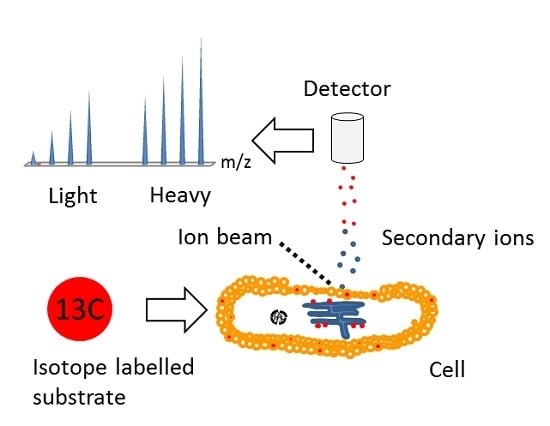
Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS): A New Tool for the Analysis of Toxicological Effects on Single Cell Level
Abstract
:1. Introduction
2. Cryopreservation of Cells in a Native State
3. Progress of ToF-SIMS Instrumentation: New Ion Cluster Guns and High Mass Resolution
3.1. Bacterial Phenotyping
3.2. Imaging of Mammalian Cervical Carcinoma Cells
3.3. Association of Cholesterol Distribtion and Alzheimer Disease
3.4. Behaviour of Hydroxypropylcellulose during the Granulation of Pharamceuticals
4. Multi Isotope Imaging Mass Spectrometry (MIMS)
4.1. MIMS and the Immortal Strand Hypothesis
4.2. Incorporation of Trehalose into the Nucleus of Mouse Sperm Cells
4.3. Internalisation of p-boronphenylalanine by Human Glioblastoma Cells
4.4. Glutamine Metabolism in Breast Cancer Cells
4.5. Uptake of Chemotherapeutics by Breast Cancer Cells
5. New Developments towards ToF-SIMS Quantification: Some Examples
5.1. Quantification of Choline in Motor Neurons
5.2. Calcium Redistribution in Certain Cell Types is Associated with Retinal Ischemia
5.3. Conversation of Strontium from Implants to Extracellular Bone Matrix
5.4. Single Cell Analysis of Aquatic Organisms
5.5. Nickel Distribution in Plants
6. Lipidomics
6.1. Differentiation of Mammalian Cell Types
6.2. Phospholipid Composition Influences on the Communication Capacity of Neighbouring Cells
6.3. Differentiation of Macrophages Exposed against Silver Nanoparticles
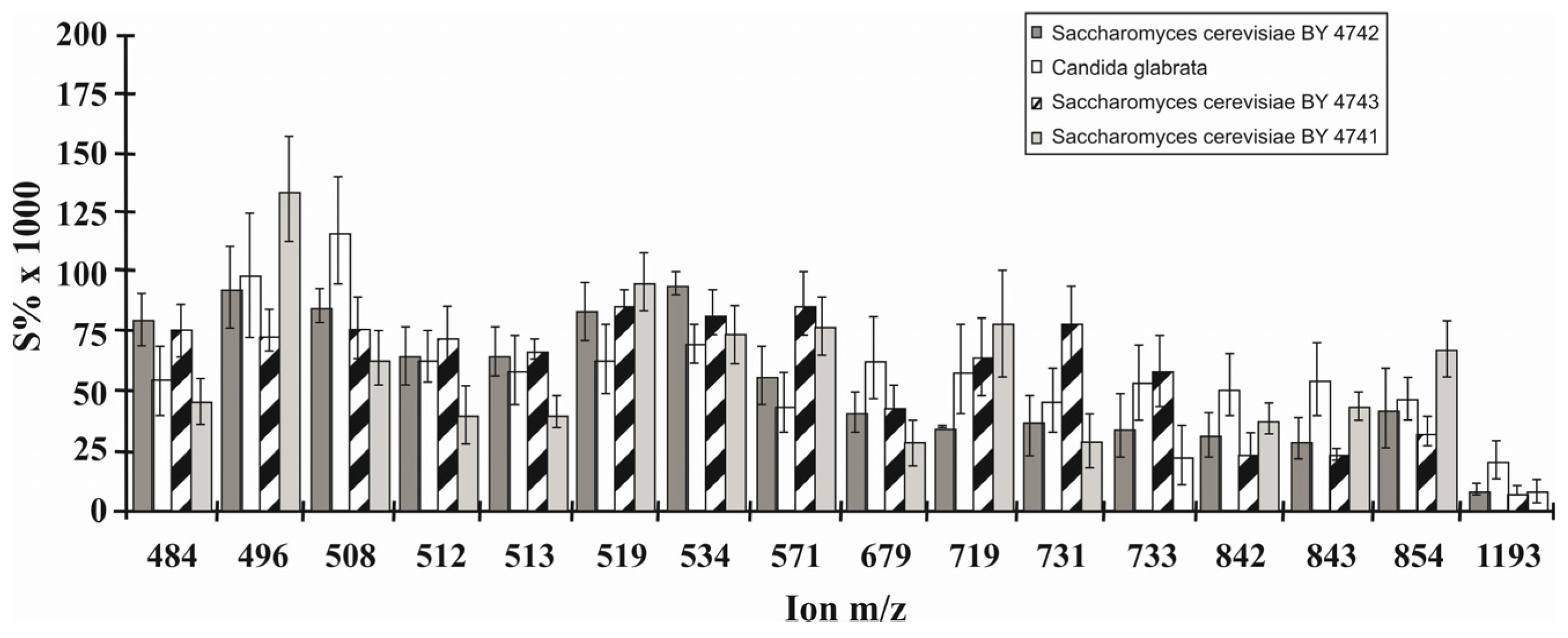
6.4. Phospholipid Pattern of Yeasts Enables for Species Discrimination

6.5. Identification of a Diatom Microalgae in a Microbial Mat
6.6. Lipid Formation Changes in the Single Cell Organism Tetrahymena Thermophila during the Mating Process
6.7. Lipid Profiling of Xenopus Laevis Embryos
6.8. Palmitoleic Acid Content of Human Breast Cancer Stem Cells is Reduced
6.9. Potential Skin Penetration of Cosmetic Ingredients
7. Conclusions
Conflicts of Interest
References
- Comolli, L.R.; Duarte, R.; Baum, D.; Luef, B.; Downing, K.H.; Larson, D.M.; Csencsits, R.; Banfield, J.F. A portable cryo-plunger for on-site intact cryogenic microscopy sample preparation in natural environments. Microsc. Res. Techniq. 2012, 75, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Berman, E.S.F.; Fortson, S.L.; Checchi, K.D.; Wu, L.; Felton, J.S.; Wu, K.J.J.; Kulp, K.S. Preparation of single cells for imaging/profiling mass spectrometry. J. Am. Soc. Mass. Spectr. 2008, 19, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Sjovall, P.; Lausmaa, J.; Nygren, H.; Carlsson, L.; Malmberg, P. Imaging of membrane lipids in single cells by imprint-imaging time-of-flight secondary ion mass spectrometry. Anal. Chem. 2003, 75, 3429–3434. [Google Scholar] [CrossRef] [PubMed]
- Malm, J.; Giannaras, D.; Riehle, M.O.; Gadegaard, N.; Sjovall, P. Fixation and drying protocols for the preparation of cell samples for time-of-flight secondary ion mass spectrometry analysis. Anal. Chem. 2009, 81, 7197–7205. [Google Scholar] [CrossRef] [PubMed]
- Winograd, N. Imaging mass spectrometry on the nanoscale with cluster ion beams. Anal. Chem. 2015, 87, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, C.M.; Roberson, S.V.; Gillen, G. Depth profiling of 4-acetamindophenol-doped poly(lactic acid) films using cluster secondary ion mass spectrometry. Anal. Chem. 2004, 76, 3199–3207. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, J.C.; Winograd, N. SIMS—A precursor and partner to contemporary mass spectrometry. Int J. Mass. Spectrom. 2015, 377, 568–579. [Google Scholar] [CrossRef]
- Holzlechner, G.; Kubicek, M.; Hutter, H.; Fleig, J. A novel ToF-SIMS operation mode for improved accuracy and lateral resolution of oxygen isotope measurements on oxides. J. Anal. Atom. Spectrom. 2013, 28, 1080–1089. [Google Scholar] [CrossRef]
- Fletcher, J.S.; Henderson, A.; Biddulph, G.X.; Vaidyanathan, S.; Lockyer, N.P.; Vickerman, J.C. Uncovering new challenges in bio-analysis with ToF-SIMS. Appl. Surf. Sci. 2008, 255, 1264–1270. [Google Scholar] [CrossRef]
- Touboul, D.; Brunelle, A.; Halgand, F.; de la Porte, S.; Laprevote, O. Lipid imaging by gold cluster time-of-flight secondary ion mass spectrometry: Application to duchenne muscular dystrophy. J. Lipid Res. 2005, 46, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, S.; Brison, J.; Castner, D.G. Exploring the surface sensitivity of ToF-secondary ion mass spectrometry by measuring the implantation and sampling depths of Bin and C60 ions in organic films. Anal. Chem. 2012, 84, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Blenkinsopp, P.; Thompson, S.; Vickerman, J.; Fletcher, J.S. A new time-of-flight SIMS instrument for 3D imaging and analysis. Surf. Interf. Anal. 2011, 43, 506–509. [Google Scholar] [CrossRef]
- Holzweber, M.; Shard, A.G.; Jungnickel, H.; Luch, A.; Unger, W.E.S. Dual beam organic depth profiling using large argon cluster ion beams. Surf. Interf. Anal. 2014, 46, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.S.; Lockyer, N.P.; Vaidyanathan, S.; Vickerman, J.C. Tof-SIMS 3D biomolecular imaging of Xenopus laevis oocytes using buckminsterfullerene (C60) primary ions. Anal. Chem. 2007, 79, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Passarelli, M.K.; Ewing, A.G.; Winograd, N. Single-cell lipidomics: Characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal. Chem. 2013, 85, 2231–2238. [Google Scholar] [CrossRef] [PubMed]
- 3D Nanosims—Label-Free Molecular Imaging. Available online: http://www.npl.co.uk/news/3d-nanosims-label-free-molecular-imaging (accessed on 28 January 2016).
- Fletcher, J.S.; Rabbani, S.; Barber, A.M.; Lockyer, N.P.; Vickerman, J.C. Comparison of C60 and GCIB primary ion beams for the analysis of cancer cells and tumour sections. Surf. Interf. Anal. 2013, 45, 273–276. [Google Scholar] [CrossRef]
- Angerer, T.B.; Pour, M.D.; Malmberg, P.; Fletcher, J.S. Improved molecular imaging in rodent brain with time-of-flight-secondary ion mass spectrometry using gas cluster ion beams and reactive vapor exposure. Anal. Chem. 2015, 87, 4305–4313. [Google Scholar] [CrossRef] [PubMed]
- Wehrli, P.M.; Lindberg, E.; Angerer, T.B.; Wold, A.E.; Gottfries, J.; Fletcher, J.S. Maximising the potential for bacterial phenotyping using time-of-flight secondary ion mass spectrometry with multivariate analysis and tandem mass spectrometry. Surf. Interf. Anal. 2014, 46, 173–176. [Google Scholar] [CrossRef]
- Lepine, F.; Milot, S.; Deziel, E.; He, J.X.; Rahme, L.G. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J. Am. Soc. Mass. Spectr. 2004, 15, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, S.; Fletcher, J.S.; Lockyer, N.P.; Vickerman, J.C. Exploring subcellular imaging on the buncher-ToF J105 3D chemical imager. Surf. Interf. Anal. 2011, 43, 380–384. [Google Scholar] [CrossRef]
- Brison, J.; Robinson, M.A.; Benoit, D.S.W.; Muramoto, S.; Stayton, P.S.; Castner, D.G. Tof-SIMS 3D imaging of native and non-native species within hela cells. Anal. Chem. 2013, 85, 10869–10877. [Google Scholar] [CrossRef] [PubMed]
- Lazar, A.N.; Bich, C.; Panchal, M.; Desbenoit, N.; Petit, V.W.; Touboul, D.; Dauphinot, L.; Marquer, C.; Laprevote, O.; Brunelle, A.; et al. Time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of alzheimer disease patients. Acta Neuropathol. 2013, 125, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Sole-Domenech, S.; Sjovall, P.; Vukojevic, V.; Fernando, R.; Codita, A.; Salve, S.; Bogdanovic, N.; Mohammed, A.H.; Hammarstrom, P.; Nilsson, K.P.; et al. Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 2013, 125, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, S.; Shimanouchi, T.; Kawashima, T.; Iwai, H. ToF-SIMS observation for evaluating the interaction between amyloid beta and lipid membranes. Anal. Bioanal. Chem. 2015, 407, 2859–2863. [Google Scholar] [CrossRef] [PubMed]
- Furudate, T.; Kurasako, Y.; Takata, E.; Morishita, T.; Miwa, A.; Suzuki, R.; Terada, K. Possibility of monitoring granulation by analyzing the amount of hydroxypropylcellulose, a binder on the surface of granules, using ToF-SIMS. Int. J. Pharmaceut. 2015, 495, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Slodzian, G.; Daigne, B.; Girard, F.; Boust, F.; Hillion, F. Scanning secondary ion analytical microscopy with parallel detection. Biol. Cell. 1992, 74, 43–50. [Google Scholar] [CrossRef]
- Boxer, S.G.; Kraft, M.L.; Weber, P.K. Advances in imaging secondary ion mass spectrometry for biological samples. Annu. Rev. Biophys. 2009, 38, 53–74. [Google Scholar] [CrossRef] [PubMed]
- Stern, R.A.; Fletcher, I.R.; Rasmussen, B.; McNaughton, N.J.; Griffin, B.J. Ion microprobe (nanosims 50) Pb-isotope geochronology at <5 µm scale. Int. J. Mass. Spectrom. 2005, 244, 125–134. [Google Scholar]
- Tendeloo, G.V.D.; Pennycook, S.J. Handbook of Nanoscopy; Wiley-VCH: Weinheim, Germany, 2012; Volume 1. [Google Scholar]
- Jiang, H.; Favaro, E.; Goulbourne, C.N.; Rakowska, P.D.; Hughes, G.M.; Ryadnov, M.G.; Fong, L.G.; Young, S.G.; Ferguson, D.J.P.; Harris, A.L.; et al. Stable isotope imaging of biological samples with high resolution secondary ion mass spectrometry and complementary techniques. Methods 2014, 68, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Peteranderl, R.; Lechene, C. Measure of carbon and nitrogen stable isotope ratios in cultured cells. J. Am. Soc. Mass. Spectr. 2004, 15, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Steinhauser, M.L.; Lechene, C.P. Quantitative imaging of subcellular metabolism with stable isotopes and multi-isotope imaging mass spectrometry. Semin. Cell. Dev. Biol. 2013, 24, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Steinhauser, M.L.; Bailey, A.P.; Senyo, S.E.; Guillermier, C.; Perlstein, T.S.; Gould, A.P.; Lee, R.T.; Lechene, C.P. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 2012, 481, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.D.; Guerquin-Kern, J.L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013, 493, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Lechene, C.P.; Lee, G.Y.; Poczatek, J.C.; Toner, M.; Biggers, J.D. 3D multi-isotope imaging mass spectrometry reveals penetration of 18O-trehalose in mouse sperm nucleus. PLoS ONE 2012. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Lorey, D.R. SIMS ion microscopy imaging of boronophenylalanine (BPA) and 13C15N-labeled phenylalanine in human glioblastoma cells: Relevance of subcellular scale observations to BPA-mediated boron neutron capture therapy of cancer. Int. J. Mass. Spectrom. 2007, 260, 90–101. [Google Scholar] [CrossRef]
- Wedlock, L.E.; Kilburn, M.R.; Liu, R.; Shaw, J.A.; Berners-Price, S.J.; Farrell, N.P. Nanosims multi-element imaging reveals internalisation and nucleolar targeting for a highly-charged polynuclear platinum compound. Chem. Commun. 2013, 49, 6944–6946. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, J.C.; Briggs, D. ToF-SIMS: Materials Analysis by Mass Spectrometry, 2nd ed.; IM Publications: Chichester, UK, 2013. [Google Scholar]
- Lee, J.L.S.; Gilmore, I.S.; Seah, M.P. Quantification and methodology issues in multivariate analysis of ToF-SIMS data for mixed organic systems. Surf. Interf. Anal. 2008, 40, 1–14. [Google Scholar] [CrossRef]
- Wilson, R.L.; Kraft, M.L. Quantifying the molar percentages of cholesterol in supported lipid membranes by time-of-flight secondary ion mass spectrometry and multivariate analysis. Anal. Chem. 2013, 85, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Hanrieder, J.; Malmberg, P.; Lindberg, O.R.; Fletcher, J.S.; Ewing, A.G. Time-of-flight secondary ion mass spectrometry based molecular histology of human spinal cord tissue and motor neurons. Anal. Chem. 2013, 85, 8741–8748. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, J.H.; Ahn, B.J.; Park, J.H.; Shon, H.K.; Yu, Y.S.; Moon, D.W.; Lee, T.G.; Kim, K.W. Label-free calcium imaging in ischemic retinal tissue by ToF-SIMS. Biophys. J. 2008, 94, 4095–4102. [Google Scholar] [CrossRef] [PubMed]
- Kokesch-Himmelreich, J.; Schumacher, M.; Rohnke, M.; Gelinsky, M.; Janek, J. ToF-SIMS analysis of osteoblast-like cells and their mineralized extracellular matrix on strontium enriched bone cements. Biointerphases 2013, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Snyder, F. Platelet-activating factor: The biosynthetic and catabolic enzymes. Biochem. J. 1995, 305, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.E.; Smith, J.A.C.; Kilburn, M.R.; Martin, B.G.H.; Hawes, C.; Grovenor, C.R.M. High-resolution elemental localization in vacuolate plant cells by nanoscale secondary ion mass spectrometry. Plant. J. 2010, 63, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.A.; Brison, J.; Robinson, M.; Graham, D.J.; Castner, D.G.; Ratner, B.D. Identifying individual cell types in heterogeneous cultures using secondary ion mass spectrometry imaging with C60 etching and multivariate analysis. Anal. Chem. 2012, 84, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Lanekoff, I.; Sjovall, P.; Ewing, A.G. Relative quantification of phospholipid accumulation in the PC12 cell plasma membrane following phospholipid incubation using ToF-SIMS imaging. Anal. Chem. 2011, 83, 5337–5343. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.; Arlinghaus, H.F.; Tentschert, J.; Jungnickel, H.; Graf, P.; Mantion, A.; Draude, F.; Galla, S.; Plendl, J.; Goetz, M.E.; et al. Application of laser postionization secondary neutral mass spectrometry/time-of-flight secondary ion mass spectrometry in nanotoxicology: Visualization of nanosilver in human macrophages and cellular responses. ACS Nano 2011, 5, 3059–3068. [Google Scholar] [CrossRef] [PubMed]
- Tentschert, J.; Draude, F.; Jungnickel, H.; Haase, A.; Mantion, A.; Galla, S.; Thunemann, A.F.; Taubert, A.; Luch, A.; Arlinghaus, H.F. ToF-SIMS analysis of cell membrane changes in functional impaired human macrophages upon nanosilver treatment. Surf. Interf. Anal. 2013, 45, 483–485. [Google Scholar] [CrossRef]
- Jungnickel, H.; Jones, E.A.; Lockyer, N.P.; Oliver, S.G.; Stephens, G.M.; Vickerman, J.C. Application of ToF-SIMS with chemometrics to discriminate between four different yeast strains from the species Candida glabrata and Saccharomyces cerevisiae. Anal. Chem. 2005, 77, 1740–1745. [Google Scholar] [CrossRef] [PubMed]
- Leefmann, T.; Heim, C.; Kryvenda, A.; Siljestrom, S.; Sjovall, P.; Thiel, V. Biomarker imaging of single diatom cells in a microbial mat using time-of-flight secondary ion mass spectrometry (ToF-SIMS). Org. Geochem. 2013, 57, 23–33. [Google Scholar] [CrossRef]
- Kurczy, M.E.; Piehowski, P.D.; van Bell, C.T.; Heien, M.L.; Winograd, N.; Ewing, A.G. Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proc. Natl. Acad. Sci. USA 2010, 107, 2751–2756. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Fletcher, J.S.; Thuret, R.; Henderson, A.; Papalopulu, N.; Vickerman, J.C.; Lockyer, N.P. Spatiotemporal lipid profiling during early embryo development of Xenopus laevis using dynamic ToF-SIMS imaging. J. Lipid Res. 2014, 55, 1970–1980. [Google Scholar] [CrossRef] [PubMed]
- Waki, M.; Ide, Y.; Ishizaki, I.; Nagata, Y.; Masaki, N.; Sugiyama, E.; Kurabe, N.; Nicolaescu, D.; Yamazaki, F.; Hayasaka, T.; et al. Single-cell time-of-flight secondary ion mass spectrometry reveals that human breast cancer stem cells have significantly lower content of palmitoleic acid compared to their counterpart non-stem cancer cells. Biochimie 2014, 107, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Kezutyte, T.; Desbenoit, N.; Brunelle, A.; Briedis, V. Studying the penetration of fatty acids into human skin by ex vivo ToF-SIMS imaging. Biointerphases 2013, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jungnickel, H.; Laux, P.; Luch, A. Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS): A New Tool for the Analysis of Toxicological Effects on Single Cell Level. Toxics 2016, 4, 5. https://doi.org/10.3390/toxics4010005
Jungnickel H, Laux P, Luch A. Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS): A New Tool for the Analysis of Toxicological Effects on Single Cell Level. Toxics. 2016; 4(1):5. https://doi.org/10.3390/toxics4010005
Chicago/Turabian StyleJungnickel, Harald, Peter Laux, and Andreas Luch. 2016. "Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS): A New Tool for the Analysis of Toxicological Effects on Single Cell Level" Toxics 4, no. 1: 5. https://doi.org/10.3390/toxics4010005
APA StyleJungnickel, H., Laux, P., & Luch, A. (2016). Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS): A New Tool for the Analysis of Toxicological Effects on Single Cell Level. Toxics, 4(1), 5. https://doi.org/10.3390/toxics4010005