Integration of “Omics”-Based Approaches in Environmental Risk Assessment to Establish Cause and Effect Relationships: A Review
Highlights
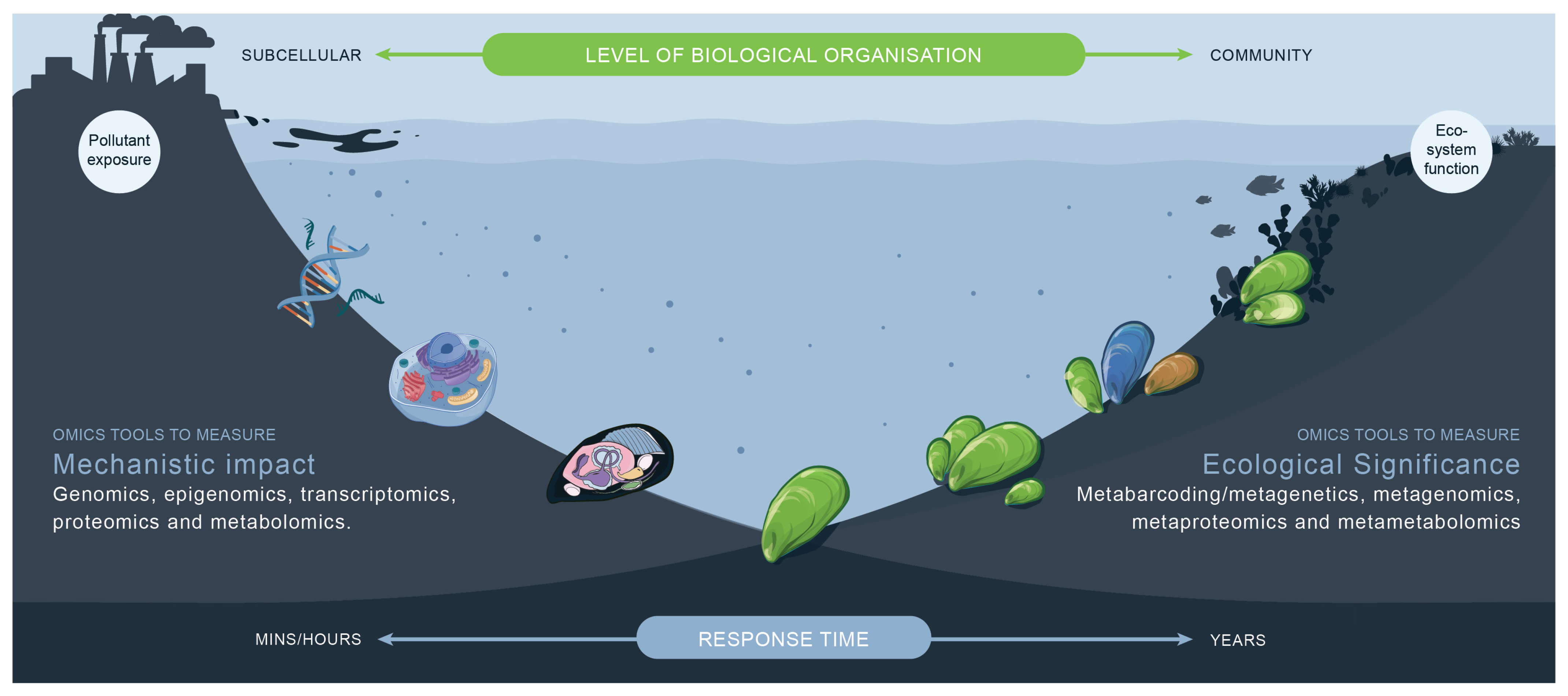
- Omics approaches can provide information to characterise mechanisms of toxicity.
- Omics generate information across different levels of biological organisation.
- Omics can lead to the identification of biomarkers to predict adverse outcomes.
- Incorporation of omics into risk assessment frameworks will require a multi-disciplinary approach.
Abstract
1. Introduction
2. Genomics
3. Epigenomics
4. Transcriptomics
5. Proteomics
6. Metabolomics
7. Ecogenomics
8. Metabarcoding or Metagenetics
9. Meta-Omics
10. Omics and the Development of Biomarkers
11. Integration of Omics Approaches for Ecotoxicology
12. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bell, C. Branding New Zealand: The National Green-wash. Br. Rev. N. Z. Stud. 2006, 15, 13–28. [Google Scholar]
- Gillespie, P.A.; Forrest, R.W.; Peake, B.M.; Basher, L.R.; Clement, D.M.; Dunmore, R.A.; Hicks, D.M. Spatial delineation of the depositional footprint of the Motueka River outwelling plume in Tasman Bay, New Zealand. N. Z. J. Mar. Freshw. Res. 2011, 45, 455–475. [Google Scholar] [CrossRef]
- Tremblay, L.A.; Trought, K.; Sheehan, T.J.; Holmes, R.J.P.; Barrick, A.; Young, R.G. Induction of metallothionein in the common bully (Gobiomorphus cotidianus) from the Motueka River. N. Z. J. Mar. Freshw. Res. 2021, 55, 497–503. [Google Scholar] [CrossRef]
- Wood, S.A.; Smith, K.F.; Banks, J.C.; Tremblay, L.A.; Rhodes, L.; Mountfort, D.; Cary, S.C.; Pochon, X. Molecular genetic tools for environmental monitoring of New Zealand’s aquatic habitats, past, present and the future. N. Z. J. Mar. Freshw. Res. 2013, 47, 90–119. [Google Scholar] [CrossRef]
- Kumar, A.; Xagoraraki, I. Pharmaceuticals, personal care products and endocrine-disrupting chemicals in U.S. surface and finished drinking waters: A proposed ranking system. Sci. Total Environ. 2010, 408, 5972–5989. [Google Scholar] [CrossRef]
- Schwarzenbach, R.P.; Escher, B.I.; Fenner, K.; Hofstetter, T.B.; Johnson, C.A.; von Gunten, U.; Wehrli, B. The challenge of micropollutants in aquatic systems. Science 2006, 313, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xiang, L.L.; Leung, K.S.Y.; Elsner, M.; Zhang, Y.; Guo, Y.M.; Pan, B.; Sun, H.W.; An, T.C.; Ying, G.G.; et al. Emerging contaminants: A One Health perspective. Innovation 2024, 5, 100612. [Google Scholar] [CrossRef]
- Matlin, S.A.; Cornell, S.E.; Krief, A.; Hopf, H.; Mehta, G. Chemistry must respond to the crisis of transgression of planetary boundaries. Chem. Sci. 2022, 13, 11710–11720. [Google Scholar] [CrossRef]
- Richardson, K.; Steffen, W.; Lucht, W.; Bendtsen, J.; Cornell, S.E.; Donges, J.F.; Drüke, M.; Fetzer, I.; Bala, G.; von Bloh, W.; et al. Earth beyond six of nine planetary boundaries. Sci. Adv. 2023, 9, eadh2458. [Google Scholar] [CrossRef]
- Rockström, J.; Donges, J.F.; Fetzer, I.; Martin, M.A.; Wang-Erlandsson, L.; Richardson, K. Planetary Boundaries guide humanity’s future on Earth. Nat. Rev. Earth Environ. 2024, 5, 773–788. [Google Scholar] [CrossRef]
- Halpern, B.S.; Walbridge, S.; Selkoe, K.A.; Kappel, C.V.; Micheli, F.; D’Agrosa, C.; Bruno, J.F.; Casey, K.S.; Ebert, C.; Fox, H.E.; et al. A global map of human impact on marine ecosystems. Science 2008, 319, 948–952. [Google Scholar] [CrossRef]
- Dallas, L.J.; Jha, A.N. Applications of biological tools or biomarkers in aquatic biota: A case study of the Tamar estuary, South West England. Mar. Pollut. Bull. 2015, 95, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.K.S.; Gray, J.S. The use of biomarkers in environmental monitoring programmes. Mar. Pollut. Bull. 2003, 46, 182–186. [Google Scholar] [CrossRef]
- Van der Oost, R.; Beyer, J.; Vermeulen, N.P.E. Fish bioaccumulation and biomarkers in environmental risk assessment: A review. Environ. Toxicol. Pharmacol. 2003, 13, 57–149. [Google Scholar] [CrossRef]
- Adams, S.M.; Giesy, J.P.; Tremblay, L.A.; Eason, C.T. The use of biomarkers in ecological risk assessment: Recommendations from the Christchurch conference on Biomarkers in Ecotoxicology. Biomarkers 2001, 6, 1–6. [Google Scholar] [CrossRef]
- Boehler, S.; Strecker, R.; Heinrich, P.; Prochazka, E.; Northcott, G.L.; Ataria, J.M.; Leusch, F.D.L.; Braunbeck, T.; Tremblay, L.A. Assessment of urban stream sediment pollutants entering estuaries using chemical analysis and multiple bioassays to characterise biological activities. Sci. Total Environ. 2017, 593–594, 498–507. [Google Scholar] [CrossRef]
- Charry, M.P.; Keesing, V.; Costello, M.; Tremblay, L.A. Assessment of the ecotoxicity of urban estuarine sediment using benthic and pelagic copepod bioassays. PeerJ 2018, 6, e4936. [Google Scholar] [CrossRef] [PubMed]
- Leusch, F.D.L.; Allen, H.; De Silva, N.A.L.; Hodson, R.; Johnson, M.; Neale, P.A.; Stewart, M.; Tremblay, L.A.; Wilde, T.; Northcott, G.L. Effect-based monitoring of two rivers under urban and agricultural influence reveals a range of biological activities in sediment and water extracts. J. Environ. Manag. 2024, 351, 119692. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.A., Jr.; Chapman, P.M.; Smith, E.P. Weight-of-evidence approaches for assessing ecosystem impairment. Hum. Ecol. Risk Assess. 2002, 8, 1657–1673. [Google Scholar] [CrossRef]
- Ankley, G.T.; Bennett, R.S.; Erickson, R.J.; Hoff, D.J.; Hornung, M.W.; Johnson, R.D.; Mount, D.R.; Nichols, J.W.; Russom, C.L.; Schmieder, P.K.; et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 2010, 29, 730–741. [Google Scholar] [CrossRef]
- Cheifet, B. Where is genomics going next? Genome Biol. 2019, 20, 17. [Google Scholar] [CrossRef]
- Gutleben, J.; De Mares, M.C.; van Elsas, J.D.; Smidt, H.; Overmann, J.; Sipkema, D. The multi-omics promise in context: From sequence to microbial isolate. Crit. Rev. Microbiol. 2018, 44, 212–229. [Google Scholar] [CrossRef]
- Hudson, M.E. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Resour. 2008, 8, 3–17. [Google Scholar] [CrossRef]
- Prat, O.; Degli-Esposti, D. 6—New Challenges: Omics Technologies in Ecotoxicology. In Ecotoxicology; Gross, E., Garric, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 181–208. [Google Scholar]
- Snape, J.R.; Maund, S.J.; Pickford, D.B.; Hutchinson, T.H. Ecotoxicogenomics: The challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat. Toxicol. 2004, 67, 143–154. [Google Scholar] [CrossRef]
- Zhang, X.; Xia, P.; Wang, P.; Yang, J.; Baird, D.J. Omics Advances in Ecotoxicology. Environ. Sci. Technol. 2018, 52, 3842–3851. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apotheloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e) DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 637, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Van Aggelen, G.; Ankley, G.T.; Baldwin, W.S.; Bearden, D.W.; Benson, W.H.; Chipman, J.K.; Collette, T.W.; Craft, J.A.; Denslow, N.D.; Embry, M.R.; et al. Integrating omic technologies into aquatic ecological risk assessment and environmental monitoring: Hurdles, achievements, and future outlook. Environ. Health Perspect. 2010, 118, 1–5. [Google Scholar] [CrossRef]
- Campos, B.; Colbourne, J.K.; Brown, J.B.; Viant, M.R.; Biales, A.D.; Gallagher, K.; Henry, T.R.; Sappington, K.G.; Marshall, S.; Whale, G. How Omics Technologies can Enhance Chemical Safety Regulation: Perspectives from Academia, Government, and Industry. Environ. Toxicol. Chem. 2018, 37, 1252–1259. [Google Scholar] [CrossRef]
- Hering, D.; Borja, A.; Jones, J.I.; Pont, D.; Boets, P.; Bouchez, A.; Bruce, K.; Drakare, S.; Hänfling, B.; Kahlert, M.; et al. Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res. 2018, 138, 192–205. [Google Scholar] [CrossRef]
- Tong, W.D.; Ostroff, S.; Blais, B.; Silva, P.; Dubuc, M.; Healy, M.; Slikker, W. Genomics in the land of regulatory science. Regul. Toxicol. Pharmacol. 2015, 72, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Reyero, N.; Perkins, E.J. Systems biology: Leading the revolution in ecotoxicology. Environ. Toxicol. Chem. 2011, 30, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.A. Next generation sequencing data for use in risk assessment. Curr. Opin. Toxicol. 2019, 18, 18–26. [Google Scholar] [CrossRef]
- Breitholtz, M.; Ruden, C.; Hansson, S.O.; Bengtsson, B.E. Ten challenges for improved ecotoxicological testing in environmental risk assessment. Ecotoxicol. Environ. Saf. 2006, 63, 324–335. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.H. The molecular genetics of insecticide resistance. Genetics 2013, 194, 807–815. [Google Scholar] [CrossRef]
- Whitehead, A.; Clark, B.W.; Reid, N.M.; Hahn, M.E.; Nacci, D. When evolution is the solution to pollution: Key principles, and lessons from rapid repeated adaptation of killifish (Fundulus heteroclitus) populations. Evol. Appl. 2017, 10, 762–783. [Google Scholar] [CrossRef]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Landry, C.R.; Aubin-Horth, N. Recent advances in ecological genomics: From phenotypic plasticity to convergent and adaptive evolution and speciation. In Ecological Genomics. Advances in Experimental Medicine and Biology; Landry, C., Aubin-Horth, N., Eds.; Springer: Dordrecht, The Netherlands, 2013; Volume 781. [Google Scholar] [CrossRef]
- Bossdorf, O.; Richards, C.L.; Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 2008, 11, 106–115. [Google Scholar] [CrossRef]
- Hawes, N.A.; Tremblay, L.A.; Pochon, X.; Dunphy, B.; Fidler, A.E.; Smith, K.F. Effects of temperature and salinity stress on DNA methylation in a highly invasive marine invertebrate, the colonial ascidian Didemnum vexillum. Peerj 2018, 6, e5003. [Google Scholar] [CrossRef]
- Bromer, J.G.; Zhou, Y.P.; Taylor, M.B.; Doherty, L.; Taylor, H.S. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010, 24, 2273–2280. [Google Scholar] [CrossRef] [PubMed]
- Fragou, D.; Fragou, A.; Kouidou, S.; Njau, S.; Kovatsi, L. Epigenetic mechanisms in metal toxicity. Toxicol. Mech. Methods 2011, 21, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Jeremias, G.; Goncalves, F.J.M.; Pereira, J.L.; Asselman, J. Prospects for incorporation of epigenetic biomarkers in human health and environmental risk assessment of chemicals. Biol. Rev. 2020, 95, 822–846. [Google Scholar] [CrossRef]
- Hawes, N.A.; Fidler, A.E.; Tremblay, L.A.; Pochon, X.; Dunphy, B.J.; Smith, K.F. Understanding the role of DNA methylation in successful biological invasions: A review. Biol. Invasions 2018, 20, 2285–2300. [Google Scholar] [CrossRef]
- Skinner, M.K.; Guerrero-Bosagna, C. Environmental signals and transgenerational epigenetics. Epigenomics 2009, 1, 111–117. [Google Scholar] [CrossRef]
- Bonduriansky, R.; Crean, A.J.; Day, T. The implications of nongenetic inheritance for evolution in changing environments. Evol. Appl. 2012, 5, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.L.; Judy, J.D.; Kumar, A.; Bertsch, P.; Wang, M.-B.; Kirby, J.K. Incorporating transgenerational epigenetic inheritance into ecological risk assessment frameworks. Environ. Sci. Technol. 2017, 51, 9433–9445. [Google Scholar] [CrossRef] [PubMed]
- Head, J.A.; Dolinoy, D.C.; Basu, N. Epigenetics for ecotoxicologists. Environ. Toxicol. Chem. 2012, 31, 221–227. [Google Scholar] [CrossRef]
- Srut, M. Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden. Chemosphere 2021, 282, 131026. [Google Scholar] [CrossRef]
- Vandegehuchte, M.B.; Janssen, C.R. Epigenetics and its implications for ecotoxicology. Ecotoxicology 2011, 20, 607–624. [Google Scholar] [CrossRef]
- Plongthongkum, N.; Diep, D.H.; Zhang, K. Advances in the profiling of DNA modifications: Cytosine methylation and beyond. Nat. Rev. Genet. 2014, 15, 647–661. [Google Scholar] [CrossRef]
- Schrey, A.W.; Alvarez, M.; Foust, C.M.; Kilvitis, H.J.; Lee, J.D.; Liebl, A.L.; Martin, L.B.; Richards, C.L.; Robertson, M. Ecological Epigenetics: Beyond MS-AFLP. Integr. Comp. Biol. 2013, 53, 340–350. [Google Scholar] [CrossRef]
- Guyon, A.; Smith, K.F.; Charry, M.P.; Champeau, O.; Tremblay, L.A. Effects of chronic exposure to benzophenone and diclofenac on DNA methylation levels and reproductive success in a marine copepod. J. Xenobiot. 2018, 8, 7674. [Google Scholar] [CrossRef]
- Hofmann, G.E. Ecological Epigenetics in Marine Metazoans. Front. Mar. Sci. 2017, 4, 4. [Google Scholar] [CrossRef]
- Oppold, A.; Kreß, A.; Vanden Bussche, J.; Diogo, J.B.; Kuch, U.; Oehlmann, J.; Vandegehuchte, M.B.; Müller, R. Epigenetic alterations and decreasing insecticide sensitivity of the Asian tiger mosquito Aedes albopictus. Ecotoxicol. Environ. Saf. 2015, 122, 45–53. [Google Scholar] [CrossRef]
- Schrey, A.W.; Coon, C.A.; Grispo, M.T.; Awad, M.; Imboma, T.; McCoy, E.D.; Mushinsky, H.R.; Richards, C.L.; Martin, L.B. Epigenetic Variation May Compensate for Decreased Genetic Variation with Introductions: A Case Study Using House Sparrows (Passer domesticus) on Two Continents. Genet. Res. Int. 2012, 2012, 979751. [Google Scholar] [CrossRef]
- Verhoeven, K.J.F.; Jansen, J.J.; van Dijk, P.J.; Biere, A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010, 185, 1108–1118. [Google Scholar] [CrossRef]
- Chatterjee, A.; Ozaki, Y.; Stockwell, P.A.; Horsfield, J.A.; Morison, I.M.; Nakagawa, S. Mapping the zebrafish brain methylome using reduced representation bisulfite sequencing. Epigenetics 2013, 8, 979–989. [Google Scholar] [CrossRef]
- Trucchi, E.; Mazzarella, A.B.; Gilfillan, G.D.; Lorenzo, M.T.; Schönswetter, P.; Paun, O. BsRADseq: Screening DNA methylation in natural populations of non-model species. Mol. Ecol. 2016, 25, 1697–1713. [Google Scholar] [CrossRef] [PubMed]
- Ramalho-Carvalho, J.; Henrique, R.; Jerónimo, C. Methylation-Specific PCR. In DNA Methylation Protocols; Tost, J., Ed.; Springer: New York, NY, USA, 2018; pp. 447–472. [Google Scholar]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, T.P.; Ravichandran, M.; Stepper, P. Synthetic epigenetics—Towards intelligent control of epigenetic states and cell identity. Clin. Epigenetics 2015, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Laufer, B.I.; Singh, S.M. Strategies for precision modulation of gene expression by epigenome editing: An overview. Epigenetics Chromatin 2015, 8, 34. [Google Scholar] [CrossRef]
- Puchta, H. Using CRISPR/Cas in three dimensions: Towards synthetic plant genomes, transcriptomes and epigenomes. Plant J. 2016, 87, 5–15. [Google Scholar] [CrossRef]
- Vojta, A.; Dobrinić, P.; Tadić, V.; Bočkor, L.; Korać, P.; Julg, B.; Klasić, M.; Zoldoš, V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef]
- Major, K.M.; DeCourten, B.M.; Li, J.; Britton, M.; Settles, M.L.; Mehinto, A.C.; Connon, R.E.; Brander, S.M. Early Life Exposure to Environmentally Relevant Levels of Endocrine Disruptors Drive Multigenerational and Transgenerational Epigenetic Changes in a Fish Model. Front. Mar. Sci. 2020, 7, 471. [Google Scholar] [CrossRef]
- Baettig, C.G.; Laroche, O.; Ockenden, A.; Smith, K.F.; Lear, G.; Tremblay, L.A. Characterization of the transcriptional effects of the plastic additive dibutyl phthalate alone and in combination with microplastic on the green-lipped mussel (Perna canaliculus). Environ. Toxicol. Chem. 2024, 43, 1604–1614. [Google Scholar] [CrossRef]
- Schirmer, K.; Fischer, B.B.; Madureira, D.J.; Pillai, S. Transcriptomics in ecotoxicology. Anal. Bioanal. Chem. 2010, 397, 917–923. [Google Scholar] [CrossRef]
- Ankley, G.T.; Daston, G.P.; Degitz, S.J.; Denslow, N.D.; Hoke, R.A.; Kennedy, S.W.; Miracle, A.L.; Perkins, E.J.; Snape, J.; Tillitt, D.E. Toxicogenomics in regulatory ecotoxicology. Environ. Sci. Technol. 2006, 40, 4055–4065. [Google Scholar] [CrossRef] [PubMed]
- Vinken, M. Omics-based input and output in the development and use of adverse outcome pathways. Curr. Opin. Toxicol. 2019, 18, 8–12. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Baettig, C.G.; Zirngibl, M.; Smith, K.F.; Lear, G.; Tremblay, L.A. Comparison between droplet digital PCR and reverse transcription-quantitative PCR methods to measure ecotoxicology biomarkers. Mar. Pollut. Bull. 2023, 190, 114829. [Google Scholar] [CrossRef]
- Fabrello, J.; Grapputo, A.; Munari, M.; Marin, M.G.; Masiero, L.; Pacchioni, B.; Millino, C.; Matozzo, V. Molecular and biochemical responses of vitellogenin in the mussel Mytilus galloprovincialis exposed to the glyphosate-based herbicide Roundup® Power 2.0. Environ. Sci. Pollut. Res. 2020, 27, 26543–26553. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Kelso, J.J.B. High-throughput DNA sequencing–concepts and limitations. Bioessays 2010, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology. Annu. Rev. Biochem. 2011, 80, 273–299. [Google Scholar] [CrossRef]
- Heijne, W.H.M.; Kienhuis, A.S.; van Ommen, B.; Stierum, R.H.; Groten, J.P. Systems toxicology: Applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev. Proteom. 2005, 2, 767–780. [Google Scholar] [CrossRef]
- Cagney, G.; Emili, A. De novo peptide sequencing and quantitative profiling of complex protein mixtures using mass-coded abundance tagging. Nat. Biotechnol. 2002, 20, 163–170. [Google Scholar] [CrossRef]
- Mann, M.; Wilm, M. Error tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 1994, 66, 4390–4399. [Google Scholar] [CrossRef]
- Altelaar, A.F.M.; Munoz, J.; Heck, A.J.R. Next-generation proteomics: Towards an integrative view of proteome dynamics. Nat. Rev. Genet. 2013, 14, 35–48. [Google Scholar] [CrossRef]
- Liang, X.; Martyniuk, C.J.; Simmons, D.B.D. Are we forgetting the “proteomics” in multi-omics ecotoxicology? Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 36, 100751. [Google Scholar] [CrossRef]
- Gouveia, D.; Almunia, C.; Cogne, Y.; Pible, O.; Degli-Esposti, D.; Salvador, A.; Cristobal, S.; Sheehan, D.; Chaumot, A.; Geffard, O.; et al. Ecotoxicoproteomics: A decade of progress in our understanding of anthropogenic impact on the environment. J. Proteom. 2019, 198, 66–77. [Google Scholar] [CrossRef]
- Trapp, J.; Armengaud, J.; Salvador, A.; Chaumot, A.; Geffard, O. Next-Generation Proteomics: Toward Customized Biomarkers for Environmental Biomonitoring. Environ. Sci. Technol. 2014, 48, 13560–13572. [Google Scholar] [CrossRef] [PubMed]
- Groh, K.J.; Dalkvist, T.; Piccapietra, F.; Behra, R.; Suter, M.J.F.; Schirmer, K. Critical influence of chloride ions on silver ion-mediated acute toxicity of silver nanoparticles to zebrafish embryos. Nanotoxicology 2015, 9, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Dreij, K.; Costa, P.M. The state-of-the art of environmental toxicogenomics: Challenges and perspectives of "omics" approaches directed to toxicant mixtures. Int. J. Environ. Res. Public Health 2019, 16, 4718. [Google Scholar] [CrossRef]
- Khoo, S.H.G.; Al-Rubeai, M. Metabolomics as a complementary tool in cell culture. Biotechnol. Appl. Biochem. 2007, 47, 71–84. [Google Scholar] [CrossRef]
- Nalbantoglu, S.; Abu-Asab, M.; Suy, S.; Collins, S.; Amri, H. Metabolomics-Based Biosignatures of Prostate Cancer in Patients Following Radiotherapy. Omics-A J. Integr. Biol. 2019, 23, 214–223. [Google Scholar] [CrossRef]
- Olesti, E.; González-Ruiz, V.; Wilks, M.F.; Boccard, J.; Rudaz, S. Approaches in metabolomics for regulatory toxicology applications. Analyst 2021, 146, 1820–1834. [Google Scholar] [CrossRef]
- German, J.B.; Gillies, L.A.; Smilowitz, J.T.; Zivkovic, A.M.; Watkins, S.M. Lipidomics and lipid profiling in metabolomics. Curr. Opin. Lipidol. 2007, 18, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, Y.; Li, M.H.; Xu, H.D.; Sheng, J.Y.; Zhang, L.; Wang, J.S. Integrated 1H NMR-based metabolomics analysis of earthworm responses to sub-lethal Pb exposure. Environ. Chem. 2016, 13, 792–803. [Google Scholar] [CrossRef]
- Melvin, S.D.; Habener, L.J.; Leusch, F.D.L.; Carroll, A.R. 1H NMR-based metabolomics reveals sub-lethal toxicity of a mixture of diabetic and lipid-regulating pharmaceuticals on amphibian larvae. Aquat. Toxicol. 2017, 184, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef]
- Van Ravenzwaay, B.; Herold, M.; Kamp, H.; Kapp, M.D.; Fabian, E.; Looser, R.; Krennrich, G.; Mellert, W.; Prokoudine, A.; Strauss, V.; et al. Metabolomics: A tool for early detection of toxicological effects and an opportunity for biology based grouping of chemicals-From QSAR to QBAR. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2012, 746, 144–150. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Melvin, S.D.; Lanctot, C.M.; Doriean, N.J.C.; Carroll, A.R.; Bennett, W.W. Untargeted NMR-based metabolomics for field-scale monitoring: Temporal reproducibility and biomarker discovery in mosquitofish (Gambusia holbrooki) from a metal(loid)-contaminated wetland. Environ. Pollut. 2018, 243, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Bundy, J.G.; Davey, M.P.; Viant, M.R. Environmental metabolomics: A critical review and future perspectives. Metabolomics 2009, 5, 3–21. [Google Scholar] [CrossRef]
- Emwas, A.H.; Luchinat, C.; Turano, P.; Tenori, L.; Roy, R.; Salek, R.M.; Ryan, D.; Merzaban, J.S.; Kaddurah-Daouk, R.; Zeri, A.C.; et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: A review. Metabolomics 2015, 11, 872–894. [Google Scholar] [CrossRef]
- Miggiels, P.; Wouters, B.; van Westen, G.J.P.; Dubbelman, A.C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trends Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Bouhifd, M.; Hartung, T.; Hogberg, H.T.; Kleensang, A.; Zhao, L. Review: Toxicometabolomics. J. Appl. Toxicol. 2013, 33, 1365–1383. [Google Scholar] [CrossRef] [PubMed]
- Peregrin-Alvarez, J.M.; Sanford, C.; Parkinson, J. The conservation and evolutionary modularity of metabolism. Genome Biol. 2009, 10, R63. [Google Scholar] [CrossRef]
- Griffiths, L.L.; Melvin, S.D.; Connolly, R.M.; Pearson, R.M.; Brown, C.J. Metabolomic indicators for low-light stress in seagrass. Ecol. Indic. 2020, 114, 106316. [Google Scholar] [CrossRef]
- Denslow, N.D.; Griffitt, R.J.; Martyniuk, C.J. Advancing the Omics in aquatic toxicology: SETAC North America 31st Annual Meeting. Ecotoxicol. Environ. Saf. 2012, 76, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Alecu, I.; Sosa-Miranda, C.D.; Sandhu, J.K.; Bennett, S.A.L.; Cuperlovic-Culf, M. Chapter 12—Cell culture metabolomics and lipidomics. In Metabolomics Perspectives – From Theory to Practical Application; Troisi, J., Ed.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 415–456. [Google Scholar]
- Martyniuk, C.J.; Simmons, D.B. Spotlight on environmental omics and toxicology: A long way in a short time. Comp. Biochem. Physiol. D Genom. Proteom. 2016, 19, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Bidwell, J.R.; Heath, A.G. An in-situ study of rock bass (Ambloplites-rupestris) physiology—Effect of season and mercury contamination. Hydrobiologia 1993, 264, 137–152. [Google Scholar] [CrossRef]
- Beale, D.J.; Jones, O.A.H.; Bose, U.; Broadbent, J.A.; Walsh, T.K.; van de Kamp, J.; Bissett, A. Omics-based ecosurveillance for the assessment of ecosystem function, health, and resilience. Emerg. Top. Life Sci. 2022, 6, 185–199. [Google Scholar] [CrossRef]
- Viant, M.R.; Ebbels, T.M.D.; Beger, R.D.; Ekman, D.R.; Epps, D.J.T.; Kamp, H.; Leonards, P.E.G.; Loizou, G.D.; MacRae, J.I.; van Ravenzwaay, B.; et al. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat. Commun. 2019, 10, 3041. [Google Scholar] [CrossRef]
- Beja, O. To BAC or not to BAC: Marine ecogenomics. Curr. Opin. Biotechnol. 2004, 15, 187–190. [Google Scholar] [CrossRef]
- Dicke, M.; van Loon, J.J.A.; de Jong, P.W. Ecogenomics benefits community ecology. Science 2004, 305, 618–619. [Google Scholar] [CrossRef]
- Maphosa, F.; de Vos, W.M.; Smidt, H. Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria. Trends Biotechnol. 2010, 28, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Ouborg, N.J.; Vriezen, W.H. An ecologist’s guide to ecogenomics. J. Ecol. 2007, 95, 8–16. [Google Scholar] [CrossRef]
- Snoeren, T.A.L.; De Jong, P.W.; Dicke, M. Ecogenomic approach to the role of herbivore-induced plant volatiles in community ecology. J. Ecol. 2007, 95, 17–26. [Google Scholar] [CrossRef]
- Faure, D.; Joly, D. 10—Structure and functioning of microbial ecosystems: Metagenomics and integration of omics. In Insight on Environmental Genomics High-Throughput Sequencing; Faure, D., Joly, D., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 103–113. [Google Scholar]
- Pattnaik, S.S.; Busi, S. Rhizospheric Fungi: Diversity and Potential Biotechnological Applications. In Recent Advancement in White Biotechnology Through Fungi, Volume 1: Diversity and Enzymes Perspectives; Yadav, A.N., Mishra, S., Singh, S., Gupta, A., Eds.; Fungal Biology-US; Springer: Cham, Switzerland, 2019; pp. 63–84. [Google Scholar]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, H.H.; Haròardóttir, S.; Ribeiro, S. Assessing the performance of short 18S rDNA markers for environmental DNA metabarcoding of marine protists. Environ. DNA 2024, 6, e580. [Google Scholar] [CrossRef]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Bescot, N.L.; Probert, I.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. DNA extraction replicates improve diversity and compositional dissimilarity in metabarcoding of eukaryotes in marine sediments. PLoS ONE 2017, 12, e0179443. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.M.; Wang, X.G.; Cheng, X.L.; Guo, J.J.; Bian, X.M.; Cai, L. Fungal communities in sediments of subtropical Chinese seas as estimated by DNA metabarcoding. Sci. Rep. 2016, 6, 26528. [Google Scholar] [CrossRef]
- Li, Y.; Hingamp, P.; Watai, H.; Endo, H.; Yoshida, T.; Ogata, H. Degenerate PCR primers to reveal the diversity of giant viruses in coastal waters. Viruses 2018, 10, 496. [Google Scholar] [CrossRef]
- Pawlowski, J.; Bruce, K.; Panksep, K.; Aguirre, F.I.; Amalfitano, S.; Apothéloz-Perret-Gentil, L.; Baussant, T.; Bouchez, A.; Carugati, L.; Cermakova, K.; et al. Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods. Sci. Total Environ. 2022, 818, 151783. [Google Scholar] [CrossRef]
- Pawlowski, J.; Esling, P.; Lejzerowicz, F.; Cedhagen, T.; Wilding, T.A. Environmental monitoring through protist next-generation sequencing metabarcoding: Assessing the impact of fish farming on benthic foraminifera communities. Mol. Ecol. Resour. 2014, 14, 1129–1140. [Google Scholar] [CrossRef]
- Girard, E.B.; Didaskalou, E.A.; Pratama, A.M.A.; Rattner, C.; Morard, R.; Renema, W. Quantitative assessment of reef foraminifera community from metabarcoding data. Mol. Ecol. Resour. 2024, 24, e14000. [Google Scholar] [CrossRef]
- Lecroq, B.; Lejzerowicz, F.; Bachar, D.; Christen, R.; Esling, P.; Baerlocher, L.; Osteras, M.; Farinelli, L.; Pawlowski, J. Ultra-deep sequencing of foraminiferal microbarcodes unveils hidden richness of early monothalamous lineages in deep-sea sediments. Proc. Natl. Acad. Sci. USA 2011, 108, 13177–13182. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Stoeck, T.; Kochems, R.; Forster, D.; Lejzerowicz, F.; Pawlowski, J. Metabarcoding of benthic ciliate communities shows high potential for environmental monitoring in salmon aquaculture. Ecol. Indic. 2018, 85, 153–164. [Google Scholar] [CrossRef]
- Zimmermann, J.; Glockner, G.; Jahn, R.; Enke, N.; Gemeinholzer, B. Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Mol. Ecol. Resour. 2015, 15, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Taberlet, P. Towards exhaustive community ecology via DNA metabarcoding. Mol. Ecol. 2023, 32, 6320–6329. [Google Scholar] [CrossRef]
- McGee, K.M.; Robinson, C.V.; Hajibabaei, M. Gaps in DNA-Based Biomonitoring Across the Globe. Front. Ecol. Evol. 2019, 7, 337. [Google Scholar] [CrossRef]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: Management and surveillance of non-indigenous species. Front. Mar. Sci. 2018, 5, 322. [Google Scholar] [CrossRef]
- Darling, J.A.; Galil, B.S.; Carvalho, G.R.; Rius, M.; Viard, F.; Piraino, S. Recommendations for developing and applying genetic tools to assess and manage biological invasions in marine ecosystems. Mar. Policy 2017, 85, 54–64. [Google Scholar] [CrossRef]
- Darling, J.A.; Pochon, X.; Abbott, C.L.; Inglis, G.J.; Zaiko, A. The risks of using molecular biodiversity data for incidental detection of species of concern. Divers. Distrib. 2020, 26, 1116–1121. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef]
- Zhang, M.L.; Zou, Y.T.; Xiao, S.; Hou, J. Environmental DNA metabarcoding serves as a promising method for aquatic species monitoring and management: A review focused on its workflow, applications, challenges and prospects. Mar. Pollut. Bull. 2023, 194, 115430. [Google Scholar] [CrossRef] [PubMed]
- Birrer, S.C.; Dafforn, K.A.; Simpson, S.L.; Kelaher, B.P.; Potts, J.; Scanes, P.; Johnston, E.L. Interactive effects of multiple stressors revealed by sequencing total (DNA) and active (RNA) components of experimental sediment microbial communities. Sci. Total Environ. 2018, 637, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Keeley, N.; Wood, S.A.; Pochon, X. Development and preliminary validation of a multi-trophic metabarcoding biotic index for monitoring benthic organic enrichment. Ecol. Indic. 2018, 85, 1044–1057. [Google Scholar] [CrossRef]
- Laroche, O.; Pochon, X.; Tremblay, L.A.; Ellis, J.I.; Lear, G.; Wood, S.A. Incorporating molecular-based functional and co-occurrence network properties into benthic marine impact assessments. FEMS Microbiol. Ecol. 2018, 94, fiy167. [Google Scholar] [CrossRef] [PubMed]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Ellis, J.I.; Lear, G.; Pochon, X. A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar. Pollut. Bull. 2018, 127, 97–107. [Google Scholar] [CrossRef]
- Compson, Z.G.; McClenaghan, B.; Singer, G.A.C.; Fahner, N.A.; Hajibabaei, M. Metabarcoding from microbes to mammals: Comprehensive bioassessment on a global scale. Front. Ecol. Evol. 2020, 8, 581835. [Google Scholar] [CrossRef]
- Cordier, T.; Alonso-Saez, L.; Apothéloz-Perret-Gentil, L.; Aylagas, E.; Bohan, D.A.; Bouchez, A.; Chariton, A.; Creer, S.; Frühe, L.; Keck, F.; et al. Ecosystems monitoring powered by environmental genomics: A review of current strategies with an implementation roadmap. Mol. Ecol. 2021, 30, 2937–2958. [Google Scholar] [CrossRef]
- Pawlowski, J.; Lejzerowicz, F.; Apotheloz-Perret-Gentil, L.; Visco, J.; Esling, P. Protist metabarcoding and environmental biomonitoring: Time for change. Eur. J. Protistol. 2016, 55, 12–25. [Google Scholar] [CrossRef]
- Takahashi, M.; Saccò, M.; Kestel, J.H.; Nester, G.; Campbell, M.A.; van der Heyde, M.; Heydenrych, M.J.; Juszkiewicz, D.J.; Nevill, P.; Dawkins, K.L.; et al. Aquatic environmental DNA: A review of the macro-organismal biomonitoring revolution. Sci. Total Environ. 2023, 873, 162322. [Google Scholar] [CrossRef]
- Dowle, E.J.; Pochon, X.; Banks, J.C.; Shearer, K.; Wood, S.A. Targeted gene enrichment and high-throughput sequencing for environmental biomonitoring: A case study using freshwater macroinvertebrates. Mol. Ecol. Resour. 2016, 16, 1240–1254. [Google Scholar] [CrossRef]
- Frühe, L.; Dully, V.; Forster, D.; Keeley, N.B.; Laroche, O.; Pochon, X.; Robinson, S.; Wilding, T.A.; Stoeck, T. Global trends of benthic bacterial diversity and community composition along organic enrichment gradients of salmon farms. Front. Microbiol. 2021, 12, 637811. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; Wood, S.A.; Keeley, N.B.; Lejzerowicz, F.; Esling, P.; Drew, J.; Pawlowski, J. Accurate assessment of the impact of salmon farming on benthic sediment enrichment using foraminiferal metabarcoding. Mar. Pollut. Bull. 2015, 100, 370–382. [Google Scholar] [CrossRef]
- He, X.P.; Sutherland, T.F.; Pawlowski, J.; Abbott, C.L. Responses of foraminifera communities to aquaculture-derived organic enrichment as revealed by environmental DNA metabarcoding. Mol. Ecol. 2019, 28, 1138–1153. [Google Scholar] [CrossRef]
- Lejzerowicz, F.; Esling, P.; Pillet, L.; Wilding, T.A.; Black, K.D.; Pawlowski, J. High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 2015, 5, 13932. [Google Scholar] [CrossRef]
- Leontidou, K.; Rubel, V.; Stoeck, T. Comparing quantile regression spline analyses and supervised machine learning for environmental quality assessment at coastal marine aquaculture installations. Peerj 2023, 11, e15425. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.B.; Rocha, A.M.; Smillie, C.S.; Olesen, S.W.; Paradis, C.; Wu, L.Y.; Campbell, J.H.; Fortney, J.L.; Mehlhorn, T.L.; Lowe, K.A.; et al. Natural bacterial communities serve as quantitative geochemical biosensors. Mbio 2015, 6, 10–1128. [Google Scholar] [CrossRef]
- Dutra, J.; García, G.; Gomes, R.; Cardoso, M.; Côrtes, A.; Silva, T.; de Jesus, L.; Rodrigues, L.; Freitas, A.; Waldow, V.; et al. Effective Biocorrosive Control in Oil Industry Facilities: 16S rRNA Gene metabarcoding for monitoring microbial communities in produced water. Microorganisms 2023, 11, 846. [Google Scholar] [CrossRef] [PubMed]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Ellis, J.I.; Lejzerowicz, F.; Pawlowski, J.; Lear, G.; Atalah, J.; Pochon, X. First evaluation of foraminiferal metabarcoding for monitoring environmental impact from an offshore oil drilling site. Mar. Environ. Res. 2016, 120, 225–235. [Google Scholar] [CrossRef]
- Lanzén, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. High-throughput metabarcoding of eukaryotic diversity for environmental monitoring of offshore oil-drilling activities. Mol. Ecol. 2016, 25, 4392–4406. [Google Scholar] [CrossRef]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Lear, G.; Ellis, J.I.; Pochon, X. Metabarcoding monitoring analysis: The pros and cons of using co-extracted environmental DNA and RNA data to assess offshore oil production impacts on benthic communities. Peerj 2017, 5, e3347. [Google Scholar] [CrossRef] [PubMed]
- Mauffrey, F.; Cordier, T.; Apothéloz-Perret-Gentil, L.; Cermakova, K.; Merzi, T.; Delefosse, M.; Blanc, P.; Pawlowski, J. Benthic monitoring of oil and gas offshore platforms in the North Sea using environmental DNA metabarcoding. Mol. Ecol. 2021, 30, 3007–3022. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, J.; Cermakova, K.; Cordier, T.; Frontalini, F.; Apothéloz-Perret-Gentil, L.; Merzi, T. Assessing the potential of nematode metabarcoding for benthic monitoring of offshore oil platforms. Sci. Total Environ. 2024, 933, 173092. [Google Scholar] [CrossRef]
- Alexander, J.B.; Marnane, M.J.; Elsdon, T.S.; Bunce, M.; Sitaworawet, P.; Songploy, S.; Chaiyakul, S.; Harvey, E.S. Using environmental DNA to better inform decision making around decommissioning alternatives for offshore oil and gas infrastructure. Sci. Total Environ. 2023, 901, 165991. [Google Scholar] [CrossRef]
- Chariton, A.A.; Stephenson, S.; Morgan, M.J.; Steven, A.D.L.; Colloff, M.J.; Court, L.N.; Hardy, C.M. Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries. Environ. Pollut. 2015, 203, 165–174. [Google Scholar] [CrossRef]
- Tagliabue, A.; Matterson, K.O.; Ponti, M.; Turicchia, E.; Abbiati, M.; Costantini, F. Sediment and bottom water eDNA metabarcoding to support coastal management. Ocean Coast. Manag. 2023, 244, 106785. [Google Scholar] [CrossRef]
- Avó, A.P.; Daniell, T.J.; Neilson, R.; Oliveira, S.; Branco, J.; Adao, H. DNA barcoding and morphological identification of benthic nematodes assemblages of estuarine intertidal sediments: Advances in molecular tools for biodiversity assessment. Front. Mar. Sci. 2017, 4, 66. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Muxika, I.; Rodríguez-Ezpeleta, N. Adapting metabarcoding-based benthic biomonitoring into routine marine ecological status assessment networks. Ecol. Indic. 2018, 95, 194–202. [Google Scholar] [CrossRef]
- Rey, A.; Viard, F.; Lizé, A.; Corre, E.; Valentini, A.; Thiriet, P. Coastal rocky reef fish monitoring in the context of the Marine Strategy Framework Directive: Environmental DNA metabarcoding complements underwater visual census. Ocean Coast. Manag. 2023, 241, 106625. [Google Scholar] [CrossRef]
- Kapshyna, I.; Veit-Koehler, G.; Hoffman, L.; Khodami, S. Impact of a coastal protection measure on sandy-beach meiofauna at Ahrenshoop (Baltic Sea, Germany): Results from metabarcoding and morphological approaches are similar. Metabarcoding Metagenomics 2024, 8, e127688. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Rodríguez-Ezpeleta, N. Environmental status assessment using DNA metabarcoding: Towards a genetics based marine biotic index (gAMBI). PLoS ONE 2014, 9, e90529. [Google Scholar] [CrossRef]
- Wangensteen, O.S.; Palacín, C.; Guardiola, M.; Turon, X. DNA metabarcoding of littoral hard-bottom communities: High diversity and database gaps revealed by two molecular markers. Peerj 2018, 6, e4705. [Google Scholar] [CrossRef] [PubMed]
- Castro-Cubillos, M.L.; Taylor, J.D.; Mastretta-Yanes, A.; Benítez-Villalobos, F.; Islas-Villanueva, V. Monitoring of benthic eukaryotic communities in two tropical coastal lagoons through eDNA metabarcoding: A spatial and temporal approximation. Sci. Rep. 2022, 12, 10089. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Zhou, X.; Li, Y.Y.; Liu, S.L.; Yang, Q.; Su, X.; Zhou, L.L.; Tang, M.; Fu, R.B.; Li, J.G.; Huang, Q.F. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. Gigascience 2013, 2, 2047-217X-2-4. [Google Scholar] [CrossRef]
- Mertes, F.; ElSharawy, A.; Sauer, S.; van Helvoort, J.; van der Zaag, P.J.; Franke, A.; Nilsson, M.; Lehrach, H.; Brookes, A.J. Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief. Funct. Genom. 2011, 10, 374–386. [Google Scholar] [CrossRef]
- Nota, K.; Orlando, L.; Marchesini, A.; Girardi, M.; Bertilsson, S.; Vernesi, C.; Parducci, L. Enriching barcoding markers in environmental samples utilizing a phylogenetic probe design: Insights from mock communities. Environ. DNA 2024, 6, e593. [Google Scholar] [CrossRef]
- Giarla, T.C.; Esselstyn, J.A. The challenges of resolving a rapid, recent radiation: Empirical and simulated phylogenomics of philippine shrews. Syst. Biol. 2015, 64, 727–740. [Google Scholar] [CrossRef]
- Horn, S. Target enrichment via DNA hybridization capture. In Ancient DNA: Methods and Protocols; Shapiro, B., Hofreiter, M., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 177–188. [Google Scholar]
- Borsetto, C.; Raguideau, S.; Travis, E.; Kim, D.W.; Lee, D.H.; Bottrill, A.; Stark, R.; Song, L.J.; Cha, C.J.; Pearson, J.; et al. Impact of sulfamethoxazole on a riverine microbiome. Water Res. 2021, 201, 117382. [Google Scholar] [CrossRef]
- Chen, W.J.; Wang, Z.G.; Xu, W.H.; Tian, R.M.; Zeng, J. Dibutyl phthalate contamination accelerates the uptake and metabolism of sugars by microbes in black soil. Environ. Pollut. 2020, 262, 114332. [Google Scholar] [CrossRef]
- Yu, K.; Yi, S.; Li, B.; Guo, F.; Peng, X.X.; Wang, Z.P.; Wu, Y.; Alvarez-Cohen, L.; Zhang, T. An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol A (BPA)-degrading microbial community. Microbiome 2019, 7, 16. [Google Scholar] [CrossRef]
- Schiml, V.C.; Delogu, F.; Kumar, P.; Kunath, B.; Batut, B.; Mehta, S.; Johnson, J.E.; Grüning, B.; Pope, P.B.; Jagtap, P.D.; et al. Integrative meta-omics in Galaxy and beyond. Environ. Microbiome 2023, 18, 56. [Google Scholar] [CrossRef]
- Segata, N.; Boernigen, D.; Tickle, T.L.; Morgan, X.C.; Garrett, W.S.; Huttenhower, C. Computational meta’omics for microbial community studies. Mol. Syst. Biol. 2013, 9, 666. [Google Scholar] [CrossRef]
- Michealsamy, A.; Thangamani, L.; Manivel, G.; Kumar, P.; Sundar, S.; Piramanayagam, S.; Natarajan, J. Current research and applications of meta-omics stratagems in bioremediation: A bird’s-eye view. J. Appl. Biotechnol. Rep. 2021, 8, 109–115. [Google Scholar] [CrossRef]
- Jeddi, M.Z.; Hopf, N.B.; Viegas, S.; Price, A.B.; Paini, A.; van Thriel, C.; Benfenati, E.; Ndaw, S.; Bessems, J.; Behnisch, P.A.; et al. Towards a systematic use of effect biomarkers in population and occupational biomonitoring. Environ. Int. 2021, 146, 106257. [Google Scholar] [CrossRef]
- Brockmeier, E.K.; Hodges, G.; Hutchinson, T.H.; Butler, E.; Hecker, M.; Tollefsen, K.E.; Garcia-Reyero, N.; Kille, P.; Becker, D.; Chipman, K.; et al. The role of omics in the application of Adverse Outcome Pathways for chemical risk assessment. Toxicol. Sci. 2017, 158, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Gatzidou, E.T.; Zira, A.N.; Theocharis, S.E. Toxicogenomics: A pivotal piece in the puzzle of toxicological research. J. Appl. Toxicol. 2007, 27, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Sauer, U.G.; Deferme, L.; Gribaldo, L.; Hackermüller, J.; Tralau, T.; van Ravenzwaay, B.; Yauk, C.; Poole, A.; Tong, W.D.; Gant, T.W. The challenge of the application of ‘omics technologies in chemicals risk assessment: Background and outlook. Regul. Toxicol. Pharmacol. 2017, 91, S14–S26. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; von Bergen, M.; Buesen, R.; Hackermuller, J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Buesen, R.; Chorley, B.N.; da Silva Lima, B.; Daston, G.; Deferme, L.; Ebbels, T.; Gant, T.W.; Goetz, A.; Greally, J.; Gribaldo, L.; et al. Applying ‘omics technologies in chemicals risk assessment: Report of an ECETOC workshop. Regul. Toxicol. Pharmacol. 2017, 91, S3–S13. [Google Scholar] [CrossRef]
- Tralau, T.; Oelgeschläger, M.; Gürtler, R.; Heinemeyer, G.; Herzler, M.; Höfer, T.; Itter, H.; Kuhl, T.; Lange, N.; Lorenz, N.; et al. Regulatory toxicology in the twenty-first century: Challenges, perspectives and possible solutions. Arch. Toxicol. 2015, 89, 823–850. [Google Scholar] [CrossRef]
- Escher, B.I.; Hackermuller, J.; Polte, T.; Scholz, S.; Aigner, A.; Altenburger, R.; Bohme, A.; Bopp, S.K.; Brack, W.; Busch, W.; et al. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ. Int. 2017, 99, 97–106. [Google Scholar] [CrossRef]
- Scala, G.; Kinaret, P.; Marwah, V.; Sund, J.; Fortino, V.; Greco, D. Multi-omics analysis of ten carbon nanomaterials effects highlights cell type specific patterns of molecular regulation and adaptation. NanoImpact 2018, 11, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ng, Q.X.; Zhang, B.; Wei, Z.; Hassan, M.; He, Y.; Ong, C.N. Employing multi-omics to elucidate the hormetic response against oxidative stress exerted by nC60 on Daphnia pulex. Environ. Pollut. 2019, 251, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Larras, F.; Billoir, E.; Scholz, S.; Tarkka, M.; Wubet, T.; Delignette-Muller, M.L.; Schmitt-Jansen, M. A multi-omics concentration-response framework uncovers novel understanding of triclosan effects in the chlorophyte Scenedesmus vacuolatus. J. Hazard. Mater. 2020, 397, 122727. [Google Scholar] [CrossRef]
- Jamers, A.; Blust, R.; De Coen, W.; Griffin, J.L.; Jones, O.A.H. An omics based assessment of cadmium toxicity in the green alga Chlamydomonas reinhardtii. Aquat. Toxicol. 2013, 126, 355–364. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.-L.; Tu, B.-J.; Yang, K.; Mo, T.-T.; Zhang, R.-Y.; Cheng, S.-Q.; Chen, C.-Z.; Jiang, X.-J.; Han, T.-L.; et al. Integrated Epigenetics, Transcriptomics, and Metabolomics to Analyze the Mechanisms of Benzo[a]pyrene Neurotoxicity in the Hippocampus. Toxicol. Sci. 2018, 166, 65–81. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, W.Y.; Liu, J.J.; Yan, W.J.; Zhang, Y.H.; Zhang, Z.; Zhang, P.D. Unraveling the impact of PFOA toxicity on Zostera marina using a multi-omics approach: Insights from growth, physiological, transcriptomic, and metabolomic signatures. J. Hazard. Mater. 2025, 486, 137024. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17, S15. [Google Scholar] [CrossRef]
- Buescher, J.M.; Driggers, E.M. Integration of omics: More than the sum of its parts. Cancer Metab. 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More is better: Recent progress in multi-omics data integration methods. Front. Genet. 2017, 8, 84. [Google Scholar] [CrossRef] [PubMed]
| Stressor(s) | Biological Group(s) | Marker(s) (Marker Region) | Application(s)/Country | Reference |
|---|---|---|---|---|
| Salmon Aquaculture | Bacteria | 16S (V4 region) ribosomal DNA (rDNA) | Monitoring benthic impacts/New Zealand | [149] |
| Bacteria | 16S (V4) rDNA | Monitoring benthic impacts/Norway | [132] | |
| Bacteria | 16S (V3-V4) rDNA | Monitoring benthic impacts/international | [150] | |
| Foraminifera | 18S (27F) rDNA | Monitoring benthic impacts/New Zealand | [151] | |
| Foraminifera | 18S (27F) rDNA | Monitoring benthic impacts/Norway | [146] | |
| Foraminifera | 18S (27F) rDNA | Monitoring benthic impacts/Canada | [152] | |
| Ciliates | 18S (V9) rDNA | Monitoring benthic impacts/Scotland | [132] | |
| Eukaryotes | 18S (V4) rDNA | Monitoring benthic impacts/Scotland | [153] | |
| Multi-trophic | 16S (V4) rDNA, 18S (27F) rDNA, 18S (V4) rDNA | Monitoring benthic impacts/New Zealand | [142] | |
| Multi-trophic | 16S (V3-V4) rDNA, 18S (V3-V4) rDNA | Monitoring benthic impacts/Norway | [154] | |
| Offshore Oil and Gas | Bacteria | 16S (V4) rDNA | Oil spill impacts/Gulf of Mexico | [155] |
| Bacteria | 16S (V3-V4) rDNA | Biocorrosive control/Brazil | [156] | |
| Foraminifera | 18S (27F region) rDNA | Monitoring benthic impacts/New Zealand | [157] | |
| Prokaryotes, Eukaryotes | 18S (V4-V5) rDNA | Monitoring benthic impacts/Norway | [158] | |
| Multi-trophic | 16S (V4) rDNA, 18S (V4) rDNA | Monitoring benthic impacts/New Zealand | [159] | |
| Multi-trophic | 18S (V1-V2; V9) rDNA, COI | Monitoring benthic impacts/North Sea (Denmark) | [160] | |
| Multi-trophic | 18S (V1-V2; V9) rDNA | Monitoring benthic impacts/North Sea (Denmark) | [161] | |
| Multi-trophic | ITS2, mitochondrial 16S, COI | Monitoring old platforms/Gulf of Thailand | [162] | |
| Coastal Development | Eukaryotes | 18S (V1-V2) rDNA | Excess organic enrichment Estuaries/Australia | [163] |
| Eukaryotes | COI | Metabarcoding for management/Italy | [164] | |
| Nematodes | 18S (V1-V2) rDNA, COI | Morpho-Molecular comparison/Estuary (Portugal) | [165] | |
| Macroinvertebrates | COI | Monitoring network/Basque Coast (Spain) | [166] | |
| Teleost Fish | Mitochondrial 16S | Coastal rocky reef fish monitoring/France | [167] | |
| Meiofauna | 18S (V1-V2) rDNA | Protected sandy beach/Germany | [168] | |
| Multi-trophic | 18S rDNA and COI | Metabarcoding-based gAMBI index/Spain | [169] | |
| Multi-trophic | 18S (V7) rDNA, COI | Hard-Bottom Impacts/Spain | [170] | |
| Multi-trophic | 16S (V1-V3) rDNA, 18S (V9) rDNA | Effects of multiple stressors/Australia | [141] | |
| Multi-trophic | 18S (V4) rDNA, COI | Tropical coastal lagoon/Mexico | [171] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, K.F.; Pochon, X.; Melvin, S.D.; Wheeler, T.T.; Tremblay, L.A. Integration of “Omics”-Based Approaches in Environmental Risk Assessment to Establish Cause and Effect Relationships: A Review. Toxics 2025, 13, 714. https://doi.org/10.3390/toxics13090714
Smith KF, Pochon X, Melvin SD, Wheeler TT, Tremblay LA. Integration of “Omics”-Based Approaches in Environmental Risk Assessment to Establish Cause and Effect Relationships: A Review. Toxics. 2025; 13(9):714. https://doi.org/10.3390/toxics13090714
Chicago/Turabian StyleSmith, Kirsty F., Xavier Pochon, Steven D. Melvin, Thomas T. Wheeler, and Louis A. Tremblay. 2025. "Integration of “Omics”-Based Approaches in Environmental Risk Assessment to Establish Cause and Effect Relationships: A Review" Toxics 13, no. 9: 714. https://doi.org/10.3390/toxics13090714
APA StyleSmith, K. F., Pochon, X., Melvin, S. D., Wheeler, T. T., & Tremblay, L. A. (2025). Integration of “Omics”-Based Approaches in Environmental Risk Assessment to Establish Cause and Effect Relationships: A Review. Toxics, 13(9), 714. https://doi.org/10.3390/toxics13090714







