Evaluation of the Potential of Sewage Sludge Mycobiome to Degrade High Diclofenac and Bisphenol-A Concentrations
Abstract
1. Introduction
2. Materials and Methods
2.1. Solutions and Reagents
2.2. Sampling
2.3. Fungal Strain Isolation and Spore Collection
2.4. DNA Extraction
2.5. PCR Identification
2.6. Bacteria and Fungi Illumina Sequencing
2.7. Data and Bioinformatics Analysis
2.8. Biodegradation at Flask Scale
2.9. Residual Diclofenac and Bisphenol A Analysis
2.10. Enzymatic Analysis
3. Results and Discussion
3.1. Fungal Culturable Population
3.2. Bacterial Community Structure
3.3. Fungal Community Structure
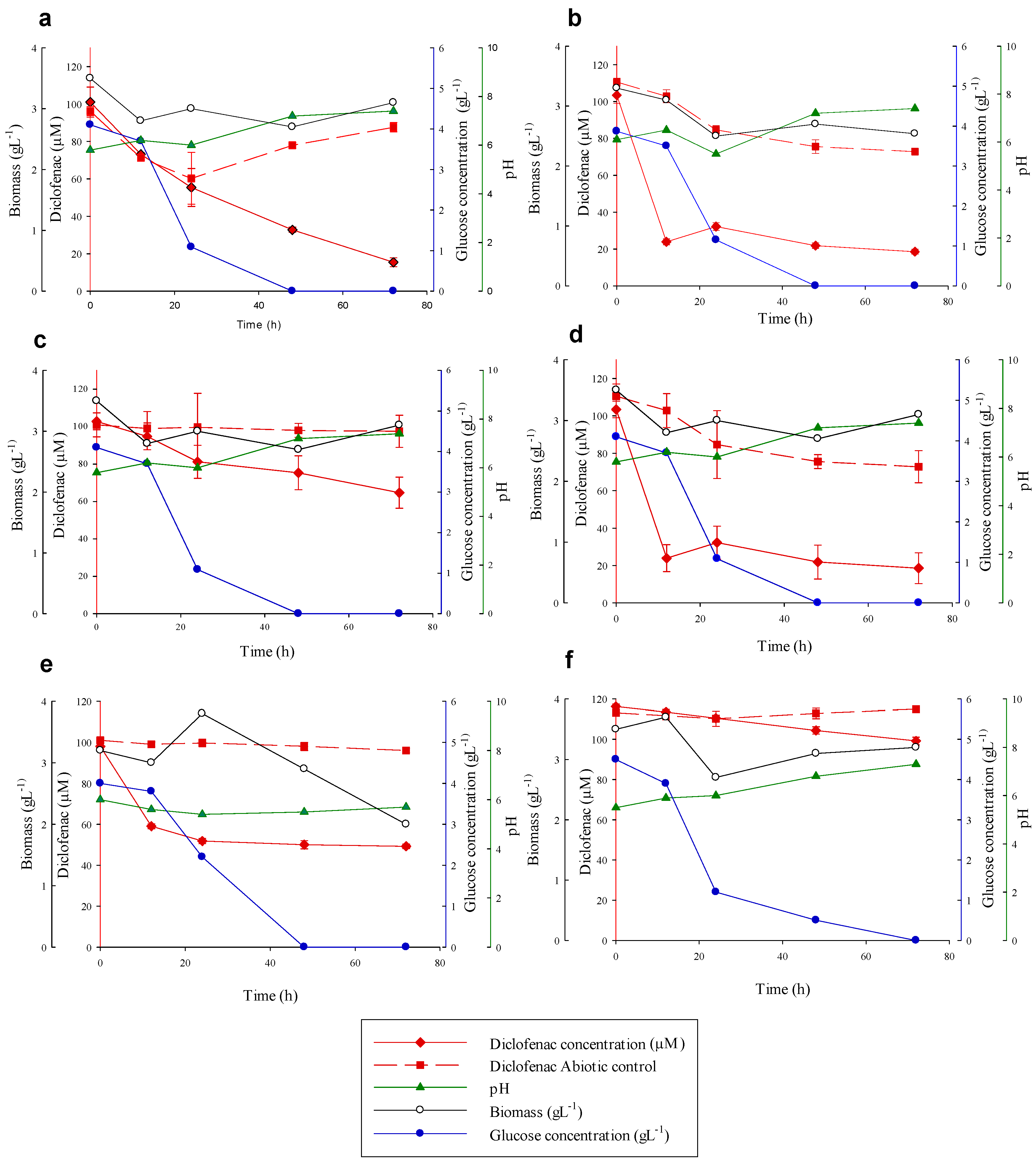
3.4. Diclofenac Degradation Experiments
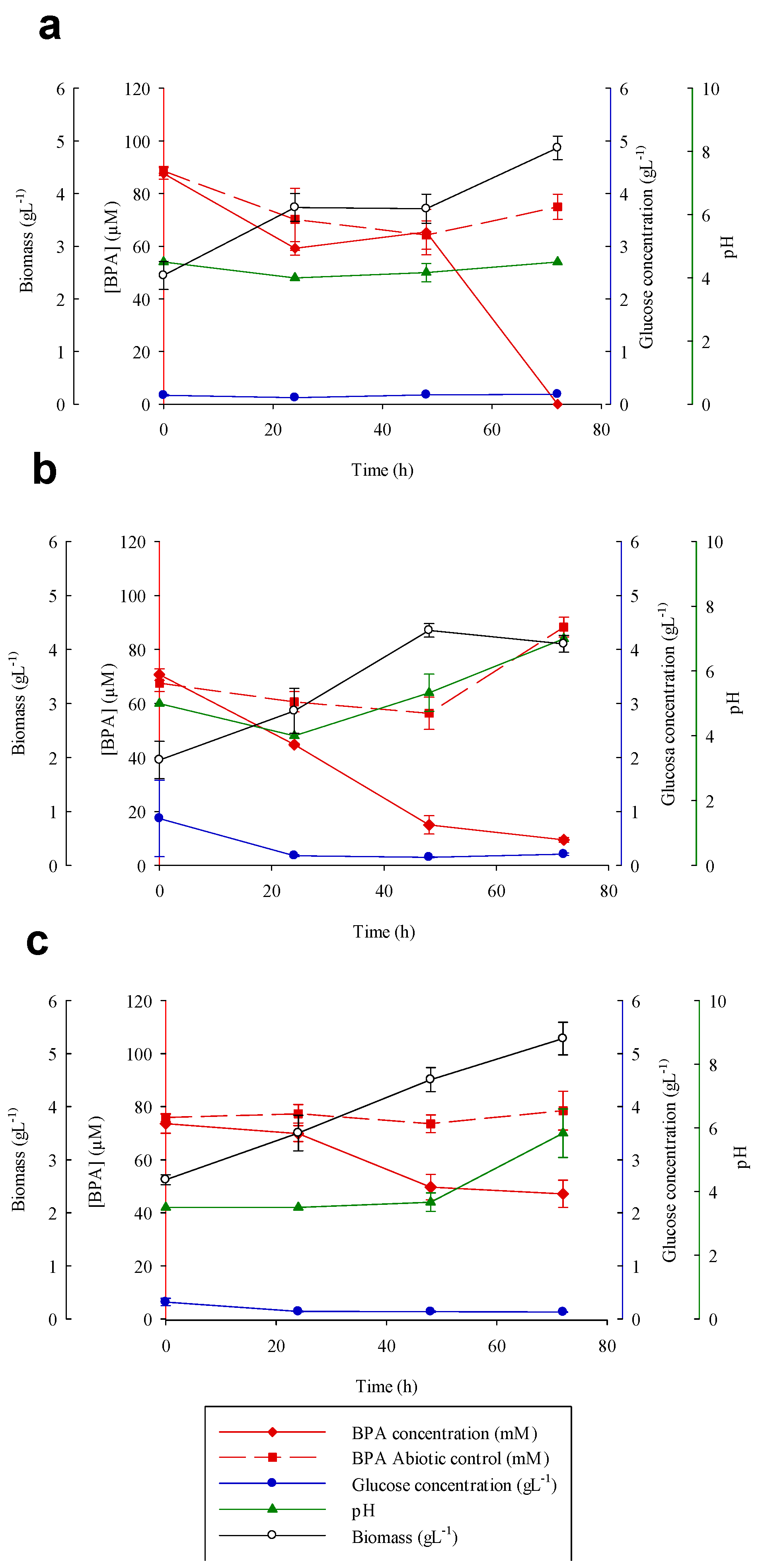
3.5. Bisphenol A Degradation Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geissen, V.; Mol, H.; Klumpp, E.; Umlauf, G.; Nadal, M.; van der Ploeg, M.; van de Zee, S.E.A.T.M.; Ritsema, C.J. Emerging pollutants in the environment: A challenge for water resource management. Int. Soil Water Conserv. Res. 2015, 3, 57–65. [Google Scholar] [CrossRef]
- Petrie, B.; Barden, R.; Kasprzyk-Hordern, B. A review on emerging contaminants in wastewaters and the environment: Current knowledge, understudied areas and recommendations for future monitoring. Water Res. 2015, 72, 3–27. [Google Scholar] [CrossRef]
- Verlicchi, P.; Galletti, A.; Petrovic, M.; Barceló, D. Hospital effluents as a source of emerging pollutants: An overview of micropollutants and sustainable treatment options. J. Hydrol. 2010, 389, 416–428. [Google Scholar] [CrossRef]
- Al Aukidy, M.; Verlicchi, P.; Jelic, A.; Petrovic, M.; Barcelò, D. Monitoring release of pharmaceutical compounds: Occurrence and environmental risk assessment of two WWTP effluents and their receiving bodies in the Po Valley, Italy. Sci. Total Environ. 2012, 438, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.T.; Hai, F.I.; Al-aboud, T.M. Chemical coagulation-based processes for trace organic contaminant removal: Current state and future potential. J. Environ. Manag. 2012, 111, 195–207. [Google Scholar] [CrossRef]
- Verlicchi, P.; Al Aukidy, M.; Zambello, E. Occurrence of pharmaceutical compounds in urban wastewater: Removal, mass load and environmental risk after a secondary treatment—A review. Sci. Total Environ. 2012, 429, 123–155. [Google Scholar] [CrossRef]
- Verlicchi, P.; Zambello, E. Pharmaceuticals and personal care products in untreated and treated sewage sludge: Occurrence and environmental risk in the case of application on soil—A critical review. Sci. Total Environ. 2015, 538, 750–767. [Google Scholar] [CrossRef]
- Pal, A.; He, Y.; Jekel, M.; Reinhard, M.; Gin, K.Y.-H. Emerging contaminants of public health significance as water quality indicator compounds in the urban water cycle. Environ. Int. 2014, 71, 46–62. [Google Scholar] [CrossRef]
- Tran, N.H.; Reinhard, M.; Gin, K.Y.-H. Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions—A review. Water Res. 2018, 133, 182–207. [Google Scholar] [CrossRef] [PubMed]
- Grelska, A.; Noszczyńska, M. White rot fungi can be a promising tool for removal of bisphenol A, bisphenol S, and nonylphenol from wastewater. Environ. Sci. Pollut. Res. 2020, 27, 39958–39976. [Google Scholar] [CrossRef] [PubMed]
- Vieno, N.; Sillanpää, M. Fate of diclofenac in municipal wastewater treatment plant—A review. Environ. Int. 2014, 69, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Schug, T.T.; Birnbaum, L.S. Human Health Effects of Bisphenol A. In Toxicants in Food Packaging and Household Plastics: Exposure and Health Risks to Consumers; Snedeker, S.M., Ed.; Springer: London, UK, 2014; pp. 1–29. [Google Scholar]
- Larsen, G.D. Transgenerational effects of BPA. Lab. Animal 2015, 44, 194. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Flores, F.G.; Isac-García, J.; Dobado, J.A. Emerging Pollutants: Origin, Structure and Properties. In Emerging Pollutants: Origin, Structure and Properties; Wiley-VCH: Weinheim, Germany, 2017; pp. 1–498. [Google Scholar]
- Bilal, M.; Iqbal, H.M.N.; Barceló, D. Mitigation of bisphenol A using an array of laccase-based robust bio-catalytic cues—A review. Sci. Total Environ. 2019, 689, 160–177. [Google Scholar] [CrossRef] [PubMed]
- Quintana, J.B.; Weiss, S.; Reemtsma, T. Pathways and metabolites of microbial degradation of selected acidic pharmaceutical and their occurrence in municipal wastewater treated by a membrane bioreactor. Water Res. 2005, 39, 2654–2664. [Google Scholar] [CrossRef]
- Yu, J.T.; Bouwer, E.J.; Coelhan, M. Occurrence and biodegradability studies of selected pharmaceuticals and personal care products in sewage effluent. Agric. Water Manag. 2006, 86, 72–80. [Google Scholar] [CrossRef]
- Li, X.; Zhou, M.; Pan, Y. Degradation of diclofenac by H2O2 activated with pre-magnetization Fe0: Influencing factors and degradation pathways. Chemosphere 2018, 212, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Ho-Young, J.; Jin-Kyu, K.; Seung-Chan, L.; Jeong-Ann, P.; Song-Bae, K. Analysis of diclofenac removal by metal-organic framework MIL-100(Fe) using multi-parameter experiments and artificial neural network modeling. J. Taiwan Inst. Chem. Eng. 2021, 121, 257–267. [Google Scholar] [CrossRef]
- Jae-Hun, C.; Jin-Kyu, K.; Seong-Jik, P.; Chang-Gu, L. Bisphenol A degradation using waste antivirus copper film with enhanced sono-Fenton-like catalytic oxidation. Chemosphere 2021, 276, 130218. [Google Scholar] [CrossRef]
- Angosto, J.M.; Roca, M.J.; Fernández-López, J.A. Removal of Diclofenac in Wastewater Using Biosorption and Advanced Oxidation Techniques: Comparative Results. Water 2020, 12, 3567. [Google Scholar] [CrossRef]
- Wang, J.; Wang, S. Removal of pharmaceuticals and personal care products (PPCPs) from wastewater: A review. J. Environ. Manag. 2016, 182, 620–640. [Google Scholar] [CrossRef]
- Sun, C.; Li, W.; Chen, Z.; Qin, W.; Wen, X. Responses of antibiotics, antibiotic resistance genes, and mobile genetic elements in sewage sludge to thermal hydrolysis pre-treatment and various anaerobic digestion conditions. Environ. Int. 2019, 133, 105156. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Cui, G.; Li, W.; Wei, Y.; Li, F. Effect of heavy metals on the performance and bacterial profiles of activated sludge in a semi-continuous reactor. Chemosphere 2020, 241, 125035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, R.; Cao, L.; Lei, Y.; Liu, J.; Feng, J.; Fu, W.; Li, X.; Li, B. High-efficiency biodegradation of chloramphenicol by enriched bacterial consortia: Kinetics study and bacterial community characterization. J. Hazard. Mater. 2020, 384, 121344. [Google Scholar] [CrossRef]
- Sankaran, S.; Khanal, S.K.; Jasti, N.; Jin, B.; Pometto, A.L.; Van Leeuwen, J.H. Use of Filamentous Fungi for Wastewater Treatment and Production of High Value Fungal Byproducts: A Review. Crit. Rev. Environ. Sci. Technol. 2010, 40, 400–449. [Google Scholar] [CrossRef]
- Mir-Tutusaus, J.A.; Baccar, R.; Caminal, G.; Sarrà, M. Can white-rot fungi be a real wastewater treatment alternative for organic micropollutants removal? A review. Water Res. 2018, 138, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Delerue-Matos, C.; Figueiredo, S.A.; Freitas, O.M. The Use of Algae and Fungi for Removal of Pharmaceuticals by Bioremediation and Biosorption Processes: A Review. Water 2019, 11, 1555. [Google Scholar] [CrossRef]
- More, T.T.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Potential use of filamentous fungi for wastewater sludge treatment. Bioresour. Technol. 2010, 101, 7691–7700. [Google Scholar] [CrossRef]
- Cruz-Morató, C.; Lucas, D.; Llorca, M.; Rodriguez-Mozaz, S.; Gorga, M.; Petrovic, M.; Barceló, D.; Vicent, T.; Sarrà, M. Marco-Urrea, E. Hospital wastewater treatment by fungal bioreactor: Removal efficiency for pharmaceuticals and endocrine disruptor compounds. Sci. Total Environ. 2014, 493, 365–376. [Google Scholar] [CrossRef]
- Olicón-Hernández, D.R.; Camacho-Morales, R.L.; Pozo, C.; González-López, J.; Aranda, E. Evaluation of diclofenac biodegradation by the ascomycete fungus Penicillium oxalicum at flask and bench bioreactor scales. Sci. Total Environ. 2019, 662, 607–614. [Google Scholar] [CrossRef]
- Chai, W.; Handa, Y.; Suzuki, M.; Saito, M.; Kato, N.; Horiuchi, C.A. Biodegradation of bisphenol a by fungi. Appl. Biochem. Biotechnol. 2005, 120, 175–182. [Google Scholar] [CrossRef]
- Marco-Urrea, E.; García-Romera, I.; Aranda, E. Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotech. 2015, 32, 620–628. [Google Scholar] [CrossRef]
- Aracagök, Y.; Goker, H.; Cihangir, N. Biodegradation of diclofenac with fungal strains. Arch. Environ. Prot. 2018, 44, 55–62. [Google Scholar] [CrossRef]
- D’Annibale, A.; Rosetto, F.; Leonardi, V.; Federici, F.; Petruccioli, M. Role of Autochthonous Filamentous Fungi in Bioremediation of a Soil Historically Contaminated with Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2006, 72, 28–36. [Google Scholar] [CrossRef]
- Mishra, A.; Malik, A. Novel fungal consortium for bioremediation of metals and dyes from mixed waste stream. Bioresour. Technol. 2014, 171, 217–226. [Google Scholar] [CrossRef]
- Sharma, S.; Malaviya, P. Bioremediation of tannery wastewater by chromium resistant novel fungal consortium. Ecol. Eng. 2016, 91, 419–425. [Google Scholar] [CrossRef]
- Waksman, S.A. A Method for Counting the Number of Fungi in the Soil. J. Bacteriol. 1922, 7, 339–341. [Google Scholar] [CrossRef]
- Vesty, A.; Biswas, K.; Taylor, M.W.; Gear, K.; Douglas, R.G. Evaluating the Impact of DNA Extraction Method on the Representation of Human Oral Bacterial and Fungal Communities. PLoS ONE 2017, 12, e0169877. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Kirk, T.K.; Schultz, E.; Connors, W.J.; Lorenz, L.F.; Zeikus, J.G. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch. Microbiol. 1978, 117, 277–285. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Dean, J.F.D.; Eriksson, K.-E.L. Laccase-mediated formation of the phenoxazinone derivative, cinnabarinic acid. FEBS Lett. 1995, 376, 202–206. [Google Scholar] [CrossRef]
- Wariishi, H.; Valli, K.; Gold, M.H. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992, 267, 23688–23695. [Google Scholar] [CrossRef]
- Ullrich, R.; Nüske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel Haloperoxidase from the Agaric Basidiomycete Agrocybe aegerita Oxidizes Aryl Alcohols and Aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar] [CrossRef]
- Reyes-César, A.; Absalón, Á.E.; Fernández, F.J.; González, J.M.; Cortés-Espinosa, D.V. Biodegradation of a mixture of PAHs by non-ligninolytic fungal strains isolated from crude oil-contaminated soil. World J. Microbiol. Biotechnol. 2014, 30, 999–1009. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Pérez-Díaz, M.I.; Zárate-Segura, P.; Bermeo-Fernández, L.A.; Nirmalkar, K.; Bastida-González, F.; García-Mena, J.; Jan-Roblero, J.; Guerrero-Barajas, C. Bacterial consortium from hydrothermal vent sediments presents electrogenic activity achieved under sulfate reducing conditions in a microbial fuel cell. J. Environ. Health Sci. Eng. 2020, 18, 1189–1205. [Google Scholar] [CrossRef]
- Costa, J.M.; Rodriguez, R.P.; Sancinetti, G.P. Removal sulfate and metals Fe+2, Cu+2, and Zn+2 from acid mine drainage in an anaerobic sequential batch reactor. J. Environ. Chem. Eng. 2017, 5, 1985–1989. [Google Scholar] [CrossRef]
- Martins, M.; Faleiro, M.L.; Barros, R.J.; Veríssimo, A.R.; Barreiros, M.A.; Costa, M.C. Characterization and activity studies of highly heavy metal resistant sulphate-reducing bacteria to be used in acid mine drainage decontamination. J. Hazard. Mater. 2009, 166, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Militon, C.; Hamdi, O.; Michotey, V.; Fardeau, M.L.; Ollivier, B.; Bouallagui, H.; Hamdi, M.; Bonin, P. Ecological significance of Synergistetes in the biological treatment of tuna cooking wastewater by an anaerobic sequencing batch reactor. Environ. Sci. Pollut. Res. Int. 2015, 22, 18230–18238. [Google Scholar] [CrossRef] [PubMed]
- Elshahed, M.S.; Bhupathiraju, V.K.; Wofford, N.Q.; Nanny, M.A.; McInerney, M.J. Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “Syntrophus aciditrophicus” strain SB in syntrophic association with H(2)-using microorganisms. Appl. Environ. Microbiol. 2001, 67, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Grimm, F.; Franz, B.; Dahl, C. Regulation of dissimilatory sulfur oxidation in the purple sulfur bacterium allochromatium vinosum. Front. Microbiol. 2011, 2, 51. [Google Scholar] [CrossRef] [PubMed]
- Kragelund, C.; Thomsen, T.R.; Mielczarek, A.T.; Nielsen, P.H. Eikelboom’s morphotype 0803 in activated sludge belongs to the genus Caldilinea in the phylum Chloroflexi. FEMS Microbiol. Ecol. 2011, 76, 451–462. [Google Scholar] [CrossRef]
- Yoon, D.N.; Park, S.J.; Kim, S.J.; Jeon, C.O.; Chae, J.C.; Rhee, S.K. Isolation, characterization, and abundance of filamentous members of Caldilineae in activated sludge. J. Microbiol. 2010, 48, 275–283. [Google Scholar] [CrossRef]
- Ju, F.; Guo, F.; Ye, L.; Xia, Y.; Zhang, T. Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 2014, 6, 80–89. [Google Scholar] [CrossRef]
- Bogan, B.W.; Lahner, L.M.; Sullivan, W.R.; Paterek, J.R. Degradation of straight-chain aliphatic and high-molecular-weight polycyclic aromatic hydrocarbons by a strain of Mycobacterium austroafricanum. J. Appl. Microbiol. 2003, 94, 230–239. [Google Scholar] [CrossRef]
- Dudhagara, D.; Dave, B. Mycobacterium as Polycyclic Aromatic Hydrocarbons (PAHs) Degrader. In Mycobacterium—Research and Development; Ribón, W., Ed.; IntechOpen Limited: London, UK, 2018. [Google Scholar]
- Rojo, F.; Martínez, J.L. Hydrocarbon Degraders as Pathogens. In Health Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids; Goldfine, H., Ed.; Springer International Publishing: Cham, Germany, 2020; pp. 267–281. [Google Scholar]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Aranda, E.; Godoy, P.; Reina, R.; Badia-Fabregat, M.; Rosell, M.; Marco-Urrea, E.; García-Romera, I. Isolation of Ascomycota fungi with capability to transform PAHs: Insights into the biodegradation mechanisms of Penicillium oxalicum. Int. Biodeterior. Biodegrad. 2017, 122, 141–150. [Google Scholar] [CrossRef]
- Godoy, P.; Reina, R.; Calderón, A.; Wittich, R.-M.; García-Romera, I.; Aranda, E. Exploring the potential of fungi isolated from PAH-polluted soil as a source of xenobiotics-degrading fungi. Environ. Sci. Pollut. Res. 2016, 23, 20985–20996. [Google Scholar] [CrossRef] [PubMed]
- Olicón-Hernández, D.R.; Gómez-Silván, C.; Pozo, C.; Andersen, G.L.; González-Lopez, J.; Aranda, E. Penicillium oxalicum XD-3.1 removes pharmaceutical compounds from hospital wastewater and outcompetes native bacterial and fungal communities in fluidised batch bioreactors. Int. Biodeterior. Biodegrad. 2021, 158, 105179. [Google Scholar] [CrossRef]
- Assress, H.A.; Selvarajan, R.; Nyoni, H.; Ntushelo, K.; Mamba, B.B.; Msagati, T.A.M. Diversity, Co-occurrence and Implications of Fungal Communities in Wastewater Treatment Plants. Sci. Rep. 2019, 9, 14056. [Google Scholar] [CrossRef] [PubMed]
- Maza-Márquez, P.; Aranda, E.; González-López, J.; Rodelas, B. Evaluation of the Abundance of Fungi in Wastewater Treatment Plants Using Quantitative PCR (qPCR). In Quantitative Real-Time PCR: Methods and Protocols; Biassoni, R., Raso, A., Eds.; Springer: New York, NY, USA, 2020; pp. 79–94. [Google Scholar]
- Hata, T.; Kawai, S.; Okamura, H.; Nishida, T. Removal of diclofenac and mefenamic acid by the white rot fungus Phanerochaete sordida YK-624 and identification of their metabolites after fungal transformation. Biodegradation 2010, 21, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Lucero Camacho-Morales, R.; García-Fontana, C.; Fernández-Irigoyen, J.; Santamaría, E.; González-López, J.; Manzanera, M.; Aranda, E. Anthracene drives sub-cellular proteome-wide alterations in the degradative system of Penicillium oxalicum. Ecotoxicol. Environ. Safe. 2018, 159, 127–135. [Google Scholar] [CrossRef]
- Liu, L.; Wang, S.; Chen, J. Hysteretic response of Microbial Eukaryotic Communities to Gradually Decreased Nutrient Concentrations in Eutrophic Water. Microb. Ecol. 2020, 79, 815–822. [Google Scholar] [CrossRef]
- Sangale, M.K.; Shahnawaz, M.; Ade, A.B. Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene. Sci. Rep. 2019, 9, 5390. [Google Scholar] [CrossRef]
- Anasonye, F.; Winquist, E.; Räsänen, M.; Kontro, J.; Björklöf, K.; Vasilyeva, G.; Jørgensen, K.S.; Steffen, K.T.; Tuomela, M. Bioremediation of TNT contaminated soil with fungi under laboratory and pilot scale conditions. Int. Biodeterior. Biodegrad. 2015, 105, 7–12. [Google Scholar] [CrossRef]
- Launen, L.; Pinto, L.; Wiebe, C.; Kiehlmann, E.; Moore, M. The oxidation of pyrene and benzo[a]pyrene by nonbasidiomycete soil fungi. Can. J. Microbiol. 1995, 41, 477–488. [Google Scholar] [CrossRef]
- Dalecka, B.; Oskarsson, C.; Juhna, T.; Kuttava Rajarao, G. Isolation of Fungal Strains from Municipal Wastewater for the Removal of Pharmaceutical Substances. Water 2020, 12, 524. [Google Scholar] [CrossRef]
- Martínková, L.; Kotik, M.; Marková, E.; Homolka, L. Biodegradation of phenolic compounds by Basidiomycota and its phenol oxidases: A review. Chemosphere 2016, 149, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Daâssi, D.; Prieto, A.; Zouari-Mechichi, H.; Martínez, M.J.; Nasri, M.; Mechichi, T. Degradation of bisphenol A by different fungal laccases and identification of its degradation products. Int. Biodeterior. Biodegrad. 2016, 110, 181–188. [Google Scholar] [CrossRef]
- Shin, E.H.; Choi, H.T.; Song, H.G. Biodegradation of endocrine-disrupting bisphenol A by white rot fungus Irpex lacteus. J. Microbiol. Biotechnol. 2007, 17, 1147–1151. [Google Scholar]
- Hirano, T.; Honda, Y.; Watanabe, T.; Kuwahara, M. Degradation of Bisphenol A by the Lignin-Degrading Enzyme, Manganese Peroxidase, Produced by the White-rot Basidiomycete, Pleurotus ostreatus. Biosci. Biotechnol. Biochem. 2000, 64, 1958–1962. [Google Scholar] [CrossRef] [PubMed]
- Mtibaà, R.; Olicón-Hernández, D.R.; Pozo, C.; Nasri, M.; Mechichi, T.; González, J.; Aranda, E. Degradation of bisphenol A and acute toxicity reduction by different thermo-tolerant ascomycete strains isolated from arid soils. Ecotoxicol. Environ. Safe 2018, 156, 87–96. [Google Scholar] [CrossRef] [PubMed]

| Accession Number | BLAST Percent of Identity | Phylum | Isolated Fungi Strain | Total DFC Consumption | Total BPA Consumption |
|---|---|---|---|---|---|
| In correction | 99.34 | Ascomycota | Talaromyces gossypii | 84.6% | 86.6% |
| MW931877 | 96.30 | Zygomycota | Syncephalastrum monosporum | 82% | 100% |
| In correction | 99.16 | Ascomycota | Aspergillus tabacinus | 76% | - |
| MW931860 | 100 | Ascomycota | Talaromyces verruculosus | 37% | 36% |
| MT792070 | 100 | Ascomycota | Aspergillus terreus | 49.7% | - |
| MW931880 | 100 | Ascomycota | Aspergillus cejpii | 14.6% | - |
| MW931859 | 100 | Ascomycota | Galactomyces candidum | - | - |
| MW931874 | 100 | Ascomycota | Galactomyces geotrichum | - | - |
| MT792001 | 100 | Ascomycota | Saccharomycetales sp. | - | - |
| MW931860 | 100 | Ascomycota | Aspergillus cremeus | - | - |
| MT792005 | 100 | Ascomycota | Byssochlamys nivea | - | - |
| MT787659 | 100 | Ascomycota | Trichoderma asperellum | - | - |
| MT792081 | 95.34 | Ascomycota | Scedosporium aurantiacum | - | - |
| MT792234 | 100 | Ascomycota | Sporothrix mexicana | - | - |
| MT792235 | 100 | Ascomycota | Aspergillus sydowii | - | - |
| MT787565 | 100 | Mucoromyceta | Mucor circinelloides | - | - |
| MW931863 | 99.82 | Ascomycota | Talaromyces pinophilus | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conejo-Saucedo, U.; Ledezma-Villanueva, A.; Ángeles de Paz, G.; Herrero-Cervera, M.; Calvo, C.; Aranda, E. Evaluation of the Potential of Sewage Sludge Mycobiome to Degrade High Diclofenac and Bisphenol-A Concentrations. Toxics 2021, 9, 115. https://doi.org/10.3390/toxics9060115
Conejo-Saucedo U, Ledezma-Villanueva A, Ángeles de Paz G, Herrero-Cervera M, Calvo C, Aranda E. Evaluation of the Potential of Sewage Sludge Mycobiome to Degrade High Diclofenac and Bisphenol-A Concentrations. Toxics. 2021; 9(6):115. https://doi.org/10.3390/toxics9060115
Chicago/Turabian StyleConejo-Saucedo, Ulises, Alejandro Ledezma-Villanueva, Gabriela Ángeles de Paz, Mario Herrero-Cervera, Concepción Calvo, and Elisabet Aranda. 2021. "Evaluation of the Potential of Sewage Sludge Mycobiome to Degrade High Diclofenac and Bisphenol-A Concentrations" Toxics 9, no. 6: 115. https://doi.org/10.3390/toxics9060115
APA StyleConejo-Saucedo, U., Ledezma-Villanueva, A., Ángeles de Paz, G., Herrero-Cervera, M., Calvo, C., & Aranda, E. (2021). Evaluation of the Potential of Sewage Sludge Mycobiome to Degrade High Diclofenac and Bisphenol-A Concentrations. Toxics, 9(6), 115. https://doi.org/10.3390/toxics9060115







