Differential Methylation and Acetylation as the Epigenetic Basis of Resveratrol’s Anticancer Activity
Abstract
1. Introduction
2. Epigenetic Regulation by Resveratrol
3. Methylation
3.1. Breast Cancer
3.1.1. Genome-Wide Analyses
3.1.2. Effects on Tumor Suppressors
3.1.3. DNMTs as Mediators of Resveratrol Effects in Breast Cancer
3.2. Glioma
3.3. Lung Cancer
4. Acetylation
4.1. Breast Cancer
4.2. Cervical Cancer
4.3. Colon Cancer
4.4. Leukemia and Lymphoma
4.5. Prostate Cancer
5. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- Miller, N.J.; Rice-Evans, C.A. Antioxidant activity of resveratrol in red wine. Clin. Chem. 1995, 41, 1789. [Google Scholar] [PubMed]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G.; et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Farhan Asad, S.; Singh, S.; Hadi, S.M. DNA breakage by resveratrol and cu(ii): Reaction mechanism and bacteriophage inactivation. Cancer Lett. 2000, 154, 29–37. [Google Scholar] [CrossRef]
- Ahmad, A.; Syed, F.A.; Singh, S.; Hadi, S.M. Prooxidant activity of resveratrol in the presence of copper ions: Mutagenicity in plasmid DNA. Toxicol. Lett. 2005, 159, 1–12. [Google Scholar] [CrossRef]
- Khan, H.Y.; Zubair, H.; Faisal, M.; Ullah, M.F.; Farhan, M.; Sarkar, F.H.; Ahmad, A.; Hadi, S.M. Plant polyphenol induced cell death in human cancer cells involves mobilization of intracellular copper ions and reactive oxygen species generation: A mechanism for cancer chemopreventive action. Mol. Nutr. Food Res. 2014, 58, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.Y.; Zubair, H.; Ullah, M.F.; Ahmad, A.; Hadi, S.M. A prooxidant mechanism for the anticancer and chemopreventive properties of plant polyphenols. Curr. Drug Targets 2012, 13, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Oves, M.; Chibber, S.; Hadi, S.M.; Ahmad, A. Mobilization of nuclear copper by green tea polyphenol epicatechin-3-gallate and subsequent prooxidant breakage of cellular DNA: Implications for cancer chemotherapy. Int. J. Mol. Sci. 2016, 18, 34. [Google Scholar] [CrossRef] [PubMed]
- de la Lastra, C.A.; Villegas, I. Resveratrol as an antioxidant and pro-oxidant agent: Mechanisms and clinical implications. Biochem. Soc. Trans. 2007, 35, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Casey, S.C.; Amedei, A.; Aquilano, K.; Azmi, A.S.; Benencia, F.; Bhakta, D.; Bilsland, A.E.; Boosani, C.S.; Chen, S.; Ciriolo, M.R.; et al. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin. Cancer Biol. 2015, 35, S199–S223. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Kao, C.L.; Liu, C.M. The cancer prevention, anti-inflammatory and anti-oxidation of bioactive phytochemicals targeting the tlr4 signaling pathway. Int. J. Mol. Sci. 2018, 19, 2729. [Google Scholar] [CrossRef]
- Kopp, P. Resveratrol, a phytoestrogen found in red wine. A possible explanation for the conundrum of the ‘french paradox’? Eur. J. Endocrinol. 1998, 138, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Shukla, Y.; Singh, R. Resveratrol and cellular mechanisms of cancer prevention. Ann. N. Y. Acad. Sci. 2011, 1215, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Ren, J. From french paradox to cancer treatment: Anti-cancer activities and mechanisms of resveratrol. Anti-Cancer Agents Med. Chem. 2014, 14, 806–825. [Google Scholar] [CrossRef]
- McCalley, A.E.; Kaja, S.; Payne, A.J.; Koulen, P. Resveratrol and calcium signaling: Molecular mechanisms and clinical relevance. Molecules 2014, 19, 7327–7340. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Wu, S.J.; Chang, Y.T.; Tang, J.Y.; Li, K.T.; Ismail, M.; Liaw, C.C.; Li, R.N.; Chang, H.W. Activation and inhibition of atm by phytochemicals: Awakening and sleeping the guardian angel naturally. Arch. Immunol. Ther. Exp. 2015, 63, 357–366. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.R.; Nabavi, S.F.; Manayi, A.; Daglia, M.; Hajheydari, Z.; Nabavi, S.M. Resveratrol and the mitochondria: From triggering the intrinsic apoptotic pathway to inducing mitochondrial biogenesis, a mechanistic view. Biochimi. Biophys. Acta 2016, 1860, 727–745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wen, X.; Li, M.; Li, S.; Zhao, H. Targeting cancer stem cells and signaling pathways by resveratrol and pterostilbene. BioFactors 2018, 44, 61–68. [Google Scholar] [CrossRef]
- Sarkar, F.H.; Li, Y.; Wang, Z.; Kong, D. The role of nutraceuticals in the regulation of wnt and hedgehog signaling in cancer. Cancer Metast. Rev. 2010, 29, 383–394. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Lertpiriyapong, K.; Steelman, L.S.; Abrams, S.L.; Yang, L.V.; Murata, R.M.; Rosalen, P.L.; Scalisi, A.; Neri, L.M.; Cocco, L.; et al. Effects of resveratrol, curcumin, berberine and other nutraceuticals on aging, cancer development, cancer stem cells and micrornas. Aging 2017, 9, 1477–1536. [Google Scholar] [CrossRef]
- Karimi Dermani, F.; Saidijam, M.; Amini, R.; Mahdavinezhad, A.; Heydari, K.; Najafi, R. Resveratrol inhibits proliferation, invasion, and epithelial-mesenchymal transition by increasing mir-200c expression in hct-116 colorectal cancer cells. J. Cell. Biochem. 2017, 118, 1547–1555. [Google Scholar] [CrossRef]
- Xu, J.; Liu, D.; Niu, H.; Zhu, G.; Xu, Y.; Ye, D.; Li, J.; Zhang, Q. Resveratrol reverses doxorubicin resistance by inhibiting epithelial-mesenchymal transition (emt) through modulating pten/akt signaling pathway in gastric cancer. J. Exp. Clin. Cancer Res. 2017, 36, 19. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Li, Y.; Bao, B.; Kong, D.; Sarkar, F.H. Epigenetic regulation of mirna-cancer stem cells nexus by nutraceuticals. Mol. Nutr. Food Res. 2014, 58, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Vanden Berghe, W. Epigenetic impact of dietary polyphenols in cancer chemoprevention: Lifelong remodeling of our epigenomes. Pharmacol. Res. 2012, 65, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic diet: Impact on the epigenome and cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Park, L.K.; Friso, S.; Choi, S.W. Nutritional influences on epigenetics and age-related disease. Proc. Nutr. Soc. 2012, 71, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Gerhauser, C. Cancer chemoprevention and nutriepigenetics: State of the art and future challenges. Top. Curr. Chem. 2013, 329, 73–132. [Google Scholar] [PubMed]
- Wang, Y.; Li, Y.; Liu, X.; Cho, W.C. Genetic and epigenetic studies for determining molecular targets of natural product anticancer agents. Curr. Cancer Drug Targets 2013, 13, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Vahid, F.; Zand, H.; Nosrat-Mirshekarlou, E.; Najafi, R.; Hekmatdoost, A. The role dietary of bioactive compounds on the regulation of histone acetylases and deacetylases: A review. Gene 2015, 562, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tollefsbol, T.O. Impact of epigenetic dietary components on cancer through histone modifications. Curr. Med. Chem. 2015, 22, 2051–2064. [Google Scholar] [CrossRef] [PubMed]
- Kala, R.; Tollefsbol, T.O. A novel combinatorial epigenetic therapy using resveratrol and pterostilbene for restoring estrogen receptor-alpha (eralpha) expression in eralpha-negative breast cancer cells. PLoS ONE 2016, 11, e0155057. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, G.F.S.; Silva, G.D.B.; Pavan, A.R.; Chiba, D.E.; Chin, C.M.; Dos Santos, J.L. Epigenetic regulatory mechanisms induced by resveratrol. Nutrients 2017, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Chiou, Y.S.; Ho, C.T.; Pan, M.H. Chemoprevention by resveratrol and pterostilbene: Targeting on epigenetic regulation. BioFactors 2018, 44, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bishayee, A.; Pandey, A.K. Targeting histone deacetylases with natural and synthetic agents: An emerging anticancer strategy. Nutrients 2018, 10, 731. [Google Scholar] [CrossRef]
- Stefanska, B.; Karlic, H.; Varga, F.; Fabianowska-Majewska, K.; Haslberger, A. Epigenetic mechanisms in anti-cancer actions of bioactive food components--the implications in cancer prevention. Br. J. Pharmacol. 2012, 167, 279–297. [Google Scholar]
- Lubecka, K.; Kurzava, L.; Flower, K.; Buvala, H.; Zhang, H.; Teegarden, D.; Camarillo, I.; Suderman, M.; Kuang, S.; Andrisani, O.; et al. Stilbenoids remodel the DNA methylation patterns in breast cancer cells and inhibit oncogenic notch signaling through epigenetic regulation of maml2 transcriptional activity. Carcinogenesis 2016, 37, 656–668. [Google Scholar] [CrossRef]
- Medina-Aguilar, R.; Perez-Plasencia, C.; Marchat, L.A.; Gariglio, P.; Garcia Mena, J.; Rodriguez Cuevas, S.; Ruiz-Garcia, E.; Astudillo-de la Vega, H.; Hernandez Juarez, J.; Flores-Perez, A.; et al. Methylation landscape of human breast cancer cells in response to dietary compound resveratrol. PLoS ONE 2016, 11, e0157866. [Google Scholar] [CrossRef]
- Ramaiah, M.J.; Vaishnave, S. Bmi1 and pten are key determinants of breast cancer therapy: A plausible therapeutic target in breast cancer. Gene 2018, 678, 302–311. [Google Scholar]
- Stefanska, B.; Salame, P.; Bednarek, A.; Fabianowska-Majewska, K. Comparative effects of retinoic acid, vitamin d and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br. J. Nutr. 2012, 107, 781–790. [Google Scholar] [CrossRef]
- Papoutsis, A.J.; Borg, J.L.; Selmin, O.I.; Romagnolo, D.F. Brca-1 promoter hypermethylation and silencing induced by the aromatic hydrocarbon receptor-ligand tcdd are prevented by resveratrol in mcf-7 cells. J. Nutr. Biochem. 2012, 23, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Papoutsis, A.J.; Selmin, O.I.; Borg, J.L.; Romagnolo, D.F. Gestational exposure to the ahr agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin induces brca-1 promoter hypermethylation and reduces brca-1 expression in mammary tissue of rat offspring: Preventive effects of resveratrol. Mol. Carcinog. 2015, 54, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Shoulson, R.; Chatterjee, A.; Ronghe, A.; Bhat, N.K.; Dim, D.C.; Bhat, H.K. Resveratrol inhibits estrogen-induced breast carcinogenesis through induction of nrf2-mediated protective pathways. Carcinogenesis 2014, 35, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Sharma, P.; Capalash, N. DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer. Curr. Cancer Drug Targets 2013, 13, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Kala, R.; Shah, H.N.; Martin, S.L.; Tollefsbol, T.O. Epigenetic-based combinatorial resveratrol and pterostilbene alters DNA damage response by affecting sirt1 and dnmt enzyme expression, including sirt1-dependent gamma-h2ax and telomerase regulation in triple-negative breast cancer. BMC Cancer 2015, 15, 672. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tollefsbol, T.O. Combinational proanthocyanidins and resveratrol synergistically inhibit human breast cancer cells and impact epigenetic(-)mediating machinery. Int. J. Mol. Sci. 2018, 19, 2204. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Qin, W.; Zhang, K.; Rottinghaus, G.E.; Chen, Y.C.; Kliethermes, B.; Sauter, E.R. Trans-resveratrol alters mammary promoter hypermethylation in women at increased risk for breast cancer. Nutr. Cancer 2012, 64, 393–400. [Google Scholar] [CrossRef]
- Qin, W.; Zhang, K.; Clarke, K.; Weiland, T.; Sauter, E.R. Methylation and mirna effects of resveratrol on mammary tumors vs. Normal tissue. Nutr. Cancer 2014, 66, 270–277. [Google Scholar] [CrossRef]
- Beetch, M.; Lubecka, K.; Kristofzski, H.; Suderman, M.; Stefanska, B. Subtle alterations in DNA methylation patterns in normal cells in response to dietary stilbenoids. Mol. Nutr. Food Res. 2018, 2018, e1800193. [Google Scholar] [CrossRef]
- Mirza, S.; Sharma, G.; Parshad, R.; Gupta, S.D.; Pandya, P.; Ralhan, R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J. Breast Cancer 2013, 16, 23–31. [Google Scholar] [CrossRef]
- Huang, H.; Lin, H.; Zhang, X.; Li, J. Resveratrol reverses temozolomide resistance by downregulation of mgmt in t98g glioblastoma cells by the nf-kappab-dependent pathway. Oncol. Rep. 2012, 27, 2050–2056. [Google Scholar] [PubMed]
- Yang, H.C.; Wang, J.Y.; Bu, X.Y.; Yang, B.; Wang, B.Q.; Hu, S.; Yan, Z.Y.; Gao, Y.S.; Han, S.Y.; Qu, M.Q. Resveratrol restores sensitivity of glioma cells to temozolamide through inhibiting the activation of wnt signaling pathway. J. Cell. Physiol. 2018, 2018. [Google Scholar] [CrossRef]
- Koval, A.; Katanaev, V.L. Dramatic dysbalancing of the wnt pathway in breast cancers. Sci. Rep. 2018, 8, 7329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; He, W.; Gao, X.; Li, B.; Mei, C.; Xu, R.; Chen, H. Resveratrol overcomes gefitinib resistance by increasing the intracellular gefitinib concentration and triggering apoptosis, autophagy and senescence in pc9/g nsclc cells. Sci. Rep 2015, 5, 17730. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, X.; Li, N.; Liu, H.; Dong, Q.; Zhang, J.; Yang, C.; Liu, Y.; Liang, Q.; Zhang, S.; et al. Resveratrol inhibits hexokinases ii mediated glycolysis in non-small cell lung cancer via targeting akt signaling pathway. Exp. Cell Res. 2016, 349, 320–327. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Cao, N.; Li, Z.; Han, J.; Li, L. Resveratrol, an activator of sirt1, induces protective autophagy in non-small-cell lung cancer via inhibiting akt/mtor and activating p38-mapk. OncoTargets Ther. 2018, 11, 7777–7786. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Gadgeel, S.M. Lung cancer and personalized medicine: Novel therapies and clinical management. Preface. Adv. Exp. Med. Biol. 2016, 890, v–vi. [Google Scholar]
- Ahmad, A. Epigenetics in personalized management of lung cancer. Adv Exp Med Biol 2016, 890, 111–122. [Google Scholar]
- Fudhaili, A.; Yoon, N.A.; Kang, S.; Ryu, J.; Jeong, J.Y.; Lee, D.H.; Kang, S.S. Resveratrol epigenetically regulates the expression of zinc finger protein 36 in nonsmall cell lung cancer cell lines. Oncol. Rep. 2019, 41, 1377–1386. [Google Scholar]
- Gupta, G.; Bebawy, M.; Pinto, T.J.A.; Chellappan, D.K.; Mishra, A.; Dua, K. Role of the tristetraprolin (zinc finger protein 36 homolog) gene in cancer. Crit. Rev. Eukaryot. Gene. Expr. 2018, 28, 217–221. [Google Scholar] [CrossRef]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Berger, A.; Bocker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Schleicher, S.; Mayer, M.; Weiss, T.S.; et al. Resveratrol as a pan-hdac inhibitor alters the acetylation status of histone [corrected] proteins in human-derived hepatoblastoma cells. PLoS ONE 2013, 8, e73097. [Google Scholar]
- Dutra, L.A.; Heidenreich, D.; Silva, G.; Man Chin, C.; Knapp, S.; Santos, J.L.D. Dietary compound resveratrol is a pan-bet bromodomain inhibitor. Nutrients 2017, 9, 1172. [Google Scholar] [CrossRef] [PubMed]
- Sahni, J.M.; Keri, R.A. Targeting bromodomain and extraterminal proteins in breast cancer. Pharmacol. Res. 2018, 129, 156–176. [Google Scholar] [CrossRef] [PubMed]
- Garmpis, N.; Damaskos, C.; Garmpi, A.; Kalampokas, E.; Kalampokas, T.; Spartalis, E.; Daskalopoulou, A.; Valsami, S.; Kontos, M.; Nonni, A.; et al. Histone deacetylases as new therapeutic targets in triple-negative breast cancer: Progress and promises. Cancer Genom. Proteom. 2017, 14, 299–313. [Google Scholar]
- Damaskos, C.; Garmpis, N.; Valsami, S.; Kontos, M.; Spartalis, E.; Kalampokas, T.; Kalampokas, E.; Athanasiou, A.; Moris, D.; Daskalopoulou, A.; et al. Histone deacetylase inhibitors: An attractive therapeutic strategy against breast cancer. Anticancer Res. 2017, 37, 35–46. [Google Scholar] [CrossRef]
- Zucchetti, B.; Shimada, A.K.; Katz, A.; Curigliano, G. The role of histone deacetylase inhibitors in metastatic breast cancer. The Breast 2018, 43, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Chen, W.; Li, H.; Li, M.; Li, L. The histone acetylation modifications of breast cancer and their therapeutic implications. Pathol. Oncol. Res. 2018, 24, 807–813. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, P.; Herrmann, A.; Yang, C.; Xin, H.; Wang, Z.; Hoon, D.S.; Forman, S.J.; Jove, R.; Riggs, A.D.; et al. Acetylated stat3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc. Natl. Acad. Sci. USA 2012, 109, 7765–7769. [Google Scholar] [CrossRef]
- Lapidus, R.G.; Nass, S.J.; Butash, K.A.; Parl, F.F.; Weitzman, S.A.; Graff, J.G.; Herman, J.G.; Davidson, N.E. Mapping of er gene cpg island methylation-specific polymerase chain reaction. Cancer Res. 1998, 58, 2515–2519. [Google Scholar]
- Saenglee, S.; Jogloy, S.; Patanothai, A.; Leid, M.; Senawong, T. Cytotoxic effects of peanut phenolics possessing histone deacetylase inhibitory activity in breast and cervical cancer cell lines. Pharmacol. Rep. 2016, 68, 1102–1110. [Google Scholar] [PubMed]
- Parashar, G.; Capalash, N. Promoter methylation-independent reactivation of pax1 by curcumin and resveratrol is mediated by uhrf1. Clin. Exp. Med. 2016, 16, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Zhu, J.; Mayr, D.; Kuhn, C.; Schulze, S.; Hofmann, S.; Dannecker, C.; Jeschke, U.; Kost, B.P. Histone h3 acetyl k9 and histone h3 tri methyl k4 as prognostic markers for patients with cervical cancer. Int. J. Mol. Sci. 2017, 18, 477. [Google Scholar] [CrossRef] [PubMed]
- Plewka, D.; Plewka, A.; Miskiewicz, A.; Morek, M.; Bogunia, E. Nuclear factor-kappa b as potential therapeutic target in human colon cancer. J. Cancer Res. Ther. 2018, 14, 516–520. [Google Scholar] [CrossRef]
- Pu, J.; Bai, D.; Yang, X.; Lu, X.; Xu, L.; Lu, J. Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer ht29 cells through induction of mir-155. Biochem. Biophys. Res. Commun. 2012, 428, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Lee, J.E.; Lee, A.S.; Kang, K.P.; Lee, S.; Park, S.K.; Lee, S.Y.; Han, M.K.; Kim, D.H.; Kim, W. Sirt1 overexpression decreases cisplatin-induced acetylation of nf-kappab p65 subunit and cytotoxicity in renal proximal tubule cells. Biochem. Biophys. Res. Commun. 2012, 419, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Buhrmann, C.; Shayan, P.; Popper, B.; Goel, A.; Shakibaei, M. Sirt1 is required for resveratrol-mediated chemopreventive effects in colorectal cancer cells. Nutrients 2016, 8, 145. [Google Scholar] [CrossRef]
- Yaseen, A.; Chen, S.; Hock, S.; Rosato, R.; Dent, P.; Dai, Y.; Grant, S. Resveratrol sensitizes acute myelogenous leukemia cells to histone deacetylase inhibitors through reactive oxygen species-mediated activation of the extrinsic apoptotic pathway. Mol. Pharmacol. 2012, 82, 1030–1041. [Google Scholar] [CrossRef]
- Frazzi, R.; Valli, R.; Tamagnini, I.; Casali, B.; Latruffe, N.; Merli, F. Resveratrol-mediated apoptosis of hodgkin lymphoma cells involves sirt1 inhibition and foxo3a hyperacetylation. Int. J. Cancer 2013, 132, 1013–1021. [Google Scholar] [CrossRef]
- Kikuchi, H.; Mimuro, H.; Kuribayashi, F. Resveratrol strongly enhances the retinoic acid-induced superoxide generating activity via up-regulation of gp91-phox gene expression in u937 cells. Biochem. Biophys. Res. Commun. 2018, 495, 1195–1200. [Google Scholar] [CrossRef]
- Kai, L.; Samuel, S.K.; Levenson, A.S. Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting mta1/nurd complex. Int. J. Cancer 2010, 126, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wong, J.; Moreno, G.T.; Young, M.K.; Cote, J.; Wang, W. Nurd, a novel complex with both atp-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 1998, 2, 851–861. [Google Scholar] [CrossRef]
- Li, K.; Dias, S.J.; Rimando, A.M.; Dhar, S.; Mizuno, C.S.; Penman, A.D.; Lewin, J.R.; Levenson, A.S. Pterostilbene acts through metastasis-associated protein 1 to inhibit tumor growth, progression and metastasis in prostate cancer. PLoS ONE 2013, 8, e57542. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Kumar, A.; Li, K.; Tzivion, G.; Levenson, A.S. Resveratrol regulates pten/akt pathway through inhibition of mta1/hdac unit of the nurd complex in prostate cancer. Biochim. Biophys. Acta 2015, 1853, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Atarashi, K.; Murata, Y.; Yamaji, R.; Nakano, Y.; Inui, H. Inhibitory mechanisms of the transcriptional activity of androgen receptor by resveratrol: Implication of DNA binding and acetylation of the receptor. J. Steroid Biochem. Mol. Biol. 2011, 123, 65–70. [Google Scholar] [CrossRef]
- Chao, S.C.; Chen, Y.J.; Huang, K.H.; Kuo, K.L.; Yang, T.H.; Huang, K.Y.; Wang, C.C.; Tang, C.H.; Yang, R.S.; Liu, S.H. Induction of sirtuin-1 signaling by resveratrol induces human chondrosarcoma cell apoptosis and exhibits antitumor activity. Sci. Rep. 2017, 7, 3180. [Google Scholar] [CrossRef]
- Zadi Heydarabad, M.; Nikasa, M.; Vatanmakanian, M.; Azimi, A.; Farshdousti Hagh, M. Regulatory effect of resveratrol and prednisolone on mdr1 gene expression in acute lymphoblastic leukemia cell line (ccrf-cem): An epigenetic perspective. J. Cell Biochem. 2018, 119, 4890–4896. [Google Scholar] [CrossRef]
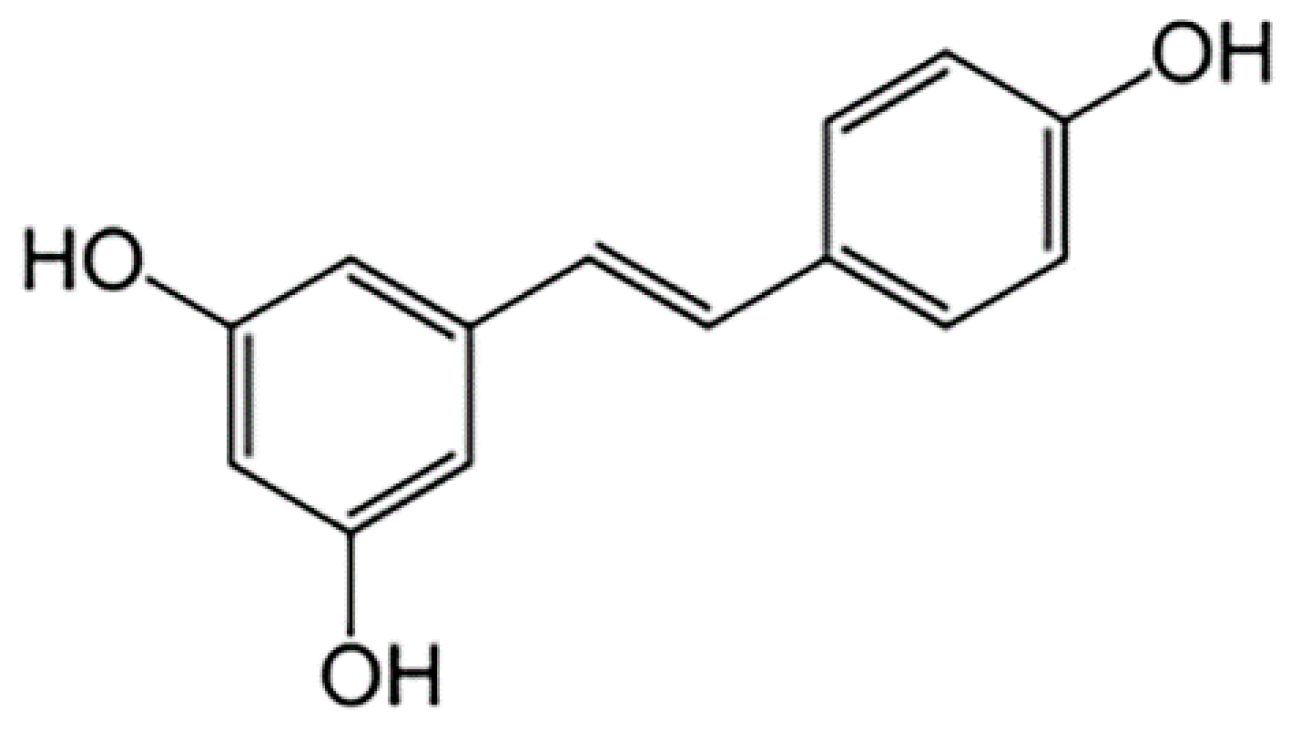
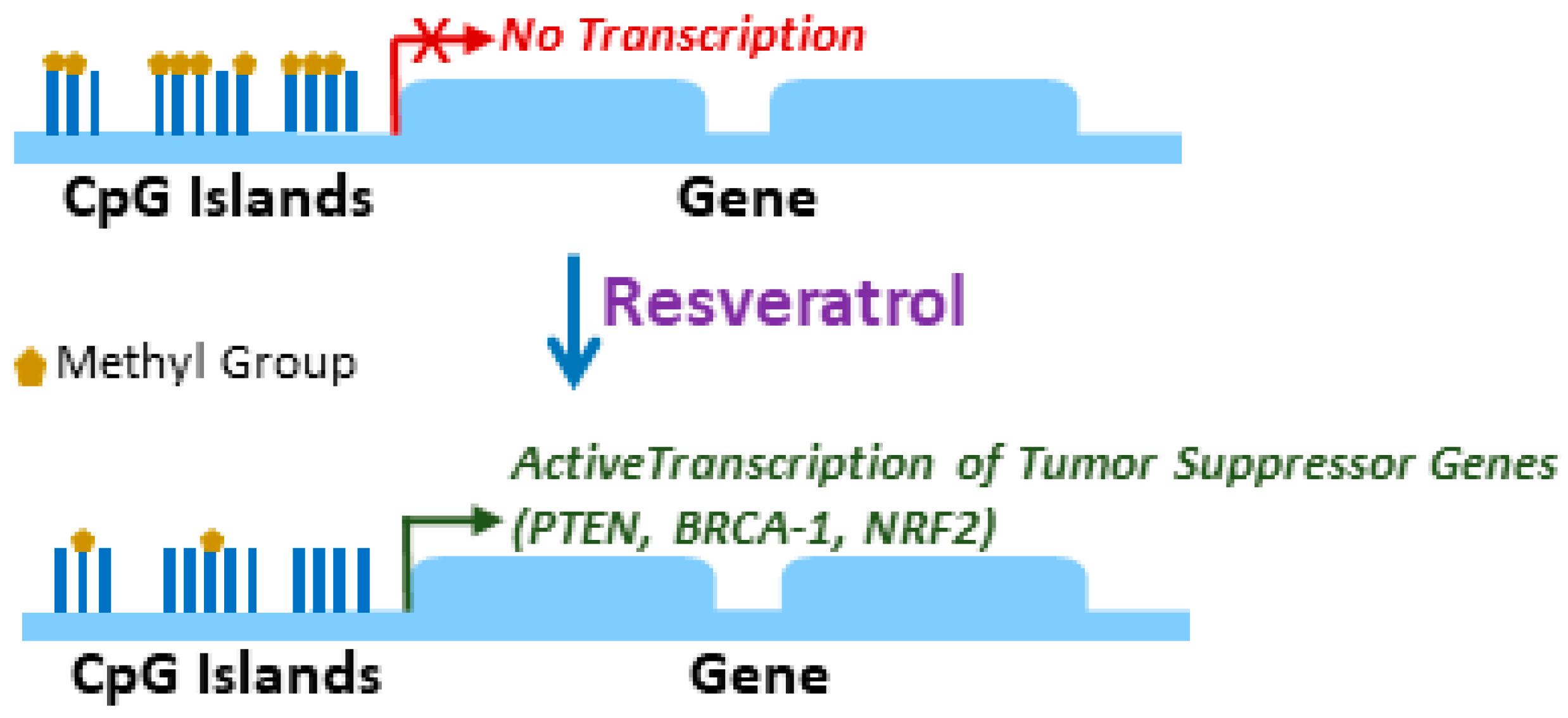
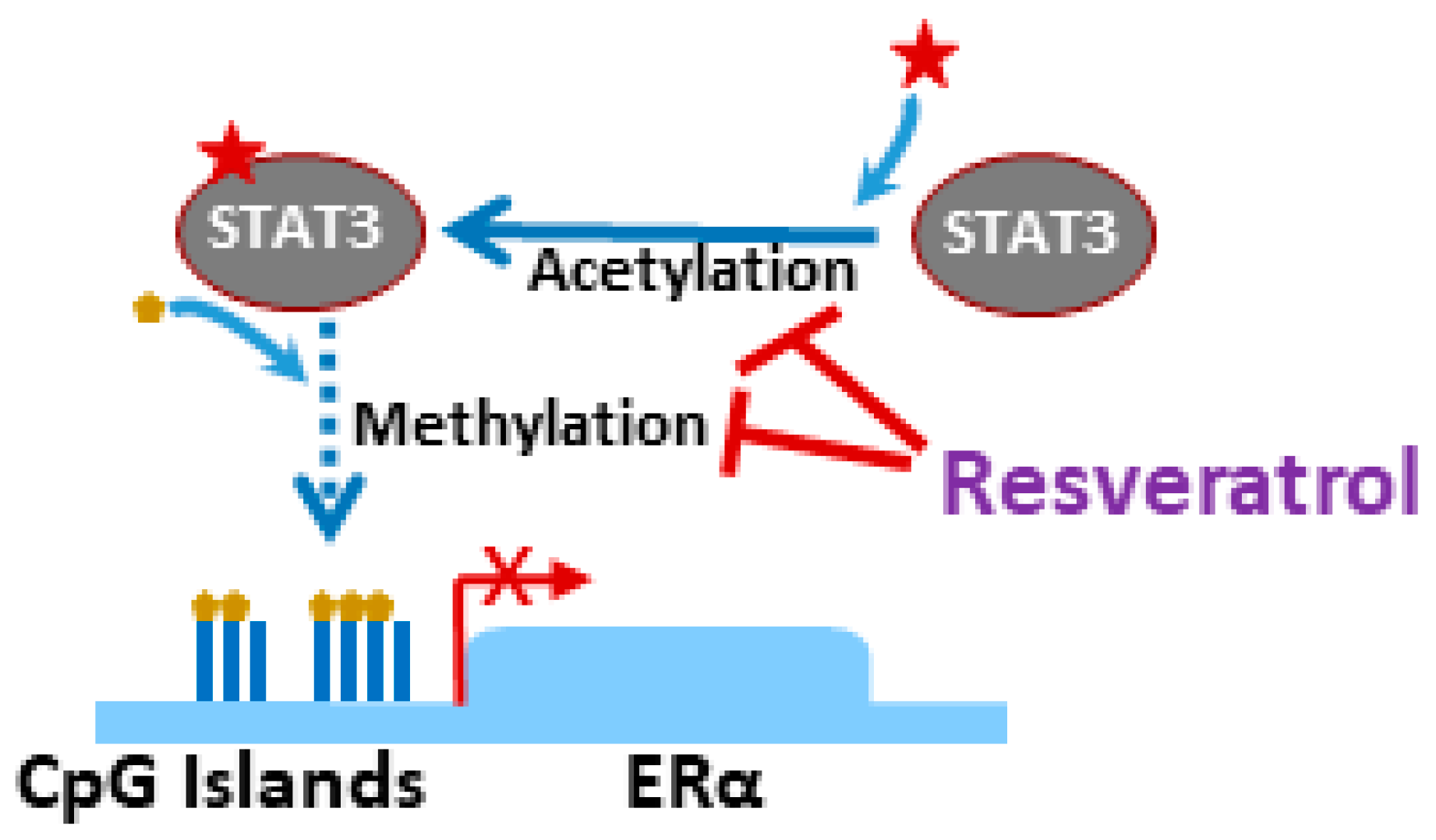
| Tumor Suppressor | Cancer Type | Effect of Resveratrol | Reference |
|---|---|---|---|
| BRCA-1 | Breast | Reduced promotor DNA methylation in vitro | [36] |
| Reduced promotor DNA methylation in vivo | [37] | ||
| NRF2 | Breast | Reduced promotor DNA methylation | [38] |
| p53 | Lymphoma | Induced acetylation | [73] |
| Prostate | [75] | ||
| PAX1 | Cervical | Regulation of histone acetylation | [66] |
| PTEN | Breast | Reduced promoter DNA methylation | [35] |
| Prostate | Acetylation and activation | [77] | |
| RASSF-1α | Breast | Reduced DNA methylation | [42] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farhan, M.; Ullah, M.F.; Faisal, M.; Farooqi, A.A.; Sabitaliyevich, U.Y.; Biersack, B.; Ahmad, A. Differential Methylation and Acetylation as the Epigenetic Basis of Resveratrol’s Anticancer Activity. Medicines 2019, 6, 24. https://doi.org/10.3390/medicines6010024
Farhan M, Ullah MF, Faisal M, Farooqi AA, Sabitaliyevich UY, Biersack B, Ahmad A. Differential Methylation and Acetylation as the Epigenetic Basis of Resveratrol’s Anticancer Activity. Medicines. 2019; 6(1):24. https://doi.org/10.3390/medicines6010024
Chicago/Turabian StyleFarhan, Mohd, Mohammad Fahad Ullah, Mohd Faisal, Ammad Ahmad Farooqi, Uteuliyev Yerzhan Sabitaliyevich, Bernhard Biersack, and Aamir Ahmad. 2019. "Differential Methylation and Acetylation as the Epigenetic Basis of Resveratrol’s Anticancer Activity" Medicines 6, no. 1: 24. https://doi.org/10.3390/medicines6010024
APA StyleFarhan, M., Ullah, M. F., Faisal, M., Farooqi, A. A., Sabitaliyevich, U. Y., Biersack, B., & Ahmad, A. (2019). Differential Methylation and Acetylation as the Epigenetic Basis of Resveratrol’s Anticancer Activity. Medicines, 6(1), 24. https://doi.org/10.3390/medicines6010024







