Amikacin Suppresses Human Breast Cancer Cell MDA-MB-231 Migration and Invasion
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Amikacin Treatment
2.2. Wound-Healing Assay
2.3. Invasion Ability Assay
2.4. Western Blotting
2.5. Transfection and siRNA Knockdown
2.6. Antibody Labeling and Images
2.7. Statistical Analysis
3. Results
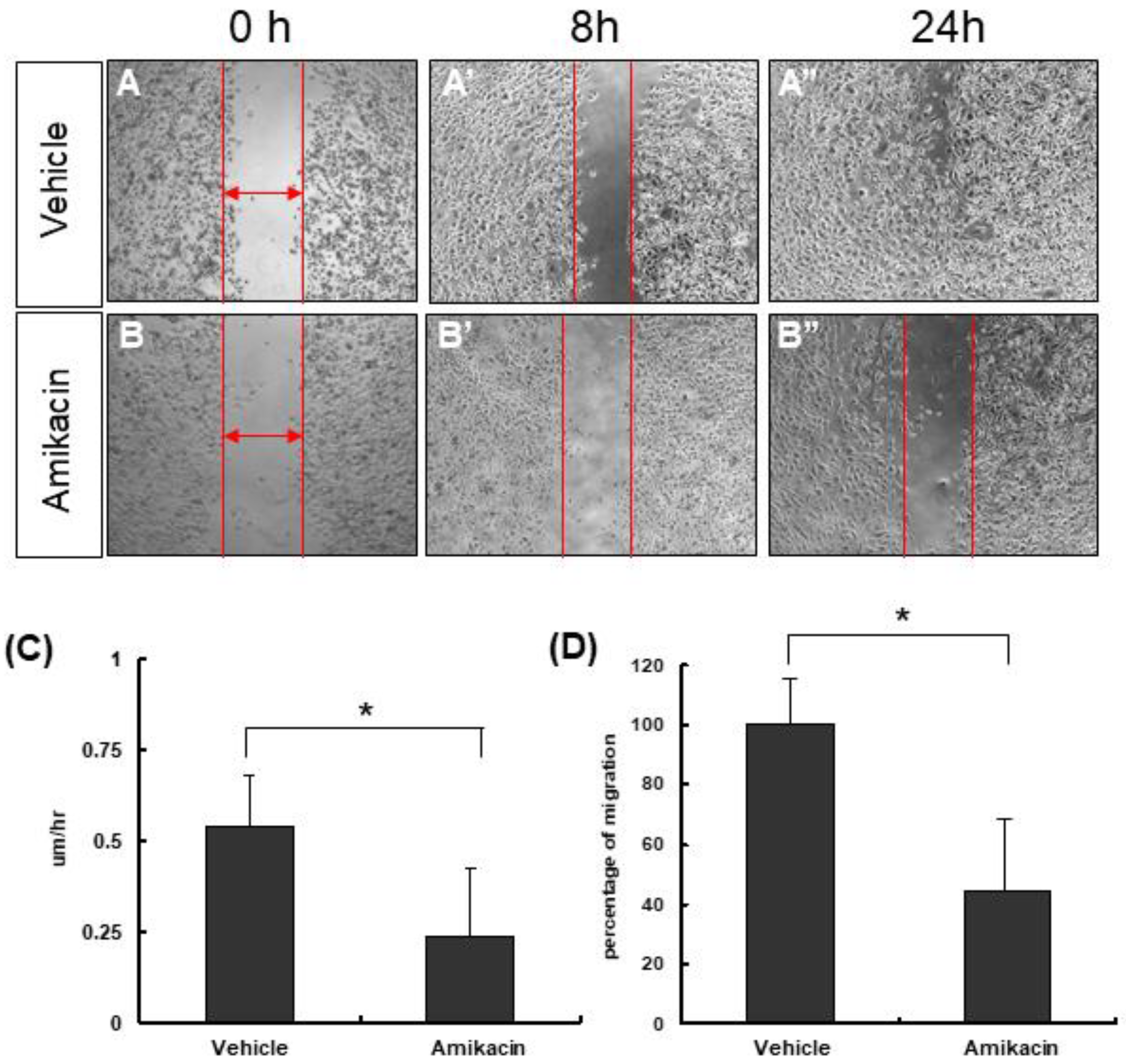
3.1. The Effects of Amikacin on MDA-MB-231 Cell Proliferation and Migration
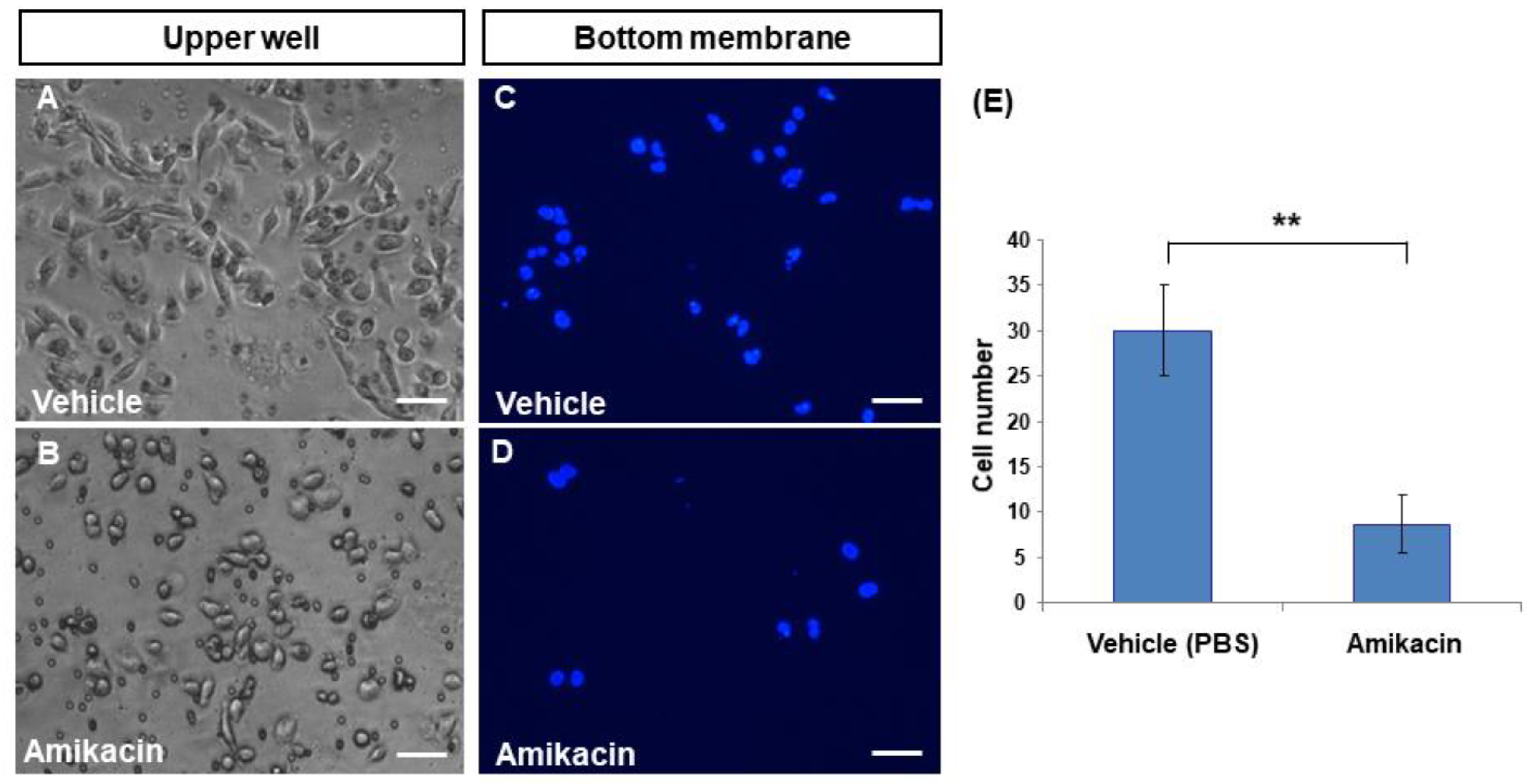
3.2. Amikacin Inhibits MDA-MB-231 Cell Invasion
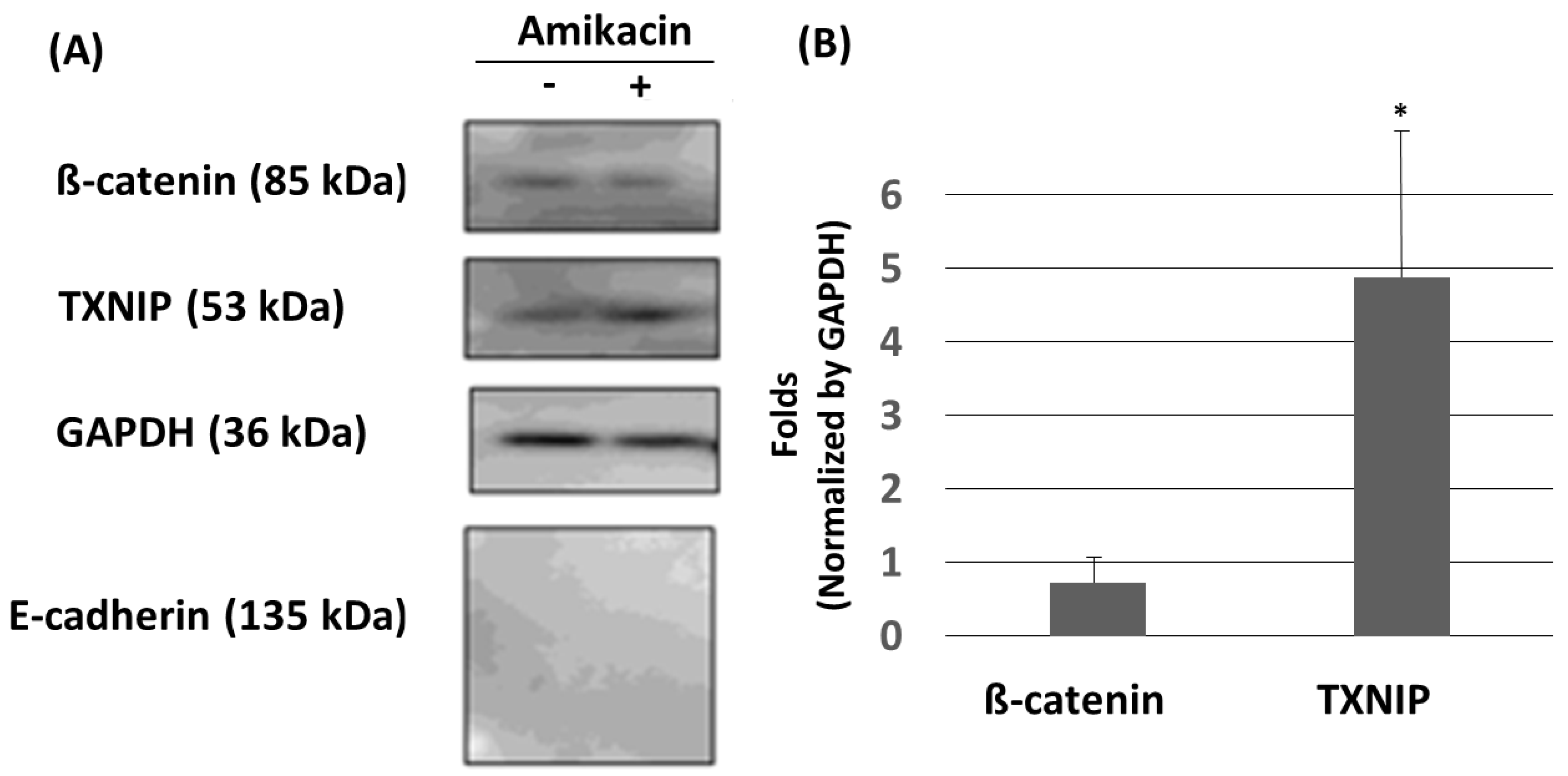
3.3. Amikacin Treatment Leads to Up-Regulation of TXNIP
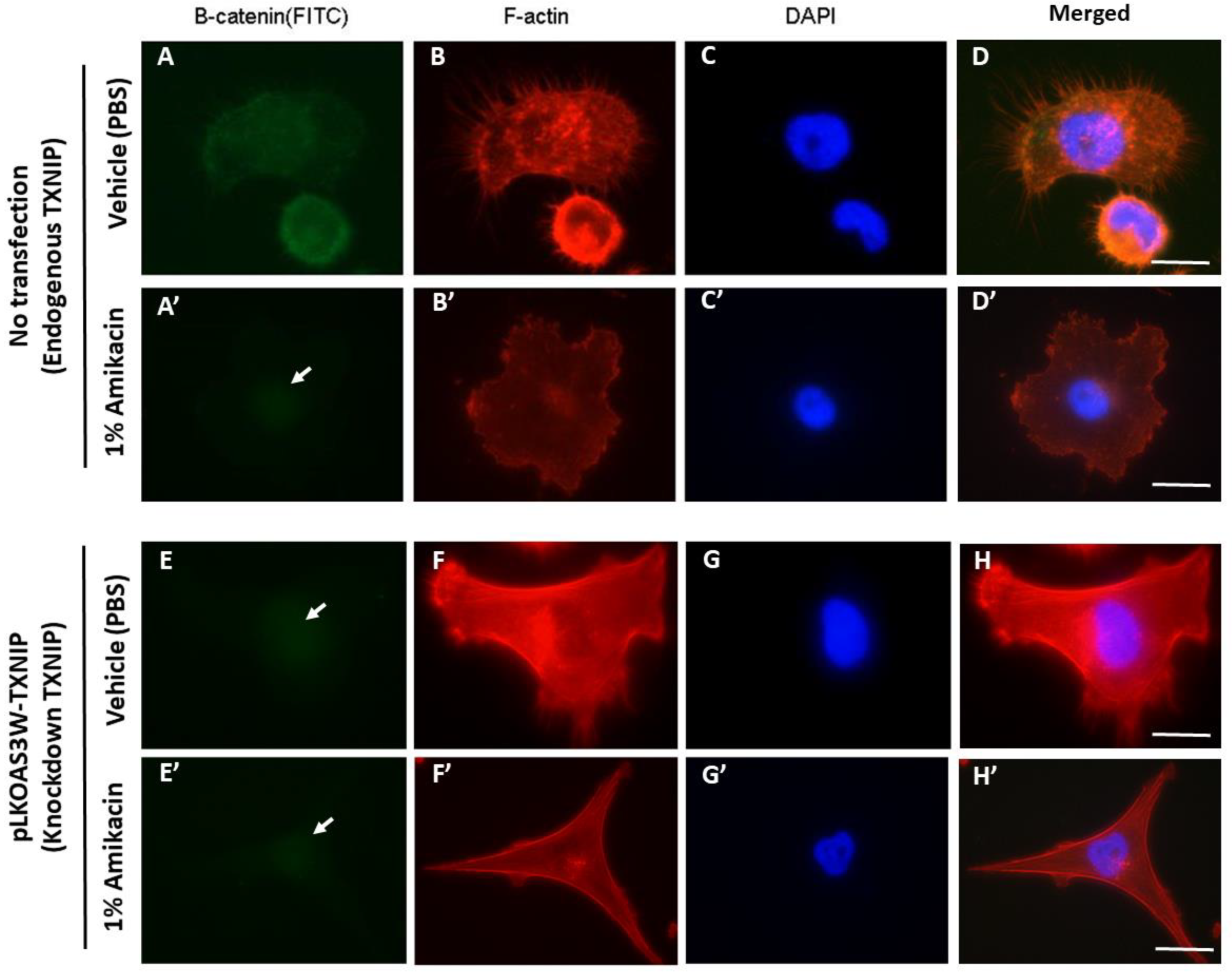
3.4. Knockdown of TXNIP Affects Cell Morphology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. Available online: http://www.cdc.gov/cancer/dataviz (accessed on 1 June 2019).
- Khosravi, S.P.; Cabezón, G.L.; Aparicio, M.I. State of art of advanced triple negative breast cancer. Breast J. 2019, 25, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Geiger, B.; Deeb, N.; Rothschild, M.F. Investigation of TXNIP (thioredoxin-interacting protein) and TRX (thioredoxin) genes for growth-related traits in pigs. Mammalian Genome 2007, 25, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, W.S.; Kim, D.O.; Byun, J.E.; Hangsak, H.; Lee, S.Y.; Song, H.Y.; Park, Y.J.; Kim, T.D.; Yoon, S.R.; et al. Macrophage migration inhibitory factor interacts with thioredoxin-interacting protein and induces NF-κB activity. Cell Signal 2017, 34, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; O’Shea, J.M.; Kaadige, M.R.; Cunha, S.; Wilde, B.R.; Cohen, A.L.; Welm, A.L.; Ayer, D.E. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc. Natl. Acad. Sci. USA 2015, 112, 5425–5430. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Zhou, L.; Liu, H.; Shan, Y.; Zhang, X. MicroRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1α pathway. Cell. Physiol. Biochem. 2018, 48, 1735–1746. [Google Scholar] [CrossRef]
- Li, J.; Yue, Z.; Xiong, W.; Sun, P.; You, K.; Wang, J. TXNIP overexpression suppresses proliferation and induces apoptosis in SMMC7221 cells through ROS generation and MAPK pathway activation. Oncol. Rep. 2017, 37, 3369–3376. [Google Scholar] [CrossRef]
- Jia, J.J.; Geng, W.S.; Wang, Z.Q.; Chen, L.; Zeng, X.S. The role of thioredoxin system in cancer: Strategy for cancer therapy. Cancer Chemother. Pharmacol. 2019, 84, 453–470. [Google Scholar] [CrossRef]
- Xiao, X.; Xu, Y.; Chen, H. Sodium butyrate-activated TRAF6-TXNIP pathway affects A549 cells proliferation and migration. Cancer Med. 2020, 9, 3477–3488. [Google Scholar] [CrossRef]
- Huczynski, A. Salinomycin: A new cancer drug candidate. Chem. Biol. Drug Des. 2012, 79, 235–238. [Google Scholar] [CrossRef]
- Kim, K.Y.; Park, K.I.; Kim, S.H.; Yu, S.N.; Park, S.G.; Kim, Y.W.; Seo, Y.K.; Ma, J.Y.; Ahn, S.C. Inhibition of autophagy promotes salinomycin-induced apoptosis via reactive oxygen species-mediated PI3K/AKT/mTOR and ERK/p38 MAPK-dependent signaling in human prostate cancer cells. Int. J. Mol. Sci. 2017, 18, 1088. [Google Scholar] [CrossRef]
- Manmuan, S.; Sakunrangsit, N.; Ketchart, W. Salinomycin overcomes acquired tamoxifen resistance through AIB1 and inhibits cancer cell invasion in endocrine resistant breast cancer. Clin. Exp. Pharmacol. Physiol. 2017, 44, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Luo, Q.; Liu, L.; Yang, X.; Zhu, S.; Song, G. Salinomycin attenuates liver cancer stem cell motility by enhancing cell stiffness and increasing F-actin formation via the FAK-ERK1/2 signaling pathway. Toxicology 2017, 384, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tian, X.; Yin, H.; Xiao, S.; Yi, S.; Zhang, Y.; Zeng, F. TXNIP induced by MondoA, rather than ChREBP, suppresses cervical cancer cell proliferation, migration and invasion. J. Biochem. 2020, 167, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Liu, J.; Shao, L. Rapamycin inhibits the proliferation and apoptosis of gastric cancer cells by down regulating the expression of surviving. Hepatogastroenterology 2011, 58, 1075–1080. [Google Scholar]
- Ristuccia, A.M.; Cunha, B.A. An overview of amikacin. Ther. Drug Monit. 1985, 7, 12–25. [Google Scholar] [CrossRef]
- Abe, J.; Yamada, Y.; Harashima, H. Validation of a strategy for cancer therapy: Delivering aminoglycoside drugs to mitochondria in HeLa cells. J. Pharm. Sci. 2016, 105, 734–740. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Belakhov, V.; Baasov, T.; Namy, O. Characterization of new-generation aminoglycoside promoting premature termination codon read through in cancer cells. RNA Biol. 2017, 14, 378–388. [Google Scholar] [CrossRef]
- Floquet, C.; Deforges, J.; Rousset, J.P.; Bidou, L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 2011, 39, 3350–3362. [Google Scholar] [CrossRef]
- Kumar, A.V.; Gassar, E.S.; Spillmann, D.; Stock, C.; Sen, Y.P.; Zhang, T.; Kuppevelt, T.H.V.; Hülsewig, C.; Koszlowski, E.O.; Pavao, M.S.G.; et al. HS3ST2 modulates breast cancer cell invasiveness via MAP kinase- and Tcf4 (Tcf7l2)-dependent regulation of protease and cadherin expression. Cancer Cell Biol. 2014, 135, 2579–2592. [Google Scholar]
- Chen, H.F.; Ma, R.R.; He, J.Y.; Zhang, H.; Liu, X.L.; Guo, X.Y.; Gao, P. Protocadherin 7 inhibits cell migration and invasion through E-cadherin in gastric cancer. Tumour Biol. 2017, 39, 1010428317697551. [Google Scholar] [CrossRef]
- Mendonsa, A.M.; Na, T.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef] [PubMed]
- Fezza, M.; Moussa, M.; Aoun, R.; Haber, R.; Hilal, G. DKK1 promotes hepatocellular carcinoma inflammation, migration and invasion: Implication of TGF-β1. PLoS ONE 2019, 14, 0223252. [Google Scholar] [CrossRef] [PubMed]
- Lombaerts, M.; Wezel, T.V.; Philippo, K.; Dierssen, J.W.F.; Zimmerman, R.M.E.; Oosting, J.; Eijk, R.V.; Eilers, P.H.; Water, B.V.D.; Cornelisse, C.J. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br. J Cancer 2006, 94, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, H.; Ma, X.; Lin, C.; Lu, L.; Liu, D.; Ma, D.; Gao, X.; Qian, X.Y. Prodigiosin inhibits proliferation, migration, and invasion of nasopharyngeal cancer cells. Cell Physiol. Biochem. 2018, 48, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, D.V.; Ward, D.A.; Barnbill, M.A. Effects of antibiotics on morphologic characteristics and migration of canine corneal epithelial cells in tissue culture. Am. J. Vet. Res. 2011, 62, 1664–1669. [Google Scholar] [CrossRef] [PubMed]
- Pilge, H.; Fröbel, J.; Höhn, L.S.; Zilkens, C.; Krauspe, R. Cefazolin irreversibly inhibits proliferation and migration of human mesenchymal stromal cells. BioMed Res. Int. 2016, 2016, 2042687. [Google Scholar] [CrossRef][Green Version]
- Arriola, J.; Foote, R.H. Effects of amikacin sulfate on the motility of stallion and bull spermatozoa at different temperatures and intervals of storage. J. Anim. Sci. 1982, 54, 1105–1110. [Google Scholar] [CrossRef]
- Zhao, L.D.; Li, L.; Wu, N.; Li, D.K.; Ren, L.L.; Guo, W.W.; Sun, J.H.; Liu, H.Z.; Chen, Z.T.; Xing, G.Q.; et al. Migration and differentiation of mouse embryonic stem cells transplanted into mature cochlea of rats with aminoglycoside-induced hearing loss. Acta Otolaryngol. 2013, 133, 136–143. [Google Scholar] [CrossRef]
- Vollmer, W. The prokaryotic cytoskeleton: A putative target for inhibitors and antibiotics? Appl. Microbiol. Biotechnol. 2006, 73, 37–47. [Google Scholar] [CrossRef]
- Ali, B.M.; Zaitone, S.A.; Shouman, S.A.; Moustafa, Y.M. Dorzolamide synergizes the antitumor activity of mitomycin C against Ehrlich’s carcinoma grown in mice: Role of thioredoxin-interacting protein. Naunyn. Schmiedebergs Arch. Pharmacol. 2015, 388, 1271–1282. [Google Scholar] [CrossRef]
- Ikeda, A.; Nemoto, K.; Yoshida, C.; Miyata, S.; Mori, J.; Soejima, S.; Yokosuka, A.; Mimaki, Y.; Ohizumi, Y.; Degawa, M. Suppressive effect of nobiletin, a citrus polymethoxyflavonoid that downregulates thioredoxin-interacting protein expression, on tunicamycin-induced apoptosis in SK-N-SH human neuroblastoma cells. Neurosci. Lett. 2013, 549, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Zhang, Z.; Gao, D.; Chan, S.; Luo, J.; Tu, Z.C.; Zhang, Z.M.; Ding, K.; Ren, X.; Lu, X. Quinolone antibiotic derivatives as new selective Axl kinase inhibitors. Eur. J. Med. Chem. 2019, 166, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, X.; Wang, T.; Xiao, J. Quinolone hybrids and their anti-cancer activities: An overview. Eur. J. Med. Chem. 2019, 165, 59–79. [Google Scholar] [CrossRef]
- Abe, A.; Yamada, H.; Moriya, S.; Miyazawa, K. The β-carboline alkaloid harmol induces cell death via autophagy but not apoptosis in human non-small cell lung cancer A549 cells. Biol. Pharm. Bull. 2012, 34, 1264–1272. [Google Scholar] [CrossRef]
- Luo, Y.; Zeng, A.; Fang, A.; Song, L.; Fan, C.; Zeng, C.; Ye, T.; Chen, H.; Tu, C.; Xie, Y. Nifuroxazide induces apoptosis, inhibits cell migration and invasion in osteosarcoma. Investig. New Drugs 2019, 37, 1006–1013. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, W.; Wang, K.; Huang, Z.; Zhang, L.; Zhang, Z.; Zhou, J.; Nice, E.C.; Huang, C. Brefeldin A inhibits colorectal cancer growth by triggering Bip/Akt-regulated autophagy. FASEB. J. 2019, 33, 5520–5534. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-H.; Chen, Y.-H.; Shen, W.-H. Amikacin Suppresses Human Breast Cancer Cell MDA-MB-231 Migration and Invasion. Toxics 2020, 8, 108. https://doi.org/10.3390/toxics8040108
Wang Y-H, Chen Y-H, Shen W-H. Amikacin Suppresses Human Breast Cancer Cell MDA-MB-231 Migration and Invasion. Toxics. 2020; 8(4):108. https://doi.org/10.3390/toxics8040108
Chicago/Turabian StyleWang, Yun-Hsin, Yau-Hung Chen, and Wen-Hao Shen. 2020. "Amikacin Suppresses Human Breast Cancer Cell MDA-MB-231 Migration and Invasion" Toxics 8, no. 4: 108. https://doi.org/10.3390/toxics8040108
APA StyleWang, Y.-H., Chen, Y.-H., & Shen, W.-H. (2020). Amikacin Suppresses Human Breast Cancer Cell MDA-MB-231 Migration and Invasion. Toxics, 8(4), 108. https://doi.org/10.3390/toxics8040108




