Rapid Diagnosis of Ductal Carcinoma In Situ and Breast Cancer Based on Raman Spectroscopy of Serum Combined with Convolutional Neural Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Raman Spectral Measurements
2.3. Spectra Preprocessing Method
2.4. Classification Model
2.4.1. K-Nearest Neighbor (KNN)
2.4.2. Random Forest (RF)
2.4.3. Support Vector Machine (SVM)
2.4.4. Convolutional Neural Networks (CNN)
3. Results and Discussions
3.1. Spectral Data Analysis
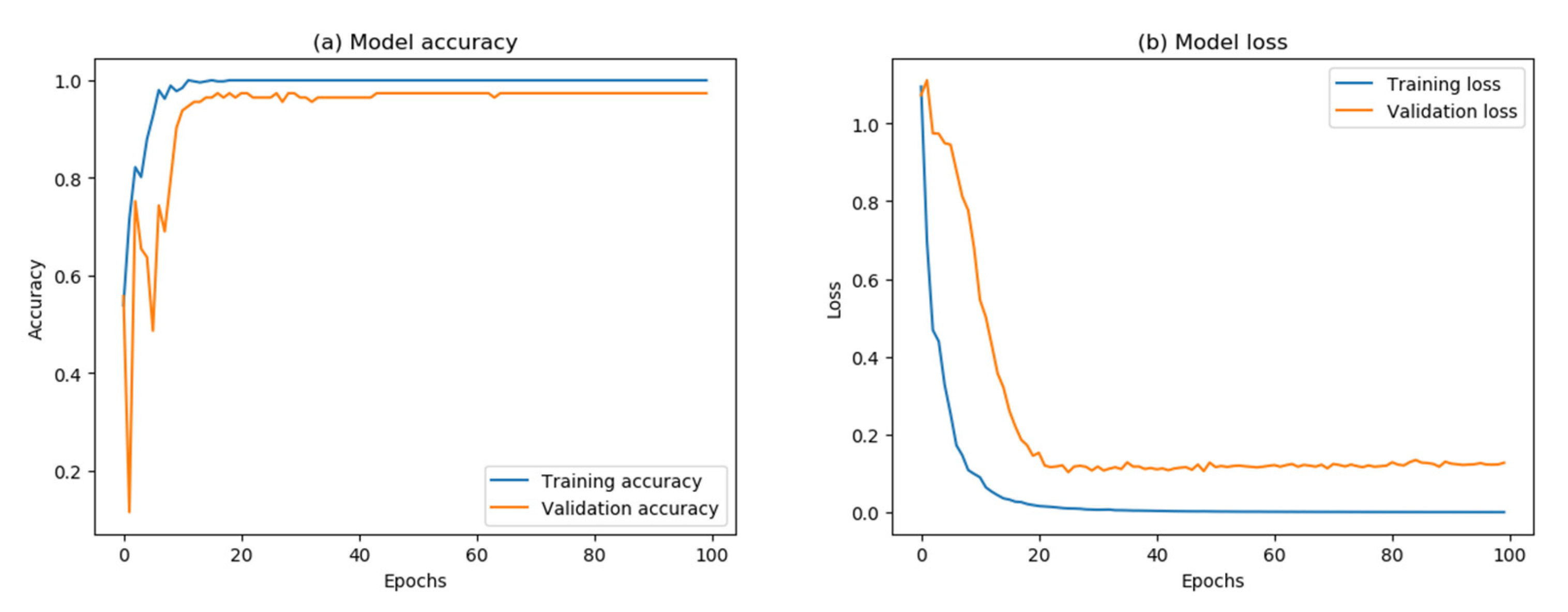
3.2. CNN Model Establishment and Classification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lazaro-Pacheco, D.; Shaaban, A.; Rehman, S.; Rehman, I. Raman spectroscopy of breast cancer. Appl. Spectrosc. Rev. 2020, 55, 439–475. [Google Scholar] [CrossRef]
- Unger-Saldaña, K. Challenges to the early diagnosis and treatment of breast cancer in developing countries. World J. Clin. Oncol. 2014, 5, 465. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.; Torre, L.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Zheng, R.; Sun, K.; Zhang, S.; Zeng, H.; Zou, X.; Chen, R. Analysis of the prevalence of malignant tumors in China in 2015. Chin. J. Oncol. 2019, 41, 19–28. [Google Scholar]
- Sakorafas, G.; Tsiotou, A. Ductal carcinoma in situ (DCIS) of the breast: Evolving perspectives. Cancer Treat. Rev. 2000, 26, 103–125. [Google Scholar] [CrossRef]
- Kuerer, H. Rational individualised selection of adjuvant therapy for ductal carcinoma in situ. Lancet Oncol. 2010, 12, 2–3. [Google Scholar] [CrossRef]
- Lari, S.; Kuerer, H. Biological markers in DCIS and risk of breast recurrence: A systematic review. J. Cancer 2011, 2, 232. [Google Scholar] [CrossRef]
- Faridah, Y. Digital versus screen film mammography: A clinical comparison. Biomed. Imaging Interv. J. 2008, 4, e31. [Google Scholar] [CrossRef][Green Version]
- Lieber, C.; Majumder, S.; Billheimer, D.; Ellis, D.; Mahadevan-Jansen, A. Raman microspectroscopy for skin cancer detection in vitro. J. Biomed. Opt. 2009, 13, 024013. [Google Scholar] [CrossRef]
- Caspers, P.; Lucassen, G.; Wolthuis, R.; Bruining, H.; Puppels, G. In vitro and in vivo Raman spectroscopy of human skin. Biospectroscopy 1998, 4, S31–S39. [Google Scholar] [CrossRef]
- Krafft, C.; Sobottka, S.; Schackert, G.; Salzer, R. Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst 2005, 130, 1070–1077. [Google Scholar] [CrossRef]
- Beljebbar, A.; Manfait, M. Fourier transform infrared microspectroscopy for cancer diagnostic of C6 glioma on animal model. Fourier Transforms-New Analytical Approaches and FTIR Strategies 2011. [Google Scholar] [CrossRef][Green Version]
- Leong, A.; Zhuang, Z. The changing role of pathology in breast cancer diagnosis; treatment. Pathobiology 2011, 78, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Blake, N.; Gaifulina, R.; Griffin, L.; Bell, I.; Thomas, G. Machine Learning of Raman Spectroscopy Data for Classifying Cancers: A Review of the Recent Literature. Diagnostics 2022, 12, 1491. [Google Scholar] [CrossRef]
- Rehman, S.; Movasaghi, Z.; Tucker, A.; Joel, S.; Darr, J.; Ruban, A.; Rehman, I. Raman spectroscopic analysis of breast cancer tissues: Identifying differences between normal, invasive ductal carcinoma and ductal carcinoma in situ of the breast tissue. J. Raman Spectrosc. Int. J. Orig. Work. All Asp. Raman Spectrosc. Incl. High. Order Process. Brillouin Rayleigh Scatt. 2007, 38, 1345–1351. [Google Scholar] [CrossRef]
- Han, B.; Du, Y.; Fu, T.; Fan, Z.; Xu, S.; Hu, C.; Bi, L.; Gao, T.; Zhang, H.; Xu, W. Differences and relationships between normal and atypical ductal hyperplasia, ductal carcinoma in situ, and invasive ductal carcinoma tissues in the breast based on raman spectroscopy. Appl. Spectrosc. 2017, 71, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Lazaro-Pacheco, D.; Shaaban, A.; Titiloye, N.; Rehman, S.; Rehman, I. Elucidating the chemical and structural composition of breast cancer using Raman micro-spectroscopy. EXCLI J. 2021, 20, 1118. [Google Scholar] [PubMed]
- Pichardo-Molina, J.; Frausto-Reyes, C.; Barbosa-García, O.; Huerta-Franco, R.; González-Trujillo, J.; Ramírez-Alvarado, C.; Gutiérrez-Juárez, G.; Medina-Gutiérrez, C. Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med. Sci. 2007, 22, 229–236. [Google Scholar] [CrossRef]
- Nargis, H.; Nawaz, H.; Bhatti, H.; Jilani, K.; Saleem, M. Comparison of surface enhanced Raman spectroscopy and Raman spectroscopy for the detection of breast cancer based on serum samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 246, 119034. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Gao, J.M.; Tang, S.Z.; Zhao, X.; Zheng, M.M.; Gong, W.; Xie, S.S.; Gao, S.Q.; Yu, Y.; Lin, J.Q. Label-free diagnosis of breast cancer based on serum protein purification assisted surface-enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 263, 120234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, C.; Peng, D.; Yi, X.; He, S.; Liu, F.; Zheng, X.; Huang, W.; Zhao, L. Label-free diagnosis of breast cancer based on serum protein purification assisted surface-enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 264, 120300. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.Q.; Yuen, C.; Padmanabhan, S.; Beuerman, R.W.; Liu, Q. Recovery of Raman spectra with low signal-to-noise ratio using Wiener estimation. Opt. Express 2014, 22, 12102–12114. [Google Scholar] [CrossRef]
- Pan, L.R.; Zhang, P.; Daengngam, C.; Peng, S.L.; Chongcheawchamnan, M. A review of artificial intelligence methods combined with Raman spectroscopy to identify the composition of substances. J. Raman Spectrosc. 2022, 53, 6–19. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.; Masson, J. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: Berlin/Heidelberg, Germany, 1999. [Google Scholar]
- Chorowski, J.; Wang, J.; Zurada, J. Review and performance comparison of SVM-and ELM-based classifiers. Neurocomputing 2014, 128, 507–516. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; FadhelFarhan, M.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Yan, H.; Yu, M.; Xia, J.; Zhu, L.; Zhang, T.; Zhu, Z. Tongue squamous cell carcinoma discrimination with Raman spectroscopy and convolutional neural networks. Vib. Spectrosc. 2019, 103, 102938. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, S.; Cui, X.; Xu, W.; Kong, C.; Zhang, Z. Identifying non-muscle-invasive and muscle-invasive bladder cancer based on blood serum surface-enhanced Raman spec-troscopy. Biomed. Opt. Express 2019, 10, 3533–3544. [Google Scholar] [CrossRef]
- Wu, Q.; Qiu, S.; Yu, Y.; Chen, W.; Lin, H.; Lin, D.; Feng, S. Assessment of the radiotherapy effect for nasopharyngeal cancer using plasma surface-enhanced Raman spectroscopy tech-nology. Biomed. Opt. Express 2018, 9, 3413–3423. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Lu, J.; Chen, Z.; Cui, X.; Chen, S.; Pei, D. Identifying benign and malignant thyroid nodules based on blood serum surface-enhanced Raman spectroscopy. Nano-Med. Nanotechnol. Biol. Med. 2021, 32, 102328. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Man, B.; Yang, C.; Zhang, C.; Liu, M.; Zhang, Y.; Liu, L.; Chen, C. Label-free and stable serum analysis based on Ag-NPs/PSi surface-enhanced Raman scattering for noninvasive lung cancer detection. Biomed. Opt. Express 2018, 9, 4345–4358. [Google Scholar] [CrossRef]
- Ullah, R.; Khan, S.; Farman, F.; Bilal, M.; Krafft, C.; Shahzad, S. Demonstrating the application of Raman spectroscopy together with chemometric technique for screening of asthma disease. Biomed. Opt. Express 2019, 10, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Bahreini, M.; Hosseinzadegan, A.; Rashidi, A.; Miri, S.; Mirzaei, H.; Hajian, P. A Raman-based serum constituents’ analysis for gastric cancer diagnosis: In vitro study. Talanta 2019, 204, 826–832. [Google Scholar] [CrossRef]
- Li, X.; Yang, T.; Li, S.; Jin, L.; Wang, D.; Guan, D. Noninvasive liver diseases detection based on serum surface enhanced Raman spectroscopy and statistical analysis. Opt. Express 2015, 23, 18361–18372. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Das, B.; Glassman, W.; Tang, G.; Yoo, K.; Zhu, H.; Akins, D.; Lubicz, S.; Cleary, J.; Prudente, R.; et al. Raman, fluorescence, and time-resolved light scattering as optical diagnostic techniques to separate diseased and normal bi-omedical media. J. Photochem. Photobiol. B Biol. 1992, 16, 187–209. [Google Scholar] [CrossRef]
- Düntsch, I.; Gediga, G. Confusion matrices and rough set data analysis. J. Phys. Conf. Ser. 2019, 1229, 012055. [Google Scholar] [CrossRef]
- Faraggi, D.; Reiser, B. Estimation of the area under the ROC curve. Stat. Med. 2002, 21, 3093–3106. [Google Scholar] [CrossRef]
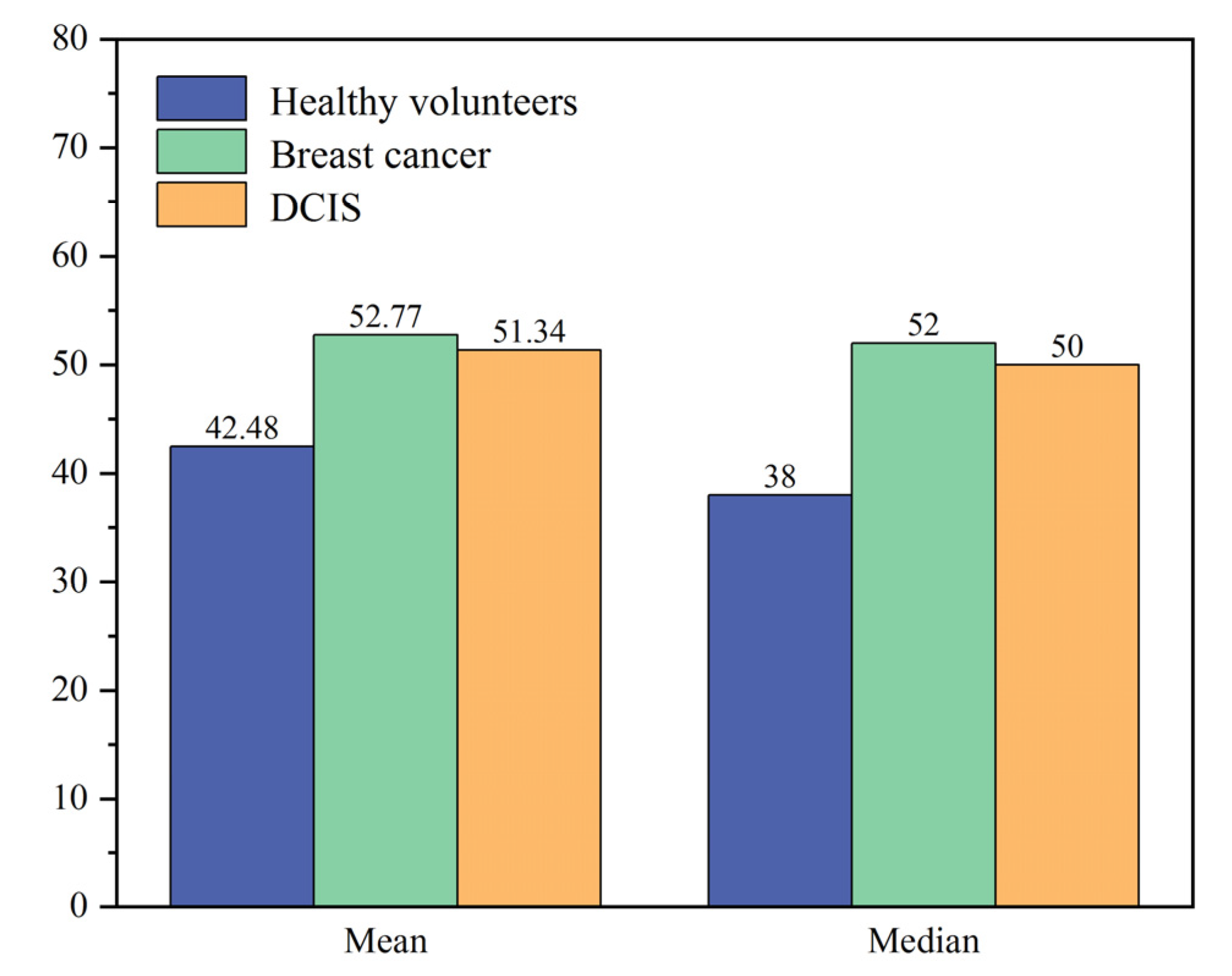
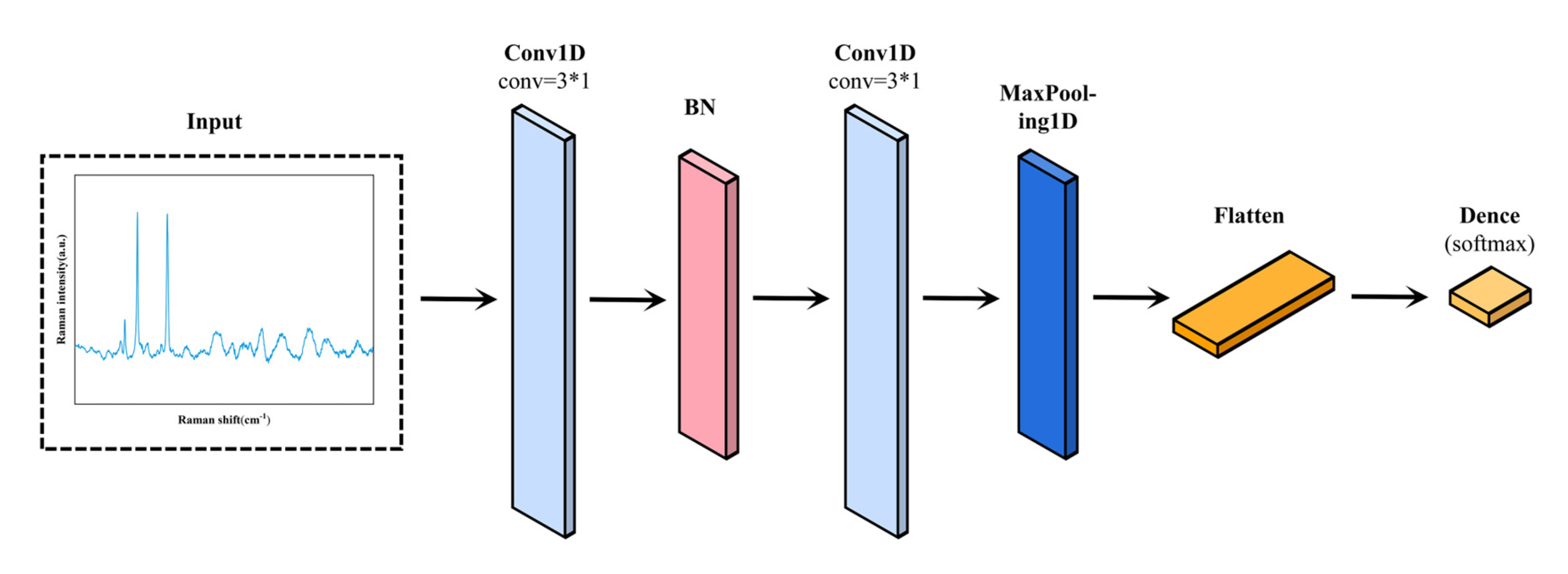



| Peaks | Major Assignments | Ref |
|---|---|---|
| 593 cm−1 | Ascorbic acid, amide-VI | Chen et al. [31] |
| 810 cm−1 | L-serine, glutathione: C-C-O stretching vibration | Wu et al. [32] |
| 950 cm−1 | α-Helix, proline, valine: C-C stretching vibration | Xia et al. [33] |
| 1004 cm−1 | Phenylalanine: C-C symmetric stretch | Zhang et al. [34] |
| 1154 cm−1 | β-carotene: C-C stretch | Ullah et al. [35] |
| 1280 cm−1 | Nucleic acid: Amide III & CH2 wagging vibration | Bahreini et al. [36] |
| 1445 cm−1 | Collagen, phospholipids: CH2 bending vibration | Zhang et al. [34] |
| 1514 cm−1 | β-carotene: C-C stretch | Ullah et al. [35] |
| 1576 cm−1 | Phenylalanine, acetoacetate, riboflavin:C-C bending | Li et al. [37] |
| 2517 cm−1 | Proteins: O-H stretch | Bahreini et al. [36] |
| 2662 cm−1 | Proteins: O-H stretch | Bahreini et al. [36] |
| Samples | Normal | Breast Cancer | DCIS | Total |
|---|---|---|---|---|
| Training set | 168 | 324 | 70 | 562 |
| Test set | 73 | 139 | 30 | 242 |
| Total | 241 | 463 | 100 | 804 |
| Parameters | Batch Size | Epoch | Learning Rate | Optimizer | Activation | Loss | Cross-Validation Accuracy (%) |
|---|---|---|---|---|---|---|---|
| Value or option | 16 | 100 | 0.001 | Adam | Tanh | Categorical_ crossentropy | 96.09 |
| KNN | RF | SVM | CNN | |||||||||
| HC | BC | DCIS | HC | BC | DCIS | HC | BC | DCIS | HC | BC | DCIS | |
| HC | 47 | 24 | 2 | 43 | 30 | 0 | 67 | 6 | 0 | 72 | 1 | 0 |
| BC | 9 | 127 | 3 | 5 | 134 | 0 | 7 | 132 | 0 | 2 | 137 | 0 |
| DCIS | 2 | 11 | 17 | 1 | 10 | 19 | 0 | 0 | 30 | 0 | 0 | 30 |
| Acc | 78.93% | 80.99% | 94.63% | 98.76% | ||||||||
| KNN | RF | SVM | CNN | |||||
| HC | BC | HC | BC | HC | BC | HC | BC | |
| HC | 53 | 20 | 50 | 23 | 69 | 4 | 71 | 2 |
| BC | 16 | 123 | 20 | 119 | 8 | 131 | 2 | 137 |
| Sensitivity | 88.49% | 85.61% | 94.25% | 98.56% | ||||
| Specificity | 72.60% | 68.49% | 94.52% | 97.26% | ||||
| Accuracy | 83.02% | 79.72% | 94.34% | 98.11% | ||||
| KNN | RF | SVM | CNN | |||||
|---|---|---|---|---|---|---|---|---|
| HC | DCIS | HC | DCIS | HC | DCIS | HC | DCIS | |
| HC | 69 | 4 | 72 | 1 | 73 | 0 | 73 | 0 |
| DCIS | 6 | 24 | 5 | 25 | 0 | 30 | 0 | 30 |
| Sensitivity | 80.00% | 83.33% | 100.00% | 100.00% | ||||
| Specificity | 94.52% | 98.63% | 100.00% | 100.00% | ||||
| Accuracy | 90.29% | 94.18% | 100.00% | 100.00% | ||||
| KNN | RF | SVM | CNN | |||||
| BC | DCIS | BC | DCIS | BC | DCIS | BC | DCIS | |
| BC | 133 | 6 | 139 | 0 | 139 | 0 | 139 | 0 |
| DCIS | 8 | 22 | 12 | 18 | 0 | 30 | 0 | 30 |
| Sensitivity | 73.33% | 60.00% | 100.00% | 100.00% | ||||
| Specificity | 95.68% | 100.00% | 100.00% | 100.00% | ||||
| Accuracy | 91.72% | 92.90% | 100.00% | 100.00% | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xie, F.; Yang, Y.; Zhao, J.; Wu, G.; Wang, S. Rapid Diagnosis of Ductal Carcinoma In Situ and Breast Cancer Based on Raman Spectroscopy of Serum Combined with Convolutional Neural Network. Bioengineering 2023, 10, 65. https://doi.org/10.3390/bioengineering10010065
Wang X, Xie F, Yang Y, Zhao J, Wu G, Wang S. Rapid Diagnosis of Ductal Carcinoma In Situ and Breast Cancer Based on Raman Spectroscopy of Serum Combined with Convolutional Neural Network. Bioengineering. 2023; 10(1):65. https://doi.org/10.3390/bioengineering10010065
Chicago/Turabian StyleWang, Xianglei, Fei Xie, Yang Yang, Jin Zhao, Guohua Wu, and Shu Wang. 2023. "Rapid Diagnosis of Ductal Carcinoma In Situ and Breast Cancer Based on Raman Spectroscopy of Serum Combined with Convolutional Neural Network" Bioengineering 10, no. 1: 65. https://doi.org/10.3390/bioengineering10010065
APA StyleWang, X., Xie, F., Yang, Y., Zhao, J., Wu, G., & Wang, S. (2023). Rapid Diagnosis of Ductal Carcinoma In Situ and Breast Cancer Based on Raman Spectroscopy of Serum Combined with Convolutional Neural Network. Bioengineering, 10(1), 65. https://doi.org/10.3390/bioengineering10010065







