Pancreatic Mass Characterization Using IVIM-DKI MRI and Machine Learning-Based Multi-Parametric Texture Analysis
Abstract
1. Introduction
- We established a novel IVIM-DKI model with a total variation penalty function to achieve improved non-invasive characterization of pancreatic masses.
- Qualitative and mean comparison between IVIM-DKI parametric maps in pancreatic masses such as PDAC, PNET, MFCP, and SPEN were evaluated.
- Cut-off values for each IVIM-DKI parameter were calculated for characterization of pancreatic masses using ROC analysis.
- We attempted to comprehensively investigate texture features of apparent diffusion coefficient (ADC), diffusion coefficient (D), pseudo-diffusion coefficient (D*), perfusion fraction (f), kurtosis (k), and combined texture features of IVIM-DKI parameters with and without ADC.
- Machine learning-based classification of pancreatic masses using ANN was used and compared with other techniques such as decision tree and ensemble.
2. Materials and Methods
2.1. Study Population
2.2. MRI Acquisition
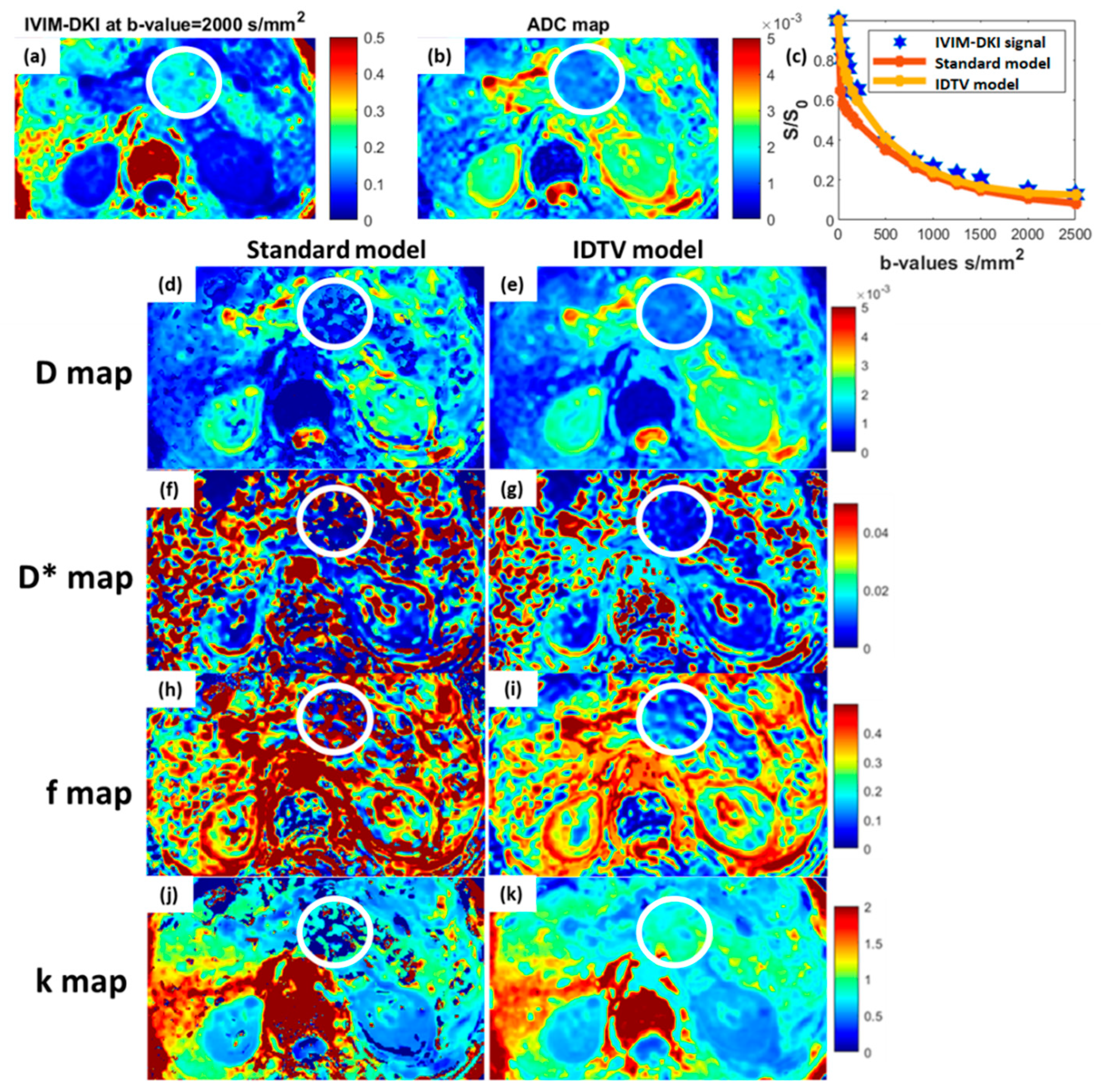
2.3. MRI Image Analysis
2.4. Localization of Region of Interest
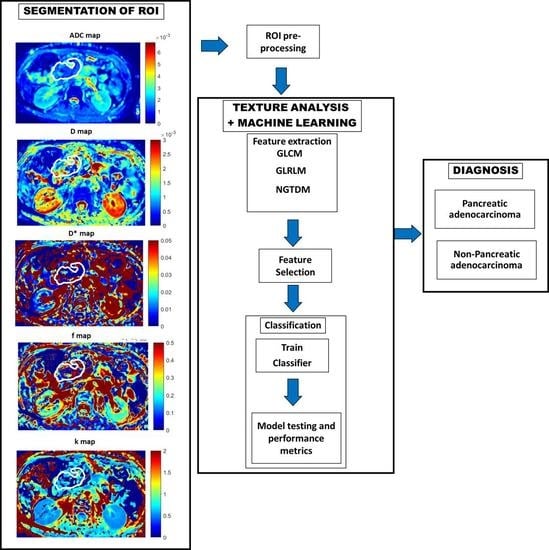
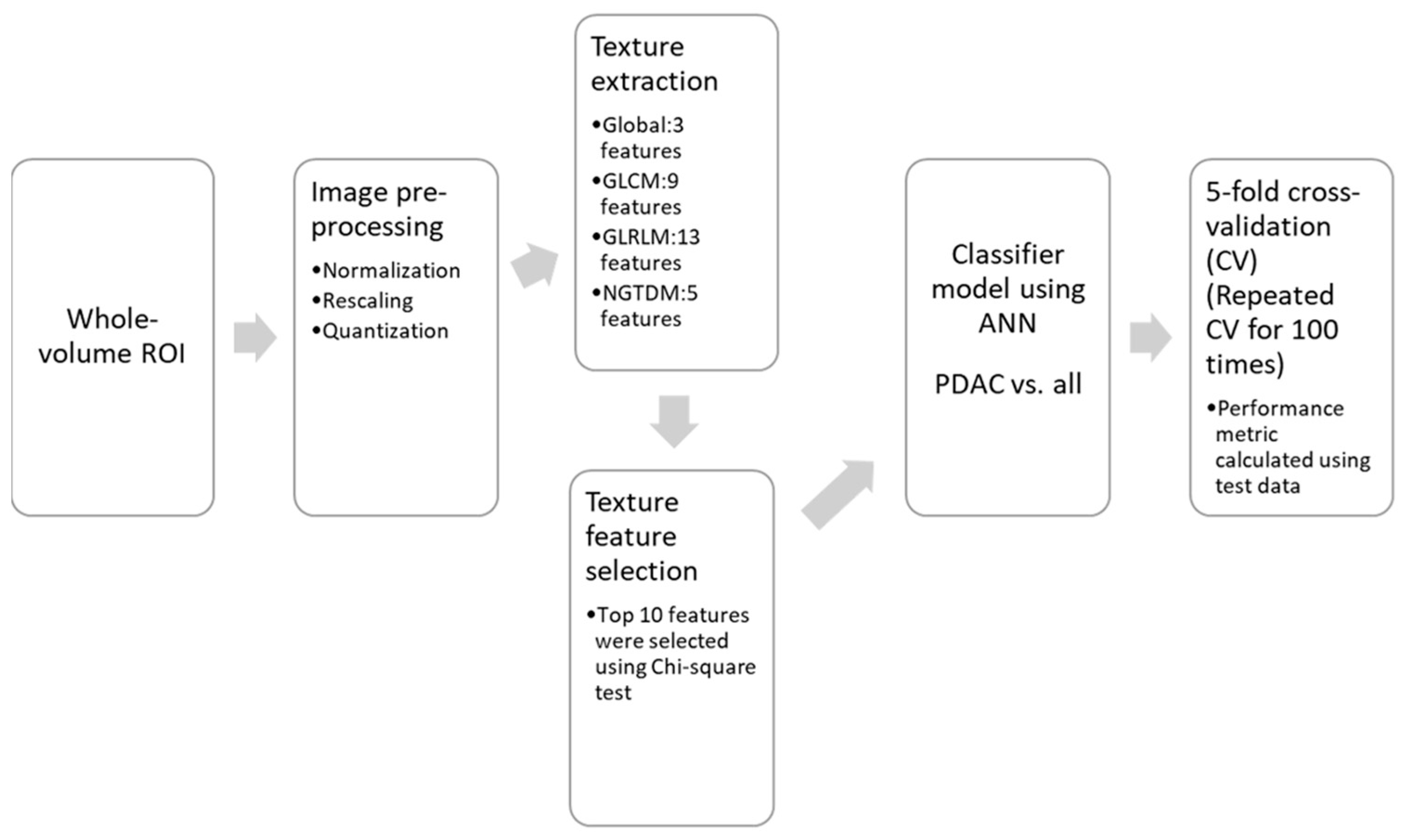
2.5. Texture Feature Calculation and Machine Learning-Based Classification
2.6. Statistical Analysis
3. Results
3.1. Patient Population and Tumor Volume
3.2. Model Performance in Pancreatic Masses
3.3. Quantitative Comparison between Subtypes of Pancreatic Masses
3.4. Differential Diagnosis of Pancreatic Masses Using ROC Analysis
3.5. Multi-Parametric Texture Analysis and Machine Learning-Based Classification of Pancreatic Masses
4. Discussion
Limitation and Future Scope
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat. Oncol. 2019, 14, 141. [Google Scholar] [CrossRef]
- Ehehalt, F.; Saeger, H.D.; Schmidt, C.M.; Grützmann, R. Neuroendocrine Tumors of the Pancreas. Oncology 2009, 14, 456–467. [Google Scholar] [CrossRef]
- Klein, A.P. Pancreatic cancer epidemiology: Understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a Biomarker for Pancreatic Cancer—A Comprehensive Review. Indian J. Surg. Oncol. 2011, 2, 88–100. [Google Scholar] [CrossRef]
- Chang, D.; Nguyen, N.Q.; Merrett, N.; Dixson, H.; Leong, R.; Biankin, A. Role of endoscopic ultrasound in pancreatic cancer. Expert Rev. Gastroenterol. Hepatol. 2009, 3, 293–303. [Google Scholar] [CrossRef]
- Garcea, G.; Ong, S.; Rajesh, A.; Neal, C.; Pollard, C.; Berry, D.; Dennison, A. Cystic Lesions of the Pancreas. Pancreatology 2008, 8, 236–251. [Google Scholar] [CrossRef]
- Kang, J.D.; Clarke, S.E.; Costa, A.F. Factors associated with missed and misinterpreted cases of pancreatic ductal adenocarcinoma. Eur. Radiol. 2020, 31, 2422–2432. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.E.; Yaghmai, V.; Nikolaidis, P.; McCarthy, R.J.; Merrick, L.; Miller, F.H. Diffusion-weighted MR imaging in pancreatic endocrine tumors correlated with histopathologic characteristics. J. Magn. Reson. Imaging 2011, 33, 1071–1079. [Google Scholar] [CrossRef]
- Le Bihan, D.; Breton, E.; Lallemand, D.; Aubin, M.L.; Vignaud, J.; Laval-Jeantet, M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 1988, 168, 497–505. [Google Scholar] [CrossRef]
- Ye, C.; Xu, D.; Qin, Y.; Wang, L.; Wang, R.; Li, W.; Kuai, Z.; Zhu, Y. Estimation of intravoxel incoherent motion parameters using low b-values. PLoS ONE 2019, 14, e0211911. [Google Scholar] [CrossRef]
- Jensen, J.H.; Helpern, J.A.; Ramani, A.; Lu, H.; Kaczynski, K. Diffusional kurtosis imaging: The quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn. Reson. Med. 2005, 53, 1432–1440. [Google Scholar] [CrossRef]
- Lu, Y.; Jansen, J.F.; Mazaheri, Y.; Stambuk, H.E.; Koutcher, J.A.; Shukla-Dave, A. Extension of the intravoxel incoherent motion model to non-gaussian diffusion in head and neck cancer. J. Magn. Reson. Imaging 2012, 36, 1088–1096. [Google Scholar] [CrossRef]
- Wu, W.-C.; Yang, S.-C.; Chen, Y.-F.; Tseng, H.-M.; My, P.-C. Simultaneous assessment of cerebral blood volume and diffusion heterogeneity using hybrid IVIM and DK MR imaging: Initial experience with brain tumors. Eur. Radiol. 2017, 27, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Núñez, D.A.; Lu, Y.; Paudyal, R.; Hatzoglou, V.; Moreira, A.L.; Oh, J.H.; Stambuk, H.E.; Mazaheri, Y.; Gonen, M.; Ghossein, R.A.; et al. Quantitative Non-Gaussian Intravoxel Incoherent Motion Diffusion-Weighted Imaging Metrics and Surgical Pathology for Stratifying Tumor Aggressiveness in Papillary Thyroid Carcinomas. Tomography 2019, 5, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Malagi, A.V.; Netaji, A.; Kumar, V.; Kayal, E.B.; Khare, K.; Das, C.J.; Calamante, F.; Mehndiratta, A. IVIM-DKI for differentiation between prostate cancer and benign prostatic hyperplasia: Comparison of 1.5 T vs. 3 T MRI. Magn. Reson. Mater. Phys. Biol. Med. 2021, 35, 609–620. [Google Scholar] [CrossRef]
- Rudin, L.I.; Osher, S.; Fatemi, E. Nonlinear total variation based noise removal algorithms. Phys. D Nonlinear Phenom. 1992, 60, 259–268. [Google Scholar] [CrossRef]
- Kayal, E.B.; Kandasamy, D.; Khare, K.; Alampally, J.T.; Bakhshi, S.; Sharma, R.; Mehndiratta, A. Quantitative Analysis of Intravoxel Incoherent Motion (IVIM) Diffusion MRI using Total Variation and Huber Penalty Function. Med. Phys. 2017, 44, 5849–5858. [Google Scholar] [CrossRef]
- Khalvati, F.; Zhang, Y.; Baig, S.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A. Prognostic Value of CT Radiomic Features in Resectable Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2019, 9, 5449. [Google Scholar] [CrossRef]
- Larroza, A.; Bodí, V.; Moratal, D. Texture Analysis in Magnetic Resonance Imaging: Review and Considerations for Future Applications; IntechOpen: London, UK, 2016. [Google Scholar]
- Liang, L.; Ding, Y.; Yu, Y.; Liu, K.; Rao, S.; Ge, Y.; Zeng, M. Whole-tumour evaluation with MRI and radiomics features to predict the efficacy of S-1 for adjuvant chemotherapy in postoperative pancreatic cancer patients: A pilot study. BMC Med. Imaging 2021, 21, 75. [Google Scholar] [CrossRef]
- Deng, Y.; Ming, B.; Zhou, T.; Wu, J.-L.; Chen, Y.; Liu, P.; Zhang, J.; Zhang, S.-Y.; Chen, T.-W.; Zhang, X.-M. Radiomics Model Based on MR Images to Discriminate Pancreatic Ductal Adenocarcinoma and Mass-Forming Chronic Pancreatitis Lesions. Front. Oncol. 2021, 11, 620981. [Google Scholar] [CrossRef]
- Shi, Y.-J.; Zhu, H.-T.; Liu, Y.-L.; Wei, Y.-Y.; Qin, X.-B.; Zhang, X.-Y.; Li, X.-T.; Sun, Y.-S. Radiomics Analysis Based on Diffusion Kurtosis Imaging and T2 Weighted Imaging for Differentiation of Pancreatic Neuroendocrine Tumors from Solid Pseudopapillary Tumors. Front. Oncol. 2020, 10, 1624. [Google Scholar] [CrossRef]
- Yingwei, W.; Xinghua, Z.; Botao, W.; Ye, W.; MengQi, L.; Haiyi, W.; Huiyi, Y.; Zhiye, C.; Wang, Y.-W.; Zhang, X.-H.; et al. Value of Texture Analysis of Intravoxel Incoherent Motion Parameters in Differential Diagnosis of Pancreatic Neuroendocrine Tumor and Pancreatic Adenocarcinoma. Chin. Med. Sci. J. 2019, 34, 1–9. [Google Scholar] [CrossRef]
- Abunahel, B.M.; Pontre, B.; Kumar, H.; Petrov, M.S. Pancreas image mining: A systematic review of radiomics. Eur. Radiol. 2021, 31, 3447–3467. [Google Scholar] [CrossRef]
- Rorden, C.; Brett, M. Stereotaxic Display of Brain Lesions. Behav. Neurol. 2000, 12, 191–200. [Google Scholar] [CrossRef]
- Miller, F.H.; Wang, Y.; McCarthy, R.J.; Yaghmai, V.; Merrick, L.; Larson, A.; Berggruen, S.; Casalino, D.D.; Nikolaidis, P. Utility of Diffusion-Weighted MRI in Characterization of Adrenal Lesions. Am. J. Roentgenol. 2010, 194, W179–W185. [Google Scholar] [CrossRef]
- Rosenkrantz, A.B.; Oei, M.; Babb, J.S.; Ba, B.E.N.; Taouli, B. Diffusion-weighted imaging of the abdomen at 3.0 Tesla: Image quality and apparent diffusion coefficient reproducibility compared with 1.5 Tesla. J. Magn. Reson. Imaging 2011, 33, 128–135. [Google Scholar] [CrossRef]
- Lemke, A.; Laun, F.B.; Klau, M.; Re, T.J.; Simon, D.; Delorme, S.; Schad, L.R.; Stieltjes, B. Differentiation of Pancreas Carcinoma from Healthy Pancreatic Tissue Using Multiple b-Values. Investig. Radiol. 2009, 44, 769–775. [Google Scholar] [CrossRef]
- Kang, K.M.; Lee, J.M.; Yoon, J.H.; Kiefer, B.; Han, J.K.; Choi, B.I. Intravoxel Incoherent Motion Diffusion-weighted MR Imaging for Characterization of Focal Pancreatic Lesions. Radiology 2014, 270, 444–453. [Google Scholar] [CrossRef]
- Malagi, A.V.; Das, C.J.; Khare, K.; Calamante, F.; Mehndiratta, A. Effect of combination and number of b values in IVIM analysis with post-processing methodology: Simulation and clinical study. Magn. Reson. Mater. Phys. Biol. Med. 2019, 32, 519–527. [Google Scholar] [CrossRef]
- Vallières, M.; Kay-Rivest, E.; Perrin, L.J.; Liem, X.; Furstoss, C.; Aerts, H.J.W.L.; Khaouam, N.; Nguyen-Tan, P.F.; Wang, C.-S.; Sultanem, K.; et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Pereira, S.P.; Oldfield, L.; Ney, A.; Hart, P.A.; Keane, M.G.; Pandol, S.J.; Li, D.; Greenhalf, W.; Jeon, C.Y.; Koay, E.J.; et al. Early detection of pancreatic cancer. Lancet Gastroenterol. Hepatol. 2020, 5, 698–710. [Google Scholar] [CrossRef] [PubMed]
- Malagi, A.V.; Kandasamy, D.; Khare, K.; Pushpam, D.; Kumar, R.; Bakhshi, S.; Mehndiratta, A. Qualitative and quantitative comparison between IVIM-DKI and PET/CT imaging in lymphoma. In Proceedings of the 29nd Annual Meeting of ISMRM 2021, Virtual, 15–20 May 2021; p. 3665. [Google Scholar]
- Baidya Kayal, E.; Khare, K.; Sharma, R.; Bakhshi, S.; Kandasamy, D.; Mehndiratta, A. Evaluating Reproducibility and Repeatability of Penalty Function Based Methods for Quantitative Intravoxel Incoherent Motion Analysis. In Proceedings of the 31st Joint Annual Meeting of ISMRM-ESMRMB 2022, London, UK, 7–12 May 2022; p. 0115. [Google Scholar]
- Kim, B.; Lee, S.S.; Sung, Y.S.; Cheong, H.; Byun, J.H.; Kim, H.J.; Kim, J.H. Intravoxel incoherent motion diffusion-weighted imaging of the pancreas: Characterization of benign and malignant pancreatic pathologies. J. Magn. Reson. Imaging 2017, 45, 260–269. [Google Scholar] [CrossRef]
- De Robertis, R.; Cardobi, N.; Ortolani, S.; Martini, P.T.; Stemmer, A.; Grimm, R.; Gobbo, S.; Butturini, G.; D’Onofrio, M. Intravoxel incoherent motion diffusion-weighted MR imaging of solid pancreatic masses: Reliability and usefulness for characterization. Abdom. Imaging 2019, 44, 131–139. [Google Scholar] [CrossRef]
- Mayer, P.; Jiang, Y.; Kuder, T.A.; Bergmann, F.; Khristenko, E.; Steinle, V.; Kaiser, J.; Hackert, T.; Kauczor, H.-U.; Klauß, M.; et al. Diffusion Kurtosis Imaging—A Superior Approach to Assess Tumor–Stroma Ratio in Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 1656. [Google Scholar] [CrossRef]
- Li, J.; Liang, L.; Yu, H.; Shen, Y.; Hu, Y.; Hu, D.; Tang, H.; Li, Z. Whole-tumor histogram analysis of non-Gaussian distribution DWI parameters to differentiation of pancreatic neuroendocrine tumors from pancreatic ductal adenocarcinomas. Magn. Reson. Imaging 2018, 55, 52–59. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Sansone, M.; Grassi, R.; Maio, F.; Palaia, R.; Tatangelo, F.; Botti, G.; Grimm, R.; Curley, S.; et al. Magnetic resonance imaging in the assessment of pancreatic cancer with quantitative parameter extraction by means of dynamic contrast-enhanced magnetic resonance imaging, diffusion kurtosis imaging and intravoxel incoherent motion diffusion-weighted imaging. Ther. Adv. Gastroenterol. 2020, 13, 1756284819885052. [Google Scholar] [CrossRef]
- Klau, M.; Lemke, A.; Grünberg, K.; Simon, D.; Re, T.J.; Wente, M.N.; Laun, F.B.; Kauczor, H.-U.; Delorme, S.; Grenacher, L.; et al. Intravoxel Incoherent Motion MRI for the Differentiation Between Mass Forming Chronic Pancreatitis and Pancreatic Carcinoma. Investig. Radiol. 2011, 46, 57–63. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, S.S.; Kim, N.; Kim, E.; Kim, Y.J.; Yun, S.-C.; Kuhn, B.; Kim, I.S.; Park, S.H.; Kim, S.Y.; et al. Intravoxel Incoherent Motion Diffusion-weighted MR Imaging of the Liver: Effect of Triggering Methods on Regional Variability and Measurement Repeatability of Quantitative Parameters. Radiology 2014, 274, 405–415. [Google Scholar] [CrossRef]
- Suda, K.; Takase, M.; Fukumura, Y.; Kashiwagi, S. Pathology of autoimmune pancreatitis and tumor-forming pancreatitis. J. Gastroenterol. 2007, 42, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.K.; Sharma, R.; Kandasamy, D.; Pradhan, R.K.; Garg, P.K.; Bhalla, A.S.; Gamanagatti, S.; Srivastava, D.N.; Sahni, P.; Upadhyay, A.D. Perfusion CT—Can it resolve the pancreatic carcinoma versus mass forming chronic pancreatitis conundrum? Pancreatology 2016, 16, 979–987. [Google Scholar] [CrossRef]
- Ren, S.; Zhang, J.; Chen, J.; Cui, W.; Zhao, R.; Qiu, W.; Duan, S.; Chen, R.; Chen, X.; Wang, Z. Evaluation of Texture Analysis for the Differential Diagnosis of Mass-Forming Pancreatitis from Pancreatic Ductal Adenocarcinoma on Contrast-Enhanced CT Images. Front. Oncol. 2019, 9, 1171. [Google Scholar] [CrossRef] [PubMed]
- Federau, C.; O’Brien, K.; Meuli, R.; Hagmann, P.; Maeder, P. Measuring brain perfusion with intravoxel incoherent motion (IVIM): Initial clinical experience. J. Magn. Reson. Imaging 2014, 39, 624–632. [Google Scholar] [CrossRef]
- Mayer, P.; Fritz, F.; Koell, M.; Skornitzke, S.; Bergmann, F.; Gaida, M.M.; Hackert, T.; Maier-Hein, K.; Laun, F.B.; Kauczor, H.-U.; et al. Assessment of tissue perfusion of pancreatic cancer as potential imaging biomarker by means of Intravoxel incoherent motion MRI and CT perfusion: Correlation with histological microvessel density as ground truth. Cancer Imaging 2021, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, X. Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: A preliminary study. Diagn. Interv. Imaging 2020, 101, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, N.; Condino, S.; Carbone, M.; Cattari, N.; D’Amato, R.; Cutolo, F.; Ferrari, V. Brain Tumor and Augmented Reality: New Technologies for the Future. Int. J. Environ. Res. Public Health 2022, 19, 6347. [Google Scholar] [CrossRef]
- Lin, J.; Tao, H.; Wang, Z.; Chen, R.; Chen, Y.; Lin, W.; Li, B.; Fang, C.; Yang, J. Augmented reality navigation facilitates laparoscopic removal of foreign body in the pancreas that cause chronic complications. Surg. Endosc. 2022, 36, 6326–6330. [Google Scholar] [CrossRef]



| MRI Technique | Advantages | Disadvantages |
|---|---|---|
| DWI/ADC | DWI allows for the qualitative and quantitative evaluation of tissue diffusivity without the need for contrast agents. ADC is quantified using a simplistic model and provides quantifiable measures of tumor cellularity. | ADC can be affected by perfusion signals from flowing blood effects that can cause overestimations. ADC becomes more sensitive to tissue microscopic characteristics with higher b-values due to non-Gaussian water diffusion. |
| IVIM/D/D*/f | Diffusion and perfusion components of tissue can be evaluated independently. | D* and f can be overestimated due to non-Gaussian water diffusion at high b-values and long scan times. |
| DKI/D/k | Accurate representation of water interactions inside tumor. | DKI model cannot eliminate the perfusion effect at low b-values. |
| IVIM-DKI/D/D*/f/k | IVIM-DKI incorporates diffusion kurtosis analysis to assess intravoxel incoherent motion in tumor tissues with restricted diffusion. | Low SNR with noisy parametric maps and long scan times. |
| Pancreatic Masses | No. of Patients | Age (Mean ± SD) | Gender Ratio (F:M) |
|---|---|---|---|
| PDAC | 25 | 57.9 ± 11 | 2:23 |
| pNET | 13 | 41.7 ± 13.9 | 4:9 |
| MFCP | 6 | 46.4 ± 15.1 | 0:6 |
| SPEN | 4 | 29.5 ± 5.1 | 3:1 |
| Parameters | PDAC | pNET | MFCP | SPEN | p-Value |
|---|---|---|---|---|---|
| ADC † | 1.7 ± 0.5 | 1.5 ± 0.4 | 1.7 ± 0.3 | 1.4 ± 0.3 | 0.3 |
| D † | 1.5 ± 0.4 | 1.2 ± 0.4 | 1.5 ± 0.3 | 1.3 ± 0.3 | 0.1 |
| D* † | 41.6 ± 16.8 | 69.5 ± 36.4 | 50.6 ± 13.7 | 46.4 ± 27.7 | 0.07 |
| f | 0.17 ± 0.06 | 0.23 ± 0.05 | 0.21 ± 0.05 | 0.19 ± 0.05 | 0.02 |
| k | 0.7 ± 0.2 | 1 ± 0.4 | 0.7 ± 0.2 | 0.9 ± 0.3 | 0.1 |
| Parameters | Threshold | Accuracy % | Sensitivity % | Specificity % | F1_Score % | AUC (CI) |
|---|---|---|---|---|---|---|
| PDAC vs. MFCP | ||||||
| f | 0.20496 | 77 | 83 | 76 | 59 | 0.77 (0.59–0.96) |
| PDAC vs. pNET | ||||||
| D* † | 0.06373 | 88 | 63 | 96 | 71 | 0.73 (0.54–0.91) |
| f | 0.20424 | 76 | 75 | 76 | 60 | 0.73 (0.54–0.91) |
| pNET vs. MFCP | ||||||
| ADC † | 0.00158 | 79 | 83 | 75 | 77 | 0.79 (0.55–1) |
| D † | 0.00137 | 79 | 83 | 75 | 77 | 0.76 (0.51–1) |
| PDAC vs. Non-PDAC | Features Selected | Accuracy % | Precision % | Specificity % | F1 Score % | AUC | Classification Error % | |
|---|---|---|---|---|---|---|---|---|
| ADC | All features (30 features) | All features | 82 ± 2.3 | 81.4 ± 2.0 | 91.4 ± 1.6 | 78 ± 2.5 | 0.81 ± 0.09 | 18.1 ± 1.9 |
| Chi-square (Top 10 features) | f27, f30, f14, f6, f5, f4, f18, f7, f28, f9 | 75 ± 2.9 | 74.1 ± 2.1 | 86.9 ± 2.3 | 69.2 ± 2.9 | 0.73 ± 0.08 | 25.1 ± 2.0 | |
| IVIM-DKI | All features (30 × 4 features) | All features | 90.5 ± 1.7 | 90.4 ± 1.7 | 93.6 ± 0.9 | 89.5 ± 2.1 | 0.92 ± 0.06 | 9.5 ± 1.7 |
| Chi-square (Top 10 features) | f_parameter_f21, D*_parameter_f29, f_parameter_f13, f_parameter_f16, D*_parameter_f5, f_parameter_f9, f_parameter_f29, k_parameter_f12, k_parameter_f9, D_parameter_f9 | 84.3 ± 1.3 | 84.2 ± 1.2 | 89 ± 1.1 | 82.6 ± 1.5 | 0.84 ± 0.06 | 15.7 ± 1.2 | |
| ADC with IVIM-DKI | All features (30 × 5 features) | All features | 90.7 ± 1.0 | 90.6 ± 1.0 | 93.8 ± 1.2 | 89.7 ± 1.2 | 0.92 ± 0.07 | 9.3 ± 1 |
| Chi-square (Top 10 features) | f_parameter_f21, D*_parameter_f29, f_parameter_f13, f_parameter_f16, D*_parameter_f5, f_parameter_f9, f_parameter_f29, k_parameter_f12, k_parameter_f9, D_parameter_f9 | 84.3 ± 1.3 | 84.2 ± 1.2 | 89.0 ± 1.1 | 82.6 ± 1.5 | 0.84 ± 0.06 | 15.7 ± 1.2 | |
| D | All features (30 features) | All features | 79.5 ± 2.1 | 78.8 ± 1.7 | 89.7 ± 1.6 | 74.8 ± 2.1 | 0.78 ± 0.09 | 20.6 ± 1.7 |
| Chi-square (Top 10 features) | f9, f1, f27, f25, f10, f17, f19, f13, f16, f24 | 73.6 ± 1.9 | 72.6 ± 1.6 | 87.4 ± 1 | 66.7 ± 2.2 | 0.71 ± 0.09 | 26.6 ± 1.5 | |
| D* | All features (30 features) | All features | 87.8 ± 1.0 | 87.5 ± 0.9 | 92.6 ± 1.0 | 85.9 ± 0.9 | 0.89 ± 0.07 | 12.3 ± 0.9 |
| Chi-square (Top 10 features) | f29, f5, f22, f27, f26, f25, f30, f11, f2, f1 | 81.8 ± 1.8 | 81.3 ± 1.2 | 90 ± 1.6 | 78.3 ± 1.4 | 0.80 ± 0.08 | 18.2 ± 1.1 | |
| f | All features (30 features) | All features | 90.3 ± 1.8 | 90.2 ± 1.8 | 93.2 ± 1.4 | 89.4 ± 2.0 | 0.90 ± 0.05 | 9.7 ± 1.7 |
| Chi-square (Top 10 features) | f21, f13, f16, f9, f29, f12, f2, f1, f23, f7 | 84.9 ± 1.4 | 85.1 ± 1.5 | 88.7 ± 1.1 | 83.8 ± 1.5 | 0.85 ± 0.06 | 15.0 ± 1.5 | |
| k | All features (30 features) | All features | 85.9 ± 2.0 | 85.6 ± 1.6 | 91.9 ± 2.1 | 83.5 ± 1.6 | 0.85 ± 0.07 | 14.2 ± 1.6 |
| Chi-square (Top 10 features) | f12, f9, f18, f20, f1, f4, f22, f28, f2, f19 | 81.2 ± 0.8 | 80.8 ± 1.0 | 89.1 ± 1.2 | 77.8 ± 1.7 | 0.80 ± 0.08 | 18.9 ± 0.9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malagi, A.V.; Shivaji, S.; Kandasamy, D.; Sharma, R.; Garg, P.; Gupta, S.D.; Gamanagatti, S.; Mehndiratta, A. Pancreatic Mass Characterization Using IVIM-DKI MRI and Machine Learning-Based Multi-Parametric Texture Analysis. Bioengineering 2023, 10, 83. https://doi.org/10.3390/bioengineering10010083
Malagi AV, Shivaji S, Kandasamy D, Sharma R, Garg P, Gupta SD, Gamanagatti S, Mehndiratta A. Pancreatic Mass Characterization Using IVIM-DKI MRI and Machine Learning-Based Multi-Parametric Texture Analysis. Bioengineering. 2023; 10(1):83. https://doi.org/10.3390/bioengineering10010083
Chicago/Turabian StyleMalagi, Archana Vadiraj, Sivachander Shivaji, Devasenathipathy Kandasamy, Raju Sharma, Pramod Garg, Siddhartha Datta Gupta, Shivanand Gamanagatti, and Amit Mehndiratta. 2023. "Pancreatic Mass Characterization Using IVIM-DKI MRI and Machine Learning-Based Multi-Parametric Texture Analysis" Bioengineering 10, no. 1: 83. https://doi.org/10.3390/bioengineering10010083
APA StyleMalagi, A. V., Shivaji, S., Kandasamy, D., Sharma, R., Garg, P., Gupta, S. D., Gamanagatti, S., & Mehndiratta, A. (2023). Pancreatic Mass Characterization Using IVIM-DKI MRI and Machine Learning-Based Multi-Parametric Texture Analysis. Bioengineering, 10(1), 83. https://doi.org/10.3390/bioengineering10010083