigation of the ammonia exhaust gas fuel reforming. Int. J. Hydrogen Energy 2013, 38, 9907–9917. [Google Scholar] [CrossRef]
- Palys, M.J.; Daoutidis, P. Using hydrogen and ammonia for renewable energy storage: A geographically comprehensive techno-economic study. Comput. Chem. Eng. 2020, 136, 106785. [Google Scholar] [CrossRef]
- Al-Aboosi, F.Y.; El-Halwagi, M.M.; Moore, M.; Nielsen, R.B. Renewable ammonia as an alternative fuel for the shipping industry. Curr. Opin. Chem. Eng. 2021, 31, 100670. [Google Scholar] [CrossRef]
- Green, L. An ammonia energy vector for the hydrogen economy. Int. J. Hydrogen Energy 1982, 7, 355–359. [Google Scholar] [CrossRef]
- Schrock, R.R. Reduction of dinitrogen. Proc. Natl. Acad. Sci. USA 2006, 103, 17087. [Google Scholar] [CrossRef]
- Shilov, A.E. Catalytic reduction of molecular nitrogen in solutions. Russ. Chem. Bull. 2003, 52, 2555–2562. [Google Scholar] [CrossRef]
- Capdevila-Cortada, M. Electrifying the Haber–Bosch. Nat. Catal. 2019, 2, 1055. [Google Scholar] [CrossRef]
- Raróg-Pilecka, W.; Miśkiewicz, E.; Szmigiel, D.; Kowalczyk, Z. Structure sensitivity of ammonia synthesis over promoted ruthenium catalysts supported on graphitised carbon. J. Catal. 2005, 231, 11–19. [Google Scholar] [CrossRef]
- Over, H. Surface Chemistry of Ruthenium Dioxide in Heterogeneous Catalysis and Electrocatalysis: From Fundamental to Applied Research. Chem. Rev. 2012, 112, 3356–3426. [Google Scholar] [CrossRef]
- Kitano, M.; Inoue, Y.; Yamazaki, Y.; Hayashi, F.; Kanbara, S.; Matsuishi, S.; Yokoyama, T.; Kim, S.W.; Hara, M.; Hosono, H. Ammonia synthesis using a stable electride as an electron donor and reversible hydrogen store. Nat. Chem. 2012, 4, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, J.; Lan, R.; Tao, S. Development and Recent Progress on Ammonia Synthesis Catalysts for Haber–Bosch Process. Adv. Energy Sustain. Res. 2020, 2, 2000043. [Google Scholar] [CrossRef]
- Ashida, Y.; Arashiba, K.; Tanaka, H.; Egi, A.; Nakajima, K.; Yoshizawa, K.; Nishibayashi, Y. Molybdenum-Catalyzed Ammonia Formation Using Simple Monodentate and Bidentate Phosphines as Auxiliary Ligands. Inorg. Chem. 2019, 58, 8927–8932. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Barros, P.; Derosa, J.; Chalkley, M.J.; Peters, J.C. Tandem electrocatalytic N2 fixation via proton-coupled electron transfer. Nature 2022, 609, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ghavam, S.; Vahdati, M.; Wilson, I.A.G.; Styring, P. Sustainable Ammonia Production Processes. Front. Energy Res. 2021, 9, 580808. [Google Scholar] [CrossRef]
- Wang, M.; Khan, M.A.; Mohsin, I.; Wicks, J.; Ip, A.H.; Sumon, K.Z.; Dinh, C.-T.; Sargent, E.H.; Gates, I.D.; Kibria, G.; et al. Can sustainable ammonia synthesis pathways compete with fossil-fuel based Haber–Bosch processes? Energy Environ. Sci. 2021, 14, 2535–2548. [Google Scholar] [CrossRef]
- Palys, M.J.; Wang, H.; Zhang, Q.; Daoutidis, P. Renewable ammonia for sustainable energy and agriculture: Vision and systems engineering opportunities. Curr. Opin. Chem. Eng. 2021, 31, 100667. [Google Scholar] [CrossRef]
- Salmon, N.; Bañares-Alcántara, R. Green ammonia as a spatial energy vector: A review. Sustain. Energy Fuels 2021, 5, 2814–2839. [Google Scholar] [CrossRef]
- Melikoglu, M.; Lin, C.S.K.; Webb, C. Analysing global food waste problem: Pinpointing the facts and estimating the energy content. Cent. Eur. J. Eng. 2013, 3, 157–164. [Google Scholar] [CrossRef]
- Liu, C.; Hotta, Y.; Santo, A.; Hengesbaugh, M.; Watabe, A.; Totoki, Y.; Allen, D.; Bengtsson, M. Food waste in Japan: Trends, current practices and key challenges. J. Clean. Prod. 2016, 133, 557–564. [Google Scholar] [CrossRef]
- Katami, T.; Yasuhara, A.; Shibamoto, T. Formation of Dioxins from Incineration of Foods Found in Domestic Garbage. Environ. Sci. Technol. 2004, 38, 1062–1065. [Google Scholar] [CrossRef]
- Chan, L.Y.; Takahashi, M.; Lim, P.J.; Aoyama, S.; Makino, S.; Ferdinandus, F.; Ng, S.Y.C.; Arai, S.; Fujita, H.; Tan, H.C.; et al. Eurotium Cristatum Fermented Okara as a Potential Food Ingredient to Combat Diabetes. Sci. Rep. 2019, 9, 17536. [Google Scholar] [CrossRef]
- Hadj Saadoun, J.; Calani, L.; Cirlini, M.; Bernini, V.; Neviani, E.; Del Rio, D.; Galavernaa, G.; Lazzi, C. Effect of fermentation with single and co-culture of lactic acid bacteria on okara: Evaluation of bioactive compounds and volatile profiles. Food Funct. 2021, 12, 3033–3043. [Google Scholar] [CrossRef]
- Gil-Lalaguna, N.; Afailal, Z.; Aznar, M.; Fonts, I. Exploring the sustainable production of ammonia by recycling N and H in biological residues: Evolution of fuel-N during glutamic acid gasification. J. Clean. Prod. 2021, 282, 124417. [Google Scholar] [CrossRef]
- Wang, P.; Xu, P.; Wang, B.; Shen, C.; Shen, L. Green ammonia production via microalgae steam catalytic gasification process over LaFeO3 perovskite. Fuel 2022, 318, 123322. [Google Scholar] [CrossRef]
- Wang, P.; Shen, C.; Wang, B.; Xu, P.; Shen, L. Ammonia production from nitrogen-rich biomass gasification: Nitrogen transformation from model amino acids. Fuel 2022, 326, 125071. [Google Scholar] [CrossRef]
- Suryanto, B.H.R.; Du, H.-L.; Wang, D.; Chen, J.; Simonov, A.N.; MacFarlane, D.R. Challenges and prospects in the catalysis of electroreduction of nitrogen to ammonia. Nat. Catal. 2019, 2, 290–296. [Google Scholar] [CrossRef]
- Wang, L.; Xia, M.; Wang, H.; Huang, K.; Qian, C.; Maravelias, C.T.; Ozin, G.A. Greening Ammonia toward the Solar Ammonia Refinery. Joule 2018, 2, 1055–1074. [Google Scholar] [CrossRef]
- Zamri, M.; Hasmady, S.; Akhiar, A.; Ideris, F.; Shamsuddin, A.; Mofijur, M.; Fattah, I.M.R.; Mahlia, T. A comprehensive review on anaerobic digestion of organic fraction of municipal solid waste. Renew. Sustain. Energy Rev. 2021, 137, 110637. [Google Scholar] [CrossRef]
- Lin, L.; Yuan, S.; Chen, J.; Xu, Z.; Lu, X. Removal of ammonia nitrogen in wastewater by microwave radiation. J. Hazard. Mater. 2009, 161, 1063–1068. [Google Scholar] [CrossRef]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef]
- Ko, Y.-S.; Kim, J.W.; Lee, J.A.; Han, T.; Kim, G.B.; Park, J.E.; Lee, S.Y. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 2020, 49, 4615–4636. [Google Scholar] [CrossRef]
- Zhang, R.; Li, C.; Wang, J.; Yang, Y.; Yan, Y. Microbial production of small medicinal molecules and biologics: From nature to synthetic pathways. Biotechnol. Adv. 2018, 36, 2219–2231. [Google Scholar] [CrossRef]
- Montaño López, J.; Duran, L.; Avalos, J.L. Physiological limitations and opportunities in microbial metabolic engineering. Nat. Rev. Microbiol. 2022, 20, 35–48. [Google Scholar] [CrossRef]
- Costa, F.; Lago, A.; Rocha, V.; Barros, Ó.; Costa, L.; Vipotnik, Z.; Silva, B.; Tavares, T. A Review on Biological Processes for Pharmaceuticals Wastes Abatement—A Growing Threat to Modern Society. Environ. Sci. Technol. 2019, 53, 7185–7202. [Google Scholar] [CrossRef]
- Lopes, M.S.G. Engineering biological systems toward a sustainable bioeconomy. J. Ind. Microbiol. Biotechnol. 2015, 42, 813–838. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef]
- Rapson, T.D.; Gregg, C.M.; Allen, R.S.; Ju, H.; Doherty, C.M.; Mulet, X.; Giddey, S.; Wood, C.C. Insights into Nitrogenase Bioelectrocatalysis for Green Ammonia Production. Chemsuschem 2020, 13, 4856–4865. [Google Scholar] [CrossRef]
- Gruber, N.; Galloway, J.N. An Earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef]
- Ishizuka, J. Trends in biological nitrogen fixation research and application. In Biological Nitrogen Fixation for Sustainable Agriculture: Extended Versions of Papers Presented in the Symposium, Role of Biological Nitrogen Fixation in Sustainable Agriculture at the 13th Congress of Soil Science, Kyoto, Japan, 1990; Ladha, J.K., George, T., Bohlool, B.B., Eds.; Springer: Dordrecht, The Netherlands, 1992; pp. 197–209. [Google Scholar]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The Evolution and Future of Earth’s Nitrogen Cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Milton, R.D.; Minteer, S.D. Nitrogenase Bioelectrochemistry for Synthesis Applications. Acc. Chem. Res. 2019, 52, 3351–3360. [Google Scholar] [CrossRef] [PubMed]
- Einsle, O.; Rees, D.C. Structural Enzymology of Nitrogenase Enzymes. Chem. Rev. 2020, 120, 4969–5004. [Google Scholar] [CrossRef]
- Cai, R.; Minteer, S.D. Nitrogenase Bioelectrocatalysis: From Understanding Electron-Transfer Mechanisms to Energy Applications. ACS Energy Lett. 2018, 3, 2736–2742. [Google Scholar] [CrossRef]
- Temme, K.; Zhao, D.; Voigt, C.A. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci. USA 2012, 109, 7085–7090. [Google Scholar] [CrossRef]
- Yang, J.; Xie, X.; Wang, X.; Dixon, R.; Wang, Y.P. Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, E3718–E3725. [Google Scholar] [CrossRef]
- López-Torrejón, G.; Burén, S.; Veldhuizen, M.; Rubio, L.M. Biosynthesis of cofactor-activatable iron-only nitrogenase in Saccharomyces cerevisiae. Microb. Biotechnol. 2021, 14, 1073–1083. [Google Scholar] [CrossRef]
- Takimoto, R.; Tatemichi, Y.; Aoki, W.; Kosaka, Y.; Minakuchi, H.; Ueda, M.; Kuroda, K. A critical role of an oxygen-responsive gene for aerobic nitrogenase activity in Azotobacter vinelandii and its application to Escherichia coli. Sci. Rep. 2022, 12, 4182. [Google Scholar] [CrossRef]
- Yang, J.; Xie, X.; Xiang, N.; Tian, Z.-X.; Dixon, R.; Wang, Y.-P. Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology. Proc. Natl. Acad. Sci. USA 2018, 115, E8509–E8517. [Google Scholar] [CrossRef]
- Li, X.-X.; Liu, Q.; Liu, X.-M.; Shi, H.-W.; Chen, S.-F. Using synthetic biology to increase nitrogenase activity. Microb. Cell Fact. 2016, 15, 43. [Google Scholar] [CrossRef]
- Wang, D.; Xu, A.; Elmerich, C.; Ma, L.Z. Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions. ISME J. 2017, 11, 1602–1613. [Google Scholar] [CrossRef]
- Klimasmith, I.M.; Kent, A.D. Micromanaging the nitrogen cycle in agroecosystems. Trends Microbiol. 2022, 30, 1045–1055. [Google Scholar] [CrossRef]
- Fox, A.R.; Soto, G.; Valverde, C.; Russo, D.; Lagares, A.; Zorreguieta, Á.; Alleva, K.; Pascuan, C.; Frare, R.; Mercado-Blanco, J.; et al. Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environ. Microbiol. 2016, 18, 3522–3534. [Google Scholar] [CrossRef]
- Bhatti, M.; Feng, P.C.C.; Pitkin, J. Methods and Compositions for Improving Plant Health. U.S. Patent 8754011, 17 June 2014. [Google Scholar]
- Davis Pires, D. Pivot Bio Proven Inoculant as a Source of Nitrogen in Corn. Kans. Agric. Exp. Stn. Res. Rep. 2020, 6, 7. [Google Scholar] [CrossRef]
- Yenigün, O.; Demirel, B. Ammonia inhibition in anaerobic digestion: A review. Process Biochem. 2013, 48, 901–911. [Google Scholar] [CrossRef]
- Whelan, M.; Everitt, T.; Villa, R. A mass transfer model of ammonia volatilisation from anaerobic digestate. Waste Manag. 2010, 30, 1808–1812. [Google Scholar] [CrossRef]
- Walker, M.; Iyer, K.; Heaven, S.; Banks, C. Ammonia removal in anaerobic digestion by biogas stripping: An evaluation of process alternatives using a first order rate model based on experimental findings. Chem. Eng. J. 2011, 178, 138–145. [Google Scholar] [CrossRef]
- Choi, K.-Y.; Wernick, D.G.; Tat, C.A.; Liao, J.C. Consolidated conversion of protein waste into biofuels and ammonia using Bacillus subtilis. Metab. Eng. 2014, 23, 53–61. [Google Scholar] [CrossRef]
- Huo, Y.-X.; Cho, K.M.; Rivera, J.G.L.; Monte, E.; Shen, C.R.; Yan, Y.; Liao, J. Conversion of proteins into biofuels by engineering nitrogen flux. Nat. Biotechnol. 2011, 29, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Tatemichi, Y.; Kuroda, K.; Nakahara, T.; Ueda, M. Efficient ammonia production from food by-products by engineered Escherichia coli. AMB Express 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Mikami, Y.; Yoneda, H.; Tatsukami, Y.; Aoki, W.; Ueda, M. Ammonia production from amino acid-based biomass-like sources by engineered Escherichia coli. AMB Express 2017, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Kuroda, K.; Tatemichi, Y.; Nakahara, T.; Aoki, W.; Ueda, M. Construction of engineered yeast producing ammonia from glutamine and soybean residues (okara). AMB Express 2020, 10, 70. [Google Scholar] [CrossRef]
- Watanabe, Y.; Aoki, W.; Ueda, M. Improved ammonia production from soybean residues by cell surface-displayed l-amino acid oxidase on yeast. Biosci. Biotechnol. Biochem. 2021, 85, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Burén, S.; Jiang, X.; López-Torrejón, G.; Echavarri-Erasun, C.; Rubio, L.M. Purification and In Vitro Activity of Mitochondria Targeted Nitrogenase Cofactor Maturase NifB. Front. Plant Sci. 2017, 8, 1567. [Google Scholar] [CrossRef] [PubMed]
- Burén, S.; Pratt, K.; Jiang, X.; Guo, Y.; Jimenez-Vicente, E.; Echavarri-Erasun, C.; Dean, D.; Saaem, I.; Gordon, B.; Voigtet, C.; et al. Biosynthesis of the nitrogenase active-site cofactor precursor NifB-co in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2019, 116, 25078–25086. [Google Scholar] [CrossRef]
- Okada, S.; Gregg, C.M.; Allen, R.S.; Menon, A.; Hussain, D.; Gillespie, V.; Johnston, E.; Byrne, K.; Colgrave, M.L.; Wood, C.C. A Synthetic Biology Workflow Reveals Variation in Processing and Solubility of Nitrogenase Proteins Targeted to Plant Mitochondria, and Differing Tolerance of Targeting Sequences in a Bacterial Nitrogenase Assay. Front. Plant Sci. 2020, 11, 552160. [Google Scholar] [CrossRef]
- Smanski, M.J.; Bhatia, S.; Zhao, D.; Park, Y.; Woodruff, L.B.A.; Giannoukos, G.; Ciulla, D.; Busby, M.; Calderon, J.; Nicol, R.; et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014, 32, 1241–1249. [Google Scholar] [CrossRef]
- Xiang, N.; Guo, C.; Liu, J.; Xu, H.; Dixon, R.; Yang, J.; Wang, Y.-P. Using synthetic biology to overcome barriers to stable expression of nitrogenase in eukaryotic organelles. Proc. Natl. Acad. Sci. USA 2020, 117, 16537–16545. [Google Scholar] [CrossRef]
- Allen, R.S.; Gregg, C.M.; Okada, S.; Menon, A.; Hussain, D.; Gillespie, V.; Johnston, E.; Devilla, R.; Warden, A.C.; Taylor, M.; et al. Plant expression of NifD protein variants resistant to mitochondrial degradation. Proc. Natl. Acad. Sci. USA 2020, 117, 23165–23173. [Google Scholar] [CrossRef]
- Chrétien, D.; Bénit, P.; Ha, H.-H.; Keipert, S.; El-Khoury, R.; Chang, Y.-T.; Jastroch, M.; Jacobs, H.T.; Rustin, P.; Rak, M. Mitochondria are physiologically maintained at close to 50 °C. PLoS Biol. 2018, 16, e2003992. [Google Scholar] [CrossRef]
- Jiang, X.; Payá-Tormo, L.; Coroian, D.; García-Rubio, I.; Castellanos-Rueda, R.; Eseverri, Á.; López-Torrejón, G.; Burén, S.; Rubio, L.M. Exploiting genetic diversity and gene synthesis to identify superior nitrogenase NifH protein variants to engineer N2-fixation in plants. Commun. Biol. 2021, 4, 4. [Google Scholar] [CrossRef]
- Li, Q.; Chen, S. Transfer of nitrogen fixation (nif) genes to non-diazotrophic hosts. ChemBioChem 2020, 21, 1717–1722. [Google Scholar] [CrossRef]
- Dong, F.; Lee, Y.S.; Gaffney, E.M.; Grattieri, M.; Haddadin, H.; Minteer, S.D.; Chen, H. An engineered, non-diazotrophic cyanobacterium and its application in bioelectrochemical nitrogen fixation. Cell Rep. Phys. Sci. 2021, 2, 100444. [Google Scholar] [CrossRef]
- Santos, J.; Sousa, M.J.; Leão, C. Ammonium Is Toxic for Aging Yeast Cells, Inducing Death and Shortening of the Chronological Lifespan. PLoS ONE 2012, 7, e37090. [Google Scholar] [CrossRef]
- Ueda, M.; Tanaka, A. Cell surface engineering of yeast: Construction of arming yeast with biocatalyst. J. Biosci. Bioeng. 2000, 90, 125–136. [Google Scholar] [CrossRef]
- Takagi, T.; Yokoi, T.; Shibata, T.; Morisaka, H.; Kuroda, K.; Ueda, M. Engineered yeast whole-cell biocatalyst for direct degradation of alginate from macroalgae and production of non-commercialized useful monosaccharide from alginate. Appl. Microbiol. Biotechnol. 2016, 100, 1723–1732. [Google Scholar] [CrossRef]
- Motone, K.; Takagi, T.; Sasaki, Y.; Kuroda, K.; Ueda, M. Direct ethanol fermentation of the algal storage polysaccharide laminarin with an optimized combination of engineered yeasts. J. Biotechnol. 2016, 231, 129–135. [Google Scholar] [CrossRef]
- Brown, G.; Singer, A.; Proudfoot, M.; Skarina, T.; Kim, Y.; Chang, C.; Dementieva, I.; Kuznetsova, E.; Gonzalez, C.F.; Joachimiak, A.; et al. Functional and Structural Characterization of Four Glutaminases from Escherichia coli and Bacillus subtilis. Biochemistry 2008, 47, 5724–5735. [Google Scholar] [CrossRef]
- Pollegioni, L.; Motta, P.; Molla, G. L-amino acid oxidase as biocatalyst: A dream too far? Appl. Microbiol. Biotechnol. 2013, 97, 9323–9341. [Google Scholar] [CrossRef] [PubMed]
- Bloess, S.; Beuel, T.; Krüger, T.; Sewald, N.; Dierks, T.; von Mollard, G.F. Expression, characterization, and site-specific covalent immobilization of an L-amino acid oxidase from the fungus Hebeloma cylindrosporum. Appl. Microbiol. Biotechnol. 2019, 103, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Kozuka, K.; Minamino, Y.; Karasuda, H.; Hasebe, F.; Ito, S. Ancestral L-amino acid oxidases for deracemization and stereoinversion of amino acids. Commun. Chem. 2020, 3, 181. [Google Scholar] [CrossRef]
- Kastner, V.; Somitsch, W.; Schnitzhofer, W. The anaerobic fermentation of food waste: A comparison of two bioreactor systems. J. Clean. Prod. 2012, 34, 82–90. [Google Scholar] [CrossRef]
- Degueurce, A.; Picard, S.; Peu, P.; Trémier, A. Storage of Food Waste: Variations of Physical–Chemical Characteristics and Consequences on Biomethane Potential. Waste Biomass-Valoriz. 2020, 11, 2441–2454. [Google Scholar] [CrossRef]
- Daly, S.E.; Usack, J.G.; Harroff, L.A.; Booth, J.G.; Keleman, M.P.; Angenent, L.T. A systematic analysis of factors that affect food-waste storage: Toward maximizing lactate accumulation for resource recovery. ACS Sustain. Chem. Eng. 2020, 8, 13934–13944. [Google Scholar] [CrossRef]
- Dawson, J.C.; Huggins, D.R.; Jones, S.S. Characterizing nitrogen use efficiency in natural and agricultural ecosystems to improve the performance of cereal crops in low-input and organic agricultural systems. Field Crop. Res. 2008, 107, 89–101. [Google Scholar] [CrossRef]
- Congreves, K.A.; Otchere, O.; Ferland, D.; Farzadfar, S.; Williams, S.; Arcand, M.M. Nitrogen Use Efficiency Definitions of Today and Tomorrow. Front. Plant Sci. 2021, 12, 637108. [Google Scholar] [CrossRef]
- Galloway, J.N.; Winiwarter, W.; Leip, A.; Leach, A.; Bleeker, A.; Erisman, J.W. Nitrogen footprints: Past, present and future. Environ. Res. Lett. 2014, 9, 115003. [Google Scholar] [CrossRef]
- Silliman, B.R.; Bertness, M.D. Shoreline Development Drives Invasion of Phragmites australis and the Loss of Plant Diversity on New England Salt Marshes. Conserv. Biol. 2004, 18, 1424–1434. [Google Scholar] [CrossRef]
- Hutchins, D.A.; Capone, D.G. The marine nitrogen cycle: New developments and global change. Nat. Rev. Genet. 2022, 20, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Montzka, S.A.; Dlugokencky, E.J.; Butler, J.H. Non-CO2 greenhouse gases and climate change. Nature 2011, 476, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Oslage, H. Composition and nutritive value of single-cell protein (SCP). Anim. Feed. Sci. Technol. 1976, 1, 9–24. [Google Scholar] [CrossRef]
- Yamakawa, S.-I.; Yamada, R.; Tanaka, T.; Ogino, C.; Kondo, A. Repeated fermentation from raw starch using Saccharomyces cerevisiae displaying both glucoamylase and α-amylase. Enzym. Microb. Technol. 2012, 50, 343–347. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Breister, A.M.; Liu, Y.; Kieft, K.; Cowley, E.S.; Karaoz, U.; Anantharaman, K. METABOLIC: High-throughput profiling of microbial genomes for functional traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome 2022, 10, 33. [Google Scholar] [CrossRef]
- Kempes, C.P.; Dutkiewicz, S.; Follows, M.J. Growth, metabolic partitioning, and the size of microorganisms. Proc. Natl. Acad. Sci. USA 2012, 109, 495–500. [Google Scholar] [CrossRef]
- Calabrese, S.; Chakrawal, A.; Manzoni, S.; Van Cappellen, P. Energetic scaling in microbial growth. Proc. Natl. Acad. Sci. USA 2021, 118, e2107668118. [Google Scholar] [CrossRef]


| Metabolic Engineering | Cell Surface Engineering | |
|---|---|---|
| Reaction Place |
|
|
| pH of reaction environment |
|
|
| Effect on the enzyme |
|
|
| Chemical Method | Bacterial Method | Yeast Method | |
|---|---|---|---|
| Resource |
|
|
|
| Energy resource |
|
|
|
| ReactionEnvironment |
|
|
|
| Point of improvement for sustainable production |
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, Y.; Aoki, W.; Ueda, M. Ammonia Production Using Bacteria and Yeast toward a Sustainable Society. Bioengineering 2023, 10, 82. https://doi.org/10.3390/bioengineering10010082
Watanabe Y, Aoki W, Ueda M. Ammonia Production Using Bacteria and Yeast toward a Sustainable Society. Bioengineering. 2023; 10(1):82. https://doi.org/10.3390/bioengineering10010082
Chicago/Turabian StyleWatanabe, Yukio, Wataru Aoki, and Mitsuyoshi Ueda. 2023. "Ammonia Production Using Bacteria and Yeast toward a Sustainable Society" Bioengineering 10, no. 1: 82. https://doi.org/10.3390/bioengineering10010082
APA StyleWatanabe, Y., Aoki, W., & Ueda, M. (2023). Ammonia Production Using Bacteria and Yeast toward a Sustainable Society. Bioengineering, 10(1), 82. https://doi.org/10.3390/bioengineering10010082






