Abstract
Mercury (Hg) is a dangerous and persistent trace element. Its organic and highly toxic form, methylmercury (MeHg), easily crosses biological membranes and accumulates in biota. Nevertheless, understanding the mechanisms of dietary MeHg toxicity in fish remains a challenge. A time-course experiment was conducted with juvenile white seabreams, Diplodus sargus (Linnaeus, 1758), exposed to realistic levels of MeHg in feed (8.7 μg g−1, dry weight), comprising exposure (E; 7 and 14 days) and post-exposure (PE; 28 days) periods. Total Hg levels increased with time in gills and liver during E and decreased significantly in PE (though levels of control fish were reached only for gills), with liver exhibiting higher levels (2.7 times) than gills. Nuclear magnetic resonance (NMR)-based metabolomics revealed multiple and often differential metabolic changes between fish organs. Gills exhibited protein catabolism, disturbances in cholinergic neurotransmission, and changes in osmoregulation and lipid and energy metabolism. However, dietary MeHg exposure provoked altered protein metabolism in the liver with decreased amino acids, likely for activation of defensive strategies. PE allowed for the partial recovery of both organs, even if with occurrence of oxidative stress and changes of energy metabolism. Overall, these findings support organ-specific responses according to their sensitivity to Hg exposure, pointing out that indications obtained in biomonitoring studies may depend also on the selected organ.
1. Introduction
Mercury (Hg) is recognized as a dangerous metallic pollutant because of its persistence and high toxicity to organisms [1], and in recent years its increase in aquatic environments rekindled serious environmental and human health concerns [2]. The high affinity of Hg for cell membrane lipids [3] and its ability to interfere with cellular events such as growth, proliferation, differentiation, or damage repair processes [4] classify it as one of the most hazardous and toxic pollutants. Its presence in aquatic ecosystems is enhanced by natural events such as forest fires and geological emissions, as well as by emissions related to fossil fuel combustion, industrial applications, or mining [5,6,7].
A major concern is the ability of Hg to be easily transferred along trophic chains through bioaccumulation and biomagnification processes and then be accumulated in wildlife at high trophic levels [8,9]. In particular, in aquatic environments, its organic and highly toxic form, methylmercury (MeHg), is produced both in the water column and in sediments by certain sulfate-reducing bacteria and tends to be accumulated mainly in the tissues of aquatic organisms such as fish [10,11]. It is noteworthy that this trace element can be absorbed by aquatic organisms either aqueously, through, e.g., respiratory surfaces, or through the diet, resulting in absorption in the gastrointestinal tract [12,13]. Indeed, due to its high solubility in lipids, single valence, and small size, MeHg can easily cross biological membranes and be completely absorbed from the digestive tract [14] and readily reach the bloodstream for distribution to various organs. Furthermore, it is also known that the elimination rates of the organic form of Hg are very slow, leading to its bioconcentration within the cells of organisms, thus causing biological impairments even at very low doses [15,16].
Although Hg toxicity has been the topic of several studies [17,18,19], clarifying the mechanisms of dietary MeHg toxicity in fish remains challenging. Fish have been widely used in ecotoxicological studies as worthy sentinel organisms [6,20,21,22,23,24,25,26] due to their ability to accumulate and metabolize different contaminants in their tissues and to produce valid and measurable responses (biomarkers) when exposed to stressful conditions, such as the presence of Hg.
The white seabream, Diplodus sargus (Linnaeus, 1758), is a common demersal fish that inhabits infralittoral and circalittoral rocky habitats in the Mediterranean Sea and eastern Atlantic Ocean [27], and it has a relatively long-life span and an omnivorous benthic diet. D. sargus accumulates dangerous elements in its tissues, such as Hg [28], even if it is a medium-size fish. These remarkable features make D. sargus a valuable bioindicator species for monitoring aquatic contaminants. In previous studies [29,30], white seabream was used as animal model to shed light on MeHg neurotoxicity after dietary exposure by combining bioaccumulation levels, oxidative stress profiles, and behavior assessment. However, these conventional biochemical assays often need the support of high-throughput methods to gain more comprehensive insights into the biological effects induced by environmental pollutants on target organisms. Environmental metabolomics is regarded as a very accurate method of investigation that assesses variation in the physiological state of organisms coping with different environmental scenarios, even under experimental conditions, through the simultaneous evaluation of a large number of biomolecules [31]. Lin et al. [32] identified metabolomics as the comprehensive analysis of all low-molecular-weight (<1500 Da) endogenous metabolites that may vary according to the physiological state, developmental stage, or pathological state of cells, tissues, or organs, or of the entire organism under investigation. In particular, metabolomics based on proton nuclear magnetic resonance (1H NMR) allows for the simultaneous analysis of a wide range of metabolites involved in different metabolic pathways, thus offering numerous advantages for elucidating organism–environment interactions. In addition, it allows for the identification of novel metabolic biomarkers in response to stress in organisms provoked by changes in abiotic factors, diseases, or environmental pollutants [31,32,33,34,35,36,37,38,39,40,41].
Although previous studies have highlighted the detrimental impact of MeHg on the brain of white seabream [29,30], so far, to our knowledge, no studies have examined its effects on organs such as gills and liver through the use of environmental metabolomics. Due to certain features (i.e., large surface area in close contact with dissolved water pollutants, intense blood flow, and high cell regeneration, which are useful to indicate more recent pollutant exposures), fish gills have been considered an important target of aquatic pollution, and therefore they are frequently used in environmental biomonitoring studies [40,41,42]. Furthermore, gills are also capable of storing pollutants absorbed in the gut [43], including MeHg [17,44]. Similarly, the liver is widely used to assess the health status of fish [41,45], as it is an important organ of metabolic activity including detoxification processes. Furthermore, its important role in bioaccumulation, transformation, and Hg cycling processes confirms the importance of liver for understanding the toxic effects and metabolic malfunctions induced by possible contact with this pollutant [17,20,44].
In order to further improve the current knowledge on the effects of Hg on aquatic biota, the present study was designed to compare the cytotoxic effects of dietary MeHg on gills and liver of white seabream, D. sargus, through the evaluation of Hg bioaccumulation and metabolomic profiling. In detail, fish were exposed by diet to a realistic concentration of MeHg (8.7 μg g−1 dry feed) over 14 days. Successively, a post-exposure period of 28 days was also taken into consideration in order to estimate the recovery ability of the two selected target organs in fish.
2. Materials and Methods
2.1. Chemicals
Deuterium oxide (D2O, heavy water; 99.8 atom %D; CAS-No. 7789-20-0) for NMR spectroscopy was obtained from Armar AG Chemicals (Dottingen, Switzerland). The 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS; 97%, molecular weight: 218.32 g mol−1; CAS-No. 2039-96-5) was purchased from Sigma-Aldrich (Milan, Italy). The other chemicals required to conduct the metabolomics analysis, namely, methanol (molecular weight: 32.04 g mol−1; CAS-No. 67-56-1) and chloroform (molecular weight: 119.38 g mol−1; CAS-No. 67-66-3) were also purchased from Sigma-Aldrich (Milan, Italy). For the experimental exposure, MeHg chloride (molecular weight: 251.08 g mol−1; CAS-No. 115-09-3) was purchased from Sigma-Aldrich Chemical (Madrid, Spain). Other routine chemicals used in this work were of analytical grade and acquired from local suppliers.
2.2. Experimental Set-Up and Sampling
Juvenile white seabreams (Diplodus sargus), provided by the Aquaculture Research Station (IPMA—Olhão, Portugal), were used in the experiment. Fish wellbeing deserved permanent attention in accordance with national and international guidelines for the protection of animal welfare. Fish were allowed to acclimatize to experimental conditions and routines for two weeks prior to MeHg exposure. Water temperature, salinity, and pH were monitored daily, varying as follows, respectively: 13.5 ± 0.3 °C, 35 ± 2, and 7–8.
Fish were held in 300 L fiberglass tanks in an average density of 0.056 kg L−1 (initial fish weight: 124 ± 11 g; initial total length: 18 ± 0.6 cm) under a 10 h light:14 h dark photoperiod. Seawater was renewed daily (around 80%), and fish were fed once a day, namely, 1–2 h before water renewal. In sampling days, fish were not fed in the 12 h preceding handling that started around 09:00 am.
Fish were fed with feed (3 mm pellets) produced by SPAROS Company (Olhão, Portugal) composed by 44% protein and 16% lipids. A solution of MeHg chloride (prepared in ethanol) was added during the process of pellet production, with a homogenous distribution of the toxicant throughout the batch. MeHg levels in contaminated pellets (8.7 ± 0.5 μg g−1 dry weight) used to expose fish to this Hg form were similar to those previously detected in natural food of D. sargus (e.g., Nereis diversicolor), as previously described [29]. Control fish were fed with uncontaminated pellets, with the same size and nutritional formulation, which were produced following an identical methodology but without the addition of MeHg (residues lower than 0.01 μg g−1).
Fish were exposed to MeHg for 7 (E7) and 14 (E14) days. Then, to potentially allow recovery for 28 days, fish started to be fed with uncontaminated pellets (post-exposure; PE28) (Figure 1). Thus, the experiment had a total duration of 42 days. At each sampling time (E7, E14, and PE28), fish were sacrificed for two different analyses as follows: (i) eight fish for total Hg determinations; and (ii) six fish for metabolomics analysis (Figure 1). During sampling, fish were anesthetized with 0.2 mg L−1 tricaine methanesulfonate (MS-222; molecular weight: 261.29 g mol−1; CAS-No. 886-86-2) purchased from Sigma-Aldrich, China, and buffered with NaHCO3. Fish were then weighed, measured, sacrificed by cervical transection, and properly bled from the cardinal vein (using heparinized Pasteur pipettes). Gills and liver were excised, and the two sets of samples stored at −80 °C until further processing for Hg determination and metabolomics.
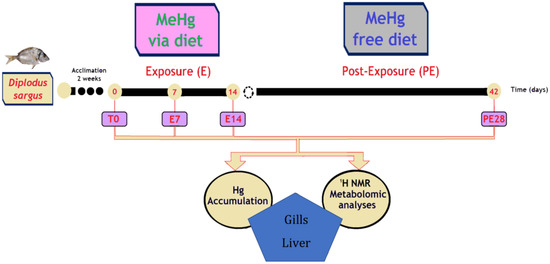
Figure 1.
Experimental design with white seabream, Diplodus sargus, comprising an acclimation time of 2 weeks (T0) followed by an exposure period to MeHg via diet for 7 (E7) and 14 days (E14). Thereafter, fish were fed with a MeHg-free diet for 28 days (Day 42; PE28) to allow recovery. In parallel, control groups of fish were also considered. At each experimental time, gills and liver were collected to measure levels of Hg and to perform a metabolomic analysis.
2.3. Total Hg Determination
Firstly, gills and liver samples were lyophilized, homogenized, and then analyzed for total Hg (tHg) in an advanced mercury analyzer (AMA) (AMA254, LECO Instruments), according to the U.S. EPA method 7473 [46]. Briefly, the initial preparation step of the analytical process of AMA was removing the moisture through a drying process to concentrate Hg in the sample. Thermal decomposition (around 750 °C) was used to pyrolytically reduce Hg in the sample to its elemental form. Elemental Hg was then trapped on a gold amalgamator and eventually liberated by heating the amalgamator. Elemental Hg was transported by a stream of oxygen and measured by atomic absorption spectrometry [47]. Certified reference materials (fish protein: DORM-4; dogfish liver: DOLT-4) from the Canadian National Research Council were used to ensure the accuracy of the procedure, and the obtained values were consistent with the certified ones.
2.4. Metabolomics Analysis
2.4.1. Extraction of Metabolites
Polar metabolites were extracted from liver and gill tissues of white seabreams (n = 6 per condition at each sampling time) by applying a “two-step” methanol/chloroform/water protocol, as reported in detail in previous works [35,40]. Briefly, frozen 100 mg sub-samples of each fish tissue were transferred in 2 mL Eppendorf tubes with the addition of cold methanol (4 mL g−1) and cold distilled water (0.85 mL g−1) in order to be homogenized using a TissueLyser LT bead mill (Qiagen, Hilden, Germany), with the inclusion of a 3.2 mm stainless steel bead in each tube. Therefore, homogenization took place for 10 min at 50 vibrations/s. The homogenates, after addition of chloroform (4 mL g−1) and distilled water (2 mL g−1), were then vortexed for 60 s to be mixed and left on ice for 10 min to partition. Afterward, samples were centrifuged for 5 min at 2000 g at 4 °C, resulting in a triphasic mixture. The upper methanol layer (600 µL) with polar metabolites was then carefully removed and transferred into clean vials to be dried in a centrifugal vacuum concentrator (Eppendorf 5301). The resulting pellet was then kept at −80 °C. Prior to NMR analysis, the dried extracts were resuspended in 600 μL of a 0.1 M sodium phosphate buffer (pH 7.0, 10% D2O) with 1 mM DSS, used as internal reference, and then transferred to a 5 mm diameter NMR tube. DSS acts as an internal standard and provides a chemical shift reference (δ = 0.0 ppm) for the NMR spectra, while D2O provides a deuterium lock for the NMR spectrometer.
2.4.2. 1H NMR-Based Metabolomics and Spectral Pre-Processing
A Varian-500 NMR spectrometer working at a spectral frequency of 499.74 MHz at 298 K was used to analyze fish tissue extracts. To obtain one-dimensional (1-D) 1H NMR spectra, a PRESAT pulse sequence was applied for suppressing the resonance of residual water, with 6 kHz spectral width and a 2.0 s relaxation delay. A total of 128 transients was collected into 16,384 data points requiring a 10 min acquisition time for each sample under investigation. All data sets were zero filled to 32,768 data points, and exponential line-broadenings of 0.5 Hz were used before application of the Fourier transformation. A Chenomx Processor, a module of Chenomx NMR Suite (version 5.1; Chenomx Inc., Edmonton, Canada) software, was then used for manually phasing, baseline-correction, and calibration (DSS at 0.0 ppm) of all 1H NMR spectra acquired. The Chenomx 500 MHz library database, another module included into the Chenomx NMR Suite software, was utilized for identifying the different peaks profiled within the acquired 1H NMR spectra from each seabream gill and liver in order to be assigned with reference to known chemical shifts and peak multiplicities. Furthermore, the use of Chenomx NMR Profiler, another module also included in the Chenomx NMR Suite software, allowed us to quantify the level of each individual metabolite detected in the 1H NMR spectra using the known concentration of the internal standard DSS, previously introduced in each sample prior to the NMR analysis [48,49]. Thereafter, the concentration of the metabolites of interest recorded in seabream gills and liver at all the experimental times was expressed as mean ± standard deviation (SD).
2.5. Data Statistical Analysis
GraphPad software (Prism 7.0, San Diego, CA, USA) was used for all the statistical analyses. Bioaccumulation data on the levels of total Hg (μg g−1) measured in seabream gills and liver were presented as mean ± SD, and statistically significant differences (p < 0.05) between the control and exposed fish at all the experimental times were established by application of a two-way analysis of variance (ANOVA) followed by the post hoc Sidak’s multiple comparisons test. Metabolomics data were expressed in mM as mean ± SD. For each fish organ, metabolite changes were calculated via the ratio between the averages of exposed fish at each sampling time and control peak areas. The metabolic dataset was tested for normality using the Shapiro–Wilk distribution test, and after confirmation of normal distribution, data homogeneity was evaluated through the Levene test. Data were analyzed by one-way ANOVA using Dunnett’s multiple comparison test, which was aimed at finding significant differences between the control and exposed fish at each experimental time. Data were considered statistically significant at p < 0.05.
3. Results
3.1. Total Hg Accumulation
The levels of total Hg (tHg) measured at all experimental times in the gills and liver of white seabreams exposed to MeHg by diet, as well as those of control fish, are depicted in Figure 2. Overall, a significant rise of tHg levels was observed in both fish organs throughout the 14-day exposure period, followed by a drop during the post-exposure time of 28 days. In particular, the highest tHg levels were detected at E14, both in the gills (3.7 μg g−1) and in the liver (10.2 μg g−1), with the latter organ depicting levels 2.7 times higher. It is worth noting that during the post-exposure period, tHg levels decreased for both organs but remained above the values recorded in the control fish.
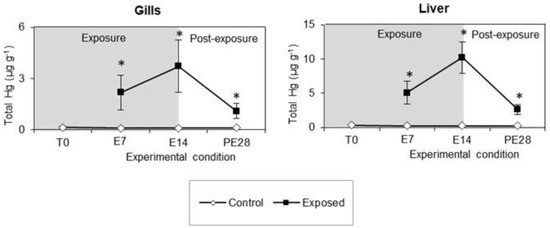
Figure 2.
Total Hg level (tHg; μg g−1) measured in the gills and liver of white seabream exposed to MeHg and in control fish at each experimental time over 14 days of exposure (7 and 14 days, corresponding to E7 and E14) and 28 days of post-exposure (day 42, corresponding to PE28). Data correspond to mean ± standard deviation (n = 8). Significant differences (Sidak’s test; p < 0.05) in relation to the control group are indicated by * for each experimental time.
3.2. Metabolomics
3.2.1. 1H NMR Spectroscopy of Gill and Liver Extracts of Unexposed Fish
Typical 1-D 1H NMR spectra of the metabolome of the gills and liver of juvenile white seabream D. sargus from the control group are shown in Figure 3. Among the several metabolites identified, it was found that gills were dominated by the osmolyte taurine, whose concentration was found to be about eight times higher than that of the other metabolites. Conversely, the metabolome of liver was characterized by a dominant presence of carbohydrates (e.g., glucose, maltose, fructose), whose concentrations were up to 60 times higher than those of the other detected metabolites. Among the other prominent classes of compounds found in the metabolome of both fish organs, amino acids (e.g., glutamate, alanine), Kreb’s cycle intermediates (e.g., succinate), and nucleotides (e.g., uracil) were observed. It is also worth noting that some metabolites such as arginine, malonate, choline, acetylcholine, betaine, and inosine were exclusively detected in the gills, whereas cholate, acetate, acetone, cystathionine, trimethylamine N-oxide (TMAO), maltose, fructose, glycogen, adenosine triphosphate/adenosine diphosphate (ATP/ADP), and niacinamide were observed only in the liver.
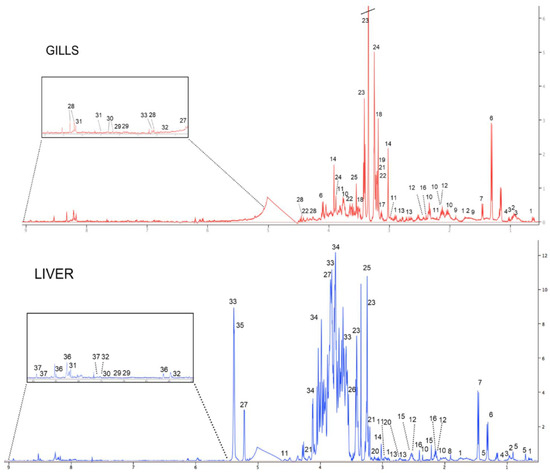
Figure 3.
Representative 1-D 500 MHz 1H NMR spectra of the gills and liver of white seabream, Diplodus sargus, from the control group: (1) DSS, (2) leucine, (3) isoleucine, (4) valine, (5) cholate, (6) lactate, (7) alanine, (8) acetate, (9) arginine, (10) glutamate, (11) glutathione, (12) glutamine, (13) aspartate, (14) creatine, (15) acetone, (16) succinate, (17) malonate, (18) choline, (19) acetylcholine, (20) cystathionine, (21) phosphocholine, (22) glycerophosphocholine, (23) taurine, (24) betaine, (25) TMAO, (26) glycine, (27) glucose, (28) inosine, (29) tyrosine, (30) phenylalanine, (31) hypoxanthine, (32) uracil, (33) maltose, (34) fructose, (35) glycogen, (36) ATP/ADP, and (37) niacinamide.
3.2.2. Metabolome of Gills in MeHg-Exposed Fish
Dietary exposure to MeHg provoked alterations in different classes of metabolites in fish gills at each experimental time (Table 1). Indeed, a significant increase in the levels of several amino acids (leucine, isoleucine, valine, alanine, tyrosine, phenylalanine) was recorded during the exposure period (E7; E14). However, it is worth noting the significant decline observed in almost all amino acids at the post-exposure time that lasted 28 days (PE28). Regarding energy metabolism, a significant notable rise in lactate was noted at all the experimental times with respect to the control, combined with a complex pattern of variation in the levels of malonate, glycogen, and glucose. Furthermore, significant increases in the levels of osmolytes (taurine, glycerophosphocholine), the neurotransmitter acetylcholine, and miscellaneous metabolites (phosphocholine, inosine) were also observed with respect to the control at all MeHg experimental times and in the post-exposure period, in which fish were fed with a MeHg-free diet.

Table 1.
Percent changes in concentrations of metabolites in the gills of white seabream (D. sargus) following MeHg-dietary exposure (E7, E14) and post-exposure (PE28) periods, in relation to control groups (n = 6) (Dunnett’s test, * p < 0.05; ↑ and ↓ indicate an increase and decrease in respect to the control group, respectively).
3.2.3. Metabolome of Liver in MeHg-Exposed Fish
Dietary exposure to MeHg provoked alterations in different classes of metabolites in fish liver at each experimental time (Table 2). In regard to amino acids, a significant increase was found in the level of alanine at all the exposure and post-exposure experimental times combined with a significant drop in the concentration of glutamine and glutamate. In terms of energy production, the concentration of glucose decreased significantly at all the experimental times, whilst the ATP/ADP ratio increased significantly. Overall, with regard to other metabolites, significant reductions in glutathione at PE28 and creatine levels at all the experimental times were detected, associated with a significant enhancement of cystathionine and niacinamide levels at both the exposure and post-exposure experimental times.

Table 2.
Percent changes in concentrations of metabolites in the liver of white seabream (D. sargus) following MeHg-dietary exposure (E7, E14) and post-exposure (PE28) periods in relation to control groups (n = 6) (Dunnett’s test, * p < 0.05; ↑ and ↓ indicate an increase and decrease in respect to the control group, respectively).
4. Discussion
To date, the use of metabolomics has proven to be an extremely effective tool for investigations of the perturbed metabolic pathways in aquatic biota triggered by a variety of pollutants [26,31,32,49,50,51], including Hg [20,40,41]. In this work, the metabolomic approach was applied on gills and liver of white seabream, D. sargus, with the aim of elucidating the cytotoxic effects of dietary MeHg and the potential recovery ability of the selected target organs during a successive post-exposure period, revealing differential impairments in various metabolic pathways.
The use of 1H NMR-based metabolomics allowed for the simultaneous detection of 27 metabolites in the gills and 31 metabolites in the liver. Interestingly, some metabolites were in common among the two fish organs, though organ-specific metabolites were also observed. In detail, the presence of metabolites involved in neurotransmission (i.e., acetylcholine, choline) and osmoregulation (betaine) was observed solely in the gills, whereas some carbohydrates and molecules involved in energy metabolism (i.e., fructose, maltose, glycogen, ATP/ADP, niacinamide), as well as ketone bodies (i.e., acetate, acetone), were predominantly recorded in the liver. The evidence of organ-specific metabolomic profiles, both in terms of presence of specific metabolites and in terms of their concentrations, as previously described in other aquatic organisms [35,52], may be explained by the differential physiological functioning of the two examined organs. In fact, the gills of fish are a multipurpose organ that, in addition to respiratory gas exchange, play prominent roles in feeding by filtering food particles, osmotic regulation, active ion transport, and nitrogenous waste excretion [35,43]. Conversely, the liver of fish is a target organ involved in a number of metabolic activities, including energy and carbohydrate metabolism [35,53], besides for playing a key role in accumulation, biotransformation, and cycling of environmental pollutants, because of its effective detoxification system useful to counteract the harmful effects of hazardous chemicals [54].
The different physiological role of the two examined organs would also explain their dissimilar capacity to accumulate Hg. In fact, in the present study it was found that the liver, responsible for detoxification of pollutants, showed more than two times higher levels of total Hg than the gills following MeHg dietary exposure. In both organs, total Hg levels tended to decrease during the post-exposure period, though concentrations did not reach baseline levels after 28 days of depuration, as they remained statistically higher than the values recorded in the control, thereby attesting to the strong persistence of Hg within biological tissues [29,55,56,57]. Comparable results were also observed in white seabream following waterborne exposure and post-exposure periods to inorganic mercury (iHg) in a study aimed at elucidating Hg toxicokinetics [58]. Overall, the differential bioaccumulation of Hg, together with the physiological specificities of the two examined fish organs, were on the basis of organ-specific impairments observed in a variety of metabolic pathways.
4.1. MeHg-Induced Metabolome Changes in Fish Gills
It is well known that free amino acids are the most abundant components in the gills of fish [38,41]. In this study, the dietary exposure of white seabreams to realistic concentration of MeHg (8.7 μg g−1) provoked, at all the selected exposure times (E7, E14), an increase in the level of branched-chain amino acids (BCAAs, including leucine, isoleucine, and valine), tyrosine, phenylalanine, and alanine, likely due to a MeHg-induced protein catabolism, followed by a noteworthy drop in their concentrations at the successive post-exposure period (PE28), perhaps associated with a possible occurrence of protein biosynthesis as a repair or cell renewal/turnover process. As is widely documented in the literature, BCAAs play a key function in the immune system by both regulating protein turnover and being precursors for the biosynthesis of new molecules (i.e., immunoglobulins, interleukins, and chemokines), which are essential for cells such as lymphocytes to correctly perform their functions [59]. Thus, the rise in BCAA levels could suggest the involvement of the immune system to counteract the detrimental impact of MeHg in the gills. The occurrence of intense protein turnover activity appears also to be confirmed by the increased levels of alanine during the exposure period. Indeed, this amino acid is implicated in nitrogen metabolism, which underlies the processes of nitrogen waste excretion occurring in the gills as a consequence of protein catabolism [43,60]. Therefore, these results hint at the existence of ammonia excretion impairment triggered by dietary exposure to MeHg, which nevertheless seemed to be slightly mitigated during the post-exposure period.
Moreover, the higher levels of tyrosine and phenylalanine, coupled with the enhancement of acetylcholine detected at all the experimental times, may be associated with alterations in the neurotransmission system. As a matter of fact, tyrosine and phenylalanine are precursors of the neurotransmitter dopamine, which is implicated in the dopaminergic system [61], while acetylcholine is the major neurotransmitter involved in the cholinergic system. It is known that both these nervous systems are strictly associated with the hypoxic signaling of fish gills [62]. Hence, the changes observed in these metabolites may be related to the disruption of the gas exchange processes [63], therefore corroborating the MeHg neurotoxic effects already documented in previous works carried out on the brain of white seabreams upon exposure to Hg forms [29,39]. Moreover, a plausible reason for the increase in acetylcholine could also be the inhibition of acetylcholine esterase (AChE) activity, already demonstrated in fish exposed to MeHg [64,65].
The depression of neuronal activity could therefore result in an alteration of the normal functioning of the gills, also supported by the increased concentration of alanine as discussed above. The increased energy demand would seem to be partly counterbalanced by the enhanced levels of lactate, which may suggest a shift towards anaerobic energy metabolism [41]. Furthermore, the increased levels of malonate, a precursor for fatty acid synthesis [66], indicate also impairments in the energy metabolism related to the highly biosynthetic activity aimed at mitigating the harmful impact caused by dietary MeHg exposure in white seabream.
Besides changes in energy metabolism, in the gills of fish the osmoregulation system appeared also to be disrupted following exposure to MeHg via diet, as supported by the increased levels of taurine and glycerophosphocholine observed during all the exposure and post-exposure times. The MeHg effect on the concentration of osmolytes confirms the high sensitivity of the osmoregulatory system in aquatic organisms exposed to Hg species and other classes of pollutants [20,67,68]. However, as already reported in previous works [20,35], it is noteworthy to highlight that the raised levels of glycerophosphocholine and phosphocholine, both precursors of phosphatidylcholine, could suggest the occurrence of phospholipid breakdown, leading to cell membrane instability, persisting even during the following 28 days when the specimens were fed by a MeHg-free diet [69].
Moreover, among the changes observed in the metabolome of fish gills, it is interesting to note the increased levels of inosine, likely due to the breakdown of adenosine, which are a sign of cellular stress, commonly occurring during high energy demand [70]. In addition, Li et al. [71] pointed out how inosine may exert a vasodilatory effect on the gill lamellae of fish. Therefore, the increase in this metabolite, persisting even during the post-exposure period, hints at the emergence of an energy deficit in the branchial tissue, probably due to the intense biosynthesis activity elicited by dietary MeHg exposure.
Finally, it is interesting to note how certain metabolites, and in particular those involved in neurotransmission, osmoregulation, and cell membrane stability, appeared to be altered even during the post-exposure period (PE28). This may reflect a higher sensitivity of certain metabolic pathways than others to MeHg exposure and emphasize the persistence of certain harmful effects triggered by MeHg, even after a considerable time beyond the exposure phase [29,72].
4.2. MeHg-Induced Metabolome Changes in Fish Liver
The MeHg dietary exposure of white seabreams provoked alterations in some metabolic pathways also at the hepatic level that, with some exceptions, appeared differently from those revealed at the gills. Among amino acids, the changes observed in the levels of alanine were comparable with those recorded in the gills. In fact, the concentrations of alanine increased during all the exposure and post-exposure times, thus confirming the impact of MeHg in nitrogen metabolism [43,64].
Contrarily to the gills, changes in the amino acids glutamate and glutamine were observed in the liver. These metabolites, which are precursors of glutathione, were found to be reduced especially during the selected exposure times. These impairments, taken together with the drop recorded in the levels of glutathione, support the involvement of the antioxidant system to face the impact triggered by MeHg exposure, as already widely reported in the literature, since it is known that a decisive way to initiate Hg toxicity in organisms is through oxidative stress [20,29,35,40,41]. Moreover, the decrease in glutathione levels could also be associated with the high MeHg reactivity towards the reduced sulphydryl group [73]. In this regard, Akiyama et al. [74] emphasized the relevance of the enzyme cystathionine γ-lyase (CSE) in producing reactive sulphur species useful in mitigating the detrimental impact of MeHg. Therefore, the enhancement of cystathionine, the main substrate of CSE, detected in the liver of fish by the metabolomics approach, could suggest the MeHg-induced inhibition of CSE.
Moreover, in the liver of seabreams dietarily exposed to MeHg, variations in the levels of glucose, ATP/ADP, creatine, and niacinamide were also noted and could be ascribed to disruptions in energy metabolism. In fact, the drop revealed in the glucose levels and the unchanged level of glycogen, despite the regular food administration to seabreams, suggest hepatic gluconeogenesis inhibition, contrarily to what was observed in the golden grey mullet collected in a highly Hg-polluted environment [20,41]. However, the increased ATP/ADP levels, coupled with increased concentrations of niacinamide, which is the NAD precursor, could indicate high energy production in the liver necessary to sustain the detoxification and repairing actions [37] to face the detrimental effects of MeHg. Furthermore, creatine is a key molecule in the transfer of energy through organs and tissues [75]. Thus, its reduction in the fish liver, despite the high ATP/ADP levels detected, could be justified by its transfer to tissues with a higher energy demand [76,77].
Contrary to the results observed in the gills, the levels of several metabolites detected at the hepatic level appeared to be altered even at PE28, indicating a higher sensitivity to MeHg of this organ than the gills, probably linked also to the higher Hg accumulation recorded in the liver. Therefore, these findings further support the occurrence of organ-specific responses elicited by fish in accordance with their sensitivity to Hg exposure, since it has been previously documented that MeHg is able to affect differently even parts of the same organ, as observed within the brain optic tectum of white seabreams [39].
5. Conclusions
In this work, the NMR-based metabolomic approach allowed us to shed light into the toxic metabolic pathways triggered by dietary MeHg exposure in juveniles of white seabream, Diplodus sargus, as well as to reveal organ-specific cytotoxicity mechanisms induced by MeHg in the gills and liver. In brief, following exposure to MeHg via diet, an intense energy catabolism associated with disruption of neurotransmission and osmoregulation systems was observed in the gills, whereas impaired antioxidant and detoxification systems were revealed in the liver, coupled with changes in the energy metabolism. Although the post-exposure period of 28 days had allowed a partial recovery of both target organs, there was still evidences of oxidative stress and changes of the energy metabolism, revealing long-term effects of dietary MeHg in fish. Overall, organ-specific cytotoxicity mechanisms of dietary MeHg exposure were discerned, pointing out the vulnerability of fish health to this Hg form at highly impacted ecosystems and the need for increased surveillance in this direction, therefore emphasizing the urgency of the further development of new remediation strategies against this persistent environmental pollutant.
Author Contributions
Conceptualization, T.C. and M.P.; methodology, T.C. and P.P.; validation, T.C., P.P. and M.P.; formal analysis, T.C., B.B., M.G., F.B. and G.D.M.; investigation, T.C., G.D.M., B.B., F.B., M.G., P.P. and M.P.; data curation, T.C. and P.P.; writing—original draft preparation, G.D.M., B.B. and T.C.; writing—review and editing, T.C., P.P. and M.P.; visualization, T.C., F.B. and G.D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Portuguese Foundation for Science and Technology (FCT, I.P.) via a researcher contract to P. Pereira (CEECIND/01144/2017), which also supported CESAM (UIDP/50017/2020+UIDB/50017/2020+LA/P/0094/2020) through National funds. Moreover, this work was also supported by the “FFABR_RD_CAPPELLO_TIZIANA_ ATENEO_2020” research funds.
Institutional Review Board Statement
This work was carried out according to the EU Directive 2010/63/EU of 22nd September on the protection of organisms used for scientific purposes, under the supervision of a team member (Mário Pacheco, University of Aveiro; Portugal) authorized by the competent authorities.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
Thanks are due to FCT/MCTES for the financial support to CESAM (UIDP/50017/2020+UIDB/50017/2020), through National funds. Pereira P. is funded by National funds (OE) through FCT (Fundação para a Ciência e a Tecnologia, I.P.), under the Scientific Employment Stimulus–Individual Call (CEECIND/01144/2017).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cao, L.; Liu, J.; Dou, S.; Huang, W. Biomagnification of methylmercury in a marine food web in Laizhou Bay (North China) and associated potential risks to public health. Mar. Pollut. Bull. 2020, 150, 110762. [Google Scholar] [CrossRef] [PubMed]
- Paduraru, E.; Iacob, D.; Rarinca, V.; Rusu, A.; Jijie, R.; Ilie, O.-D.; Ciobica, A.; Nicoara, M.; Doroftei, B. Comprehensive review regarding mercury poisoning and its complex involvement in Alzheimer’s disease. Int. J. Mol. Sci. 2022, 23, 1992. [Google Scholar] [CrossRef] [PubMed]
- Kerek, E.; Hassanin, M.; Prenner, E.J. Inorganic mercury and cadmium induce rigidity in eukaryotic lipid extracts while mercury also ruptures red blood cells. BBA–Biomembranes 2018, 1860, 710–717. [Google Scholar] [CrossRef]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadegh, M. Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef] [PubMed]
- Obrist, D.; Kirk, J.L.; Zhang, L.; Sunderland, E.M.; Jiskra, M.; Selin, N.E. A review of global environmental mercury processes in response to human and natural perturbations: Changes of emissions, climate, and land use. Ambio 2018, 47, 116–140. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Korbas, M.; Pereira, V.; Cappello, T.; Maisano, M.; Canario, J.; Almeida, A.; Pacheco, M. A multidimensional concept for mercury neuronal and sensory toxicity in fish—From toxicokinetics and biochemistry to morphometry and behavior. BBA–Gen. Subj. 2019, 1863, 129298. [Google Scholar] [CrossRef] [PubMed]
- Parrino, V.; Costa, G.; Giannetto, A.; De Marco, G.; Cammilleri, G.; Acar, Ü.; Piccione, G.; Fazio, F. Trace elements (Al, Cd, Cr, Cu, Fe, Mn, Ni, Pb and Zn) in Mytilus galloprovincialis and Tapes decussatus from Faro and Ganzirri Lakes (Sicily, Italy): Flow cytometry applied for hemocytes analysis. J. Trace Elem. Med. Biol. 2021, 68, 126870. [Google Scholar] [CrossRef]
- Polak-Juszczak, L. Distribution of organic and inorganic mercury in the tissues and organs of fish from the southern Baltic Sea. Environ. Sci. Pollut. Res. 2018, 25, 34181–34189. [Google Scholar] [CrossRef]
- Łuczyńska, J.; Łuczyński, M.J.; Nowosad, J.; Kowalska-Góralska, M.; Senze, M. Total mercury and fatty acids in selected fish species on the Polish market: A risk to human health. Int. J. Environ. Res. Public Health 2022, 19, 10092. [Google Scholar] [CrossRef]
- Mason, R.P.; Coulibaly, M.; Hansen, G.; Inman, H.; Myer, P.K.; Yao, K.M. An examination of mercury levels in the coastal environment and fish of Cote d’Ivoire. Chemosphere 2022, 300, 134609. [Google Scholar] [CrossRef]
- Olson, C.I.; Fakhraei, H.; Driscoll, C.T. Mercury emissions, atmospheric concentrations, and wet deposition across the conterminous United States: Changes over 20 years of monitoring. Environ. Sci. Technol. Lett. 2020, 7, 376–381. [Google Scholar] [CrossRef]
- Arcagni, M.; Juncos, R.; Rizzo, A.; Pavlin, M.; Fajon, V.; Arribere, M.A.; Horvat, M.; Guevara, S.R. Species- and habitat-specific bioaccumulation of total mercury and methylmercury in the food web of a deep oligotrophic lake. Sci. Total Environ. 2018, 612, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.M.; Lopez, L.; Bhavsar, S.P.; Sharma, S. What’s hot about mercury? Examining the influence of climate on mercury levels in Ontario top predator fishes. Environ. Res. 2018, 162, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, T.W. The pharmacology of mercury compounds. Annu. Rev. Pharmacol. 1972, 12, 375–406. [Google Scholar] [CrossRef]
- Taylor, D.L.; Calabrese, N.M. Mercury content of blue crabs (Callinectes sapidus) from southern New England coastal habitats: Contamination in an emergent fishery and risks to human consumers. Mar. Pollut. Bull. 2018, 126, 166–178. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Carocci, A.; Lauria, G.; Catalano, A. Mercury exposure and heart diseases. Int. J. Environ. Res. Public Health 2017, 14, 74. [Google Scholar] [CrossRef]
- Raihan, S.M.; Moniruzzaman, M.; Park, Y.; Lee, S.; Bai, S.C. Evaluation of dietary organic and inorganic mercury threshold levels on induced mercury toxicity in a marine fish model. Animals 2020, 10, 405. [Google Scholar] [CrossRef]
- Zulkipli, S.Z.; Liew, H.J.; Ando, M.; Lim, L.S.; Wang, M.; Sung, Y.Y.; Mok, W.J. A review of mercury pathological effects on organs specific of fishes. Environ. Pollut. Bioavailab. 2021, 33, 76–87. [Google Scholar] [CrossRef]
- Strungaru, S.A.; Robea, M.A.; Plavan, G.; Todirascu-Ciornea, E.; Ciobica, A.; Nicoara, M. Acute exposure to methylmercury chloride induces fast changes in swimming performance, cognitive processes and oxidative stress of zebrafish (Danio rerio) as reference model for fish community. J. Trace Elem. Med. Biol. 2018, 47, 115–123. [Google Scholar] [CrossRef]
- Brandão, F.; Cappello, T.; Raimundo, J.; Santos, M.A.; Maisano, M.; Mauceri, A.; Pacheco, M.; Pereira, P. Unravelling the mechanisms of mercury hepatotoxicity in wild fish (Liza aurata) through a triad approach: Bioaccumulation, metabolomic profiles and oxidative stress. Metallomics 2015, 7, 1352–1363. [Google Scholar] [CrossRef]
- Khadra, M.; Caron, A.; Dolors, P.; Ponton, D.E.; Rosabal, M.; Amyot, M. The fish or the egg: Maternal transfer and subcellular partitioning of mercury and selenium in Yellow Perch (Perca flavescens). Sci. Total Environ. 2019, 675, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Liu, X.; Wang, Z.; Wang, B.; Lin, C.; Xin, M.; Zhang, B.T.; Wu, T.; He, M.; Ouyang, W. Trophic transfer and dietary exposure risk of mercury in aquatic organisms from urbanized coastal ecosystems. Chemosphere 2021, 281, 130836. [Google Scholar] [CrossRef] [PubMed]
- Noureen, A.; De Marco, G.; Rehman, N.; Jabeen, F.; Cappello, T. Ameliorative hematological and histomorphological effects of dietary Trigonella foenum-graecum seeds in common carp (Cyprinus carpio) exposed to copper oxide nanoparticles. Int. J. Environ. Res. Public Health 2022, 19, 13462. [Google Scholar] [CrossRef] [PubMed]
- Parrino, V.; De Marco, G.; Minutoli, R.; Lo Paro, G.; Giannetto, A.; Cappello, T.; De Plano, L.M.; Cecchini, S.; Fazio, F. Effects of pesticides on Chelon labrosus (Risso, 1827) evaluated by enzymatic activities along the north eastern Sicilian coastlines (Italy). Eur. Zool. J. 2021, 88, 540–548. [Google Scholar] [CrossRef]
- Yuan, L.; Shi, X.; Tang, B.Z.; Wang, W.X. Real-time in vitro monitoring of the subcellular toxicity of inorganic Hg and methylmercury in zebrafish cells. Aquat. Toxicol. 2021, 236, 105859. [Google Scholar] [CrossRef]
- Zitouni, N.; Cappello, T.; Missawi, O.; Boughattas, I.; De Marco, G.; Belbekhouche, S.; Mokni, M.; Alphonse, V.; Guerbej, H.; Bousserrhine, N.; et al. Metabolomic disorders unveil hepatotoxicity of environmental microplastics in wild fish Serranus scriba (Linnaeus 1758). Sci. Total Environ. 2022, 838, 155872. [Google Scholar] [CrossRef]
- Giacalone, V.M.; Pipitone, C.; Abecasis, D.; Badalamenti, F.; D’Anna, G. Movement ecology of the white seabream Diplodus sargus across its life cycle: A review. Environ. Biol. Fish. 2022, 105, 1809–1823. [Google Scholar] [CrossRef]
- Merciai, R.; Rodriguez-Prieto, C.; Torres, J.; Casadevall, M. Bioaccumulation of mercury and other trace elements in bottom-dwelling omnivorous fishes: The case of Diplodus sargus (L.) (Osteichthyes: Sparidae). Mar. Pollut. Bull. 2018, 136, 10–21. [Google Scholar] [CrossRef]
- Cardoso, O.; Puga, S.; Brandão, F.; Canário, J.; O’Driscoll, N.J.; Santos, M.A.; Pacheco, M.; Pereira, P. Oxidative stress profiles in brain point out a higher susceptibility of fish to waterborne divalent mercury compared to dietary organic mercury. Mar. Pollut. Bull. 2017, 122, 110–121. [Google Scholar] [CrossRef]
- Puga, S.; Pereira, P.; Pinto-Ribeiro, F.; O’Driscoll, N.J.; Mann, E.; Barata, M.; Pousão-Ferreira, P.; Canário, J.; Almeida, A.; Pacheco, M. Unveiling the neurotoxicity of methylmercury in fish (Diplodus sargus) through a regional morphometric analysis of brain and swimming behavior assessment. Aquat. Toxicol. 2016, 180, 320–333. [Google Scholar] [CrossRef]
- Cappello, T. NMR-based metabolomics of aquatic organisms. eMagRes 2020, 9, 81–100. [Google Scholar] [CrossRef]
- Lin, C.Y.; Viant, M.R.; Tjeerdema, R.S. Metabolomics: Methodologies and applications in the environmental sciences. J. Pestic. Sci. 2006, 31, 245–251. [Google Scholar] [CrossRef]
- Caliani, I.; De Marco, G.; Cappello, T.; Giannetto, A.; Mancini, G.; Ancora, S.; Maisano, M.; Parrino, V.; Cappello, S.; Bianchi, N.; et al. Assessment of the effectiveness of a novel BioFilm-Membrane BioReactor oil-polluted wastewater treatment technology by applying biomarkers in the mussel Mytilus galloprovincialis. Aquat. Toxicol. 2022, 243, 106059. [Google Scholar] [CrossRef]
- Cappello, T.; Vitale, V.; Oliva, S.; Villari, V.; Mauceri, A.; Fasulo, S.; Maisano, M. Alteration of neurotransmission and skeletogenesis in sea urchin Arbacia lixula embryos exposed to copper oxide nanoparticles. Comp. Biochem. Physiol. C 2017, 199, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Cappello, T.; De Marco, G.; Oliveri Conti, G.; Giannetto, A.; Ferrante, M.; Mauceri, A.; Maisano, M. Time-dependent metabolic disorders induced by short-term exposure to polystyrene microplastics in the Mediterranean mussel Mytilus galloprovincialis. Ecotoxicol. Environ. Saf. 2021, 209, 111780. [Google Scholar] [CrossRef] [PubMed]
- De Marco, G.; Brandão, F.; Pereira, P.; Pacheco, M.; Cappello, T. Organ-specific metabolome deciphering cell pathways to cope with mercury in wild fish (golden grey mullet Chelon auratus). Animals 2022, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, T.; Li, M.; Xu, H.; Jia, A.; Zhang, J.; Wang, J. 1H NMR based metabolomics approach to study the toxic effects of dichlorvos on goldfish (Carassius auratus). Chemosphere 2015, 138, 537–545. [Google Scholar] [CrossRef]
- Missawi, O.; Venditti, M.; Cappello, T.; Zitouni, N.; Marco, G.D.E.; Boughattas, I.; Bousserrhine, N.; Belbekhouche, S.; Minucci, S.; Maisano, M.; et al. Autophagic event and metabolomic disorders unveil cellular toxicity of environmental microplastics on marine polychaete Hediste diversicolor. Environ. Pollut. 2022, 302, 119106. [Google Scholar] [CrossRef]
- Xu, H.-D.; Wang, J.-S.; Li, M.-H.; Liu, Y.; Chen, T.; Jia, A.-Q. 1H NMR based metabolomics approach to study the toxic effects of herbicide butachlor on goldfish (Carassius auratus). Aquat. Toxicol. 2015, 159, 69–80. [Google Scholar] [CrossRef]
- Cappello, T.; Brandão, F.; Guilherme, S.; Santos, M.A.; Maisano, M.; Mauceri, A.; Canário, J.; Pacheco, M.; Pereira, P. Insights into the mechanisms underlying mercury-induced oxidative stress in gills of wild fish (Liza aurata) combining 1H NMR metabolomics and conventional biochemical assays. Sci. Total Environ. 2016, 548–549, 13–24. [Google Scholar] [CrossRef]
- Cappello, T.; Pereira, P.; Maisano, M.; Mauceri, A.; Pacheco, M.; Fasulo, S. Advances in understanding the mechanisms of mercury toxicity in wild golden grey mullet (Liza aurata) by 1H NMR-based metabolomics. Environ. Pollut. 2016, 219, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Pablo, H.D.; Vale, C.; Pacheco, M. Combined use of environmental data and biomarkers in fish (Liza aurata) inhabiting a eutrophic and metal contaminated coastal system—Gills reflect environmental contamination. Mar. Environ. Res. 2010, 69, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H. The fish gill: Site of action and model for toxic effects of environmental pollutants. Environ. Health Perspect. 1987, 71, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Leaner, J.J.; Mason, R.P. Methylmercury uptake and distribution kinetics in sheepshead minnows, Cyprinodon variegatus, after exposure to CH3Hg-spiked food. Environ. Toxicol. Chem. 2004, 23, 2138–2146. [Google Scholar] [CrossRef]
- Ardeshir, R.A.; Movahedinia, A.A.; Rastgar, S. Fish liver biomarkers for heavy metal pollution: A review article. Am. J. Toxicol. 2017, 2, 1–8. [Google Scholar]
- EPA. Method 7473 (SW-846): Mercury in Solids and Solutions by Thermal Decomposition, Amalgamation, and Atomic Absorption Spectrophotometry; EPA: Washington, DC, USA, 1998. [Google Scholar]
- Costley, C.T.; Mossop, K.F.; Dean, J.R.; Garden, L.M.; Marshall, J.; Carrol, J. Determination of mercury in environmental and biological samples using pyrolysis atomic absorption spectrometry with gold amalgamation. Anal. Chim. Acta 2000, 405, 179–183. [Google Scholar] [CrossRef]
- Cappello, T.; Maisano, M.; Giannetto, A.; Natalotto, A.; Parrino, V.; Mauceri, A.; Spanò, N. Pen shell Pinna nobilis L. (Mollusca, Bivalvia) from different peculiar environments: Adaptive mechanisms of osmoregulation and neurotransmission. Eur. Zool. J. 2019, 86, 333–342. [Google Scholar] [CrossRef]
- Vignet, C.; Cappello, T.; Fu, Q.; Lajoie, K.; De Marco, G.; Clerandeau, C.; Mottaz, H.; Maisano, M.; Hollender, J.; Schirmer, K.; et al. Imidacloprid induces adverse effects on fish early life stages that are more severe in Japanese medaka (Oryzias latipes) than in zebrafish (Danio rerio). Chemosphere 2019, 225, 470–478. [Google Scholar] [CrossRef]
- Kostopoulou, S.; Ntatsi, G.; Arapis, G.; Aliferis, K.A. Assessment of the effects of metribuzin, glyphosate, and their mixtures on the metabolism of the model plant Lemna minor L. applying metabolomics. Chemosphere 2020, 239, 124582. [Google Scholar] [CrossRef]
- Tian, S.; Teng, M.; Meng, Z.; Yan, S.; Jia, M.; Li, R.; Liu, L.; Yan, J.; Zhou, Z.; Zhu, W. Toxicity effects in zebrafish embryos (Danio rerio) induced by prothioconazole. Environ. Pollut. 2019, 255, 113269. [Google Scholar] [CrossRef]
- Cappello, T.; Giannetto, A.; Parrino, V.; Maisano, M.; Oliva, S.; De Marco, G.; Guerriero, G.; Mauceri, A.; Fasulo, S. Baseline levels of metabolites in different tissues of mussel Mytilus galloprovincialis (Bivalvia: Mytilidae). Comp. Biochem. Physiol. D 2018, 26, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T.W. Glucose metabolism in fish: A review. J. Comp. Physiol. B 2012, 182, 1015–1045. [Google Scholar] [CrossRef] [PubMed]
- Ung, C.Y.; Lam, S.H.; Hlaing, M.M.; Winata, C.L.; Korzh, S.; Mathavan, S.; Gong, Z. Mercury-induced hepatotoxicity in zebrafish: In vivo mechanistic insights from transcriptome analysis, phenotype anchoring and targeted gene expression validation. BMC Genom. 2010, 11, 212. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Puga, S.; Cardoso, V.; Pinto-Ribeiro, F.; Raimundo, J.; Barata, M.; Pousão-Ferreira, P.; Pacheco, M.; Almeida, A. Inorganic mercury accumulation in brain following waterborne exposure elicits a deficit on the number of brain cells and impairs swimming behavior in fish (white seabream—Diplodus sargus). Aquat. Toxicol. 2016, 170, 400–412. [Google Scholar] [CrossRef]
- Xu, Q.; Zhao, L.; Wang, Y.; Xie, Q.; Yin, D.; Feng, X.; Wang, D. Bioaccumulation characteristics of mercury in fish in the Three Gorges Reservoir, China. Environ. Pollut. 2018, 243, 115–126. [Google Scholar] [CrossRef]
- Zheng, N.; Wang, S.; Dong, W.; Hua, X.; Li, Y.; Song, X.; Chu, Q.; Hou, S.; Li, Y. The toxicological effects of mercury exposure in marine fish. Bull. Environ. Contam. Toxicol. 2019, 102, 714–720. [Google Scholar] [CrossRef]
- Pereira, P.; Raimundo, J.; Barata, M.; Araujo, O.; Pousao-Ferreira, P.; Canario, J.; Almeida, A.; Pahceco, M. A new page on the road book of inorganic mercury in fish body—Tissue distribution and elimination following waterborne exposure and post-exposure periods. Metallomics 2015, 7, 525–535. [Google Scholar] [CrossRef]
- Calder, P.C. Branched-chain amino acids and immunity. J. Nutr. 2006, 136, 288S–293S. [Google Scholar] [CrossRef]
- Bucking, C. A broader look at ammonia production, excretion, and transport in fish: A review of impacts of feeding and the environment. J. Comp. Physiol. B 2017, 187, 1–18. [Google Scholar] [CrossRef]
- Reed, M.; Jonz, M.G. Neurochemical signalling associated with gill oxygen sensing and ventilation: A receptor focused mini-review. Front. Physiol. 2022, 13, 1396. [Google Scholar] [CrossRef]
- Pan, W.; Godoy, R.S.; Cook, D.P.; Scott, A.L.; Nurse, C.A.; Jonz, M.G. Single-cell transcriptomic analysis of neuroepithelial cells and other cell types of the gills of zebrafish (Danio rerio) exposed to hypoxia. Sci. Rep. 2022, 12, 10144. [Google Scholar] [CrossRef] [PubMed]
- Zachar, P.C.; Pan, W.; Jonz, M.G. Distribution and morphology of cholinergic cells in the branchial epithelium of zebrafish (Danio rerio). Cell Tissue Res. 2017, 367, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Scheuhammer, A.M.; Rouvinen-Watt, K.; Grochowina, N.; Klenavic, K.; Evans, R.D.; Man Chan, H. Methylmercury impairs components of the cholinergic system in captive mink (Mustela vison). Toxicol. Sci. 2006, 91, 202–209. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, T.B.; Colombi, J.S.; Ribeiro, C.A.O.; De Assis, H.C.S.; De Carvalho, C.E.V. Cholinesterase activity in methylmercury and mercury chloride exposure fish. Ecotoxicol. Environ. Contam. 2013, 8, 147–148. [Google Scholar] [CrossRef]
- Chen, H.; Diao, X.; Wang, H.; Zhou, H. An integrated metabolomic and proteomic study of toxic effects of benzo[a]pyrene on gills of the pearl oyster Pinctada martensii. Ecotoxicol. Environ. Saf. 2018, 156, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Sullivan, H.; Seligman, C.; Gilchrist, S.; Wang, B. An NMR-based metabolomics study on sea anemones Exaiptasia diaphana (Rapp, 1829) with atrazine exposure. Mol. Omi. 2021, 17, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; You, L.; Zhou, D.; Wang, Q.; Li, F.; Cong, M.; Li, L.; Zhao, J.; Liu, D.; et al. Benzo(a)pyrene-induced metabolic responses in Manila clam Ruditapes philippinarum by proton nuclear magnetic resonance (1H NMR) based metabolomics. Environ. Toxicol. Pharmacol. 2011, 32, 218–225. [Google Scholar] [CrossRef]
- Karakach, T.K.; Huenupi, E.C.; Soo, E.C.; Walter, J.A.; Afonso, L.O.B. 1H-NMR and mass spectrometric characterization of the metabolic response of juvenile Atlantic salmon (Salmo salar) to long-term handling stress. Metabolomics 2009, 5, 123–137. [Google Scholar] [CrossRef]
- Qin, H.; Yu, Z.; Zhu, Z.; Lin, Y.; Xia, J.; Jia, Y. The integrated analyses of metabolomics and transcriptomics in gill of GIFT tilapia in response to long term salinity challenge. Aquac. Fish. 2022, 7, 131–139. [Google Scholar] [CrossRef]
- Li, Q.-Q.; Xiang, Q.-Q.; Lian, L.-H.; Chen, Z.-Y.; Luo, X.; Ding, C.-Z.; Chen, L.-Q. Metabolic profiling of nanosilver toxicity in the gills of common carp. Ecotoxicol. Environ. Saf. 2021, 222, 112548. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y.; Wang, F.; Luo, Z.; Guo, S.; Strähle, U. Toxicity of mercury: Molecular evidence. Chemosphere 2020, 245, 125586. [Google Scholar] [CrossRef] [PubMed]
- Kerper, L.E.; Ballatori, N.; Clarkson, T.W. Methylmercury transport across the blood-brain barrier by an amino acid carrier. Am. J. Physiol. Integr. Comp. Physiol. 1992, 262, R761–R765. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M.; Unoki, T.; Yoshida, E.; Ding, Y.; Yamakawa, H.; Shinkai, Y.; Ishii, I.; Kumagai, Y. Repression of mercury accumulation and adverse effects of methylmercury exposure is mediated by cystathionine γ-lyase to produce reactive sulfur species in mouse brain. Toxicol. Lett. 2020, 330, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Kazak, L.; Cohen, P. Creatine metabolism: Energy homeostasis, immunity and cancer biology. Nat. Rev. Endocrinol. 2020, 16, 421–436. [Google Scholar] [CrossRef]
- Borchel, A.; Verleih, M.; Rebl, A.; Kühn, C.; Goldammer, T. Creatine metabolism differs between mammals and rainbow trout (Oncorhynchus mykiss). Springerplus 2014, 3, 510. [Google Scholar] [CrossRef]
- Gronczewska, J.; Niedźwiecka, N.; Grzyb, K.; Skorkowski, E.F. Bioenergetics of fish spermatozoa with focus on some herring (Clupea harengus) enzymes. Fish Physiol. Biochem. 2019, 45, 1615–1625. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).