Non-Lethal Concentrations of CdCl2 Cause Marked Alternations in Cellular Stress Responses within Exposed Sertoli Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Experimental Conditions
2.3. Formazan Formation Assay
2.4. Cellular ROS Assay
2.5. TUNEL Assay
2.6. RNA Extraction, cDNA Synthesis
2.7. Quantitative Polymerase Chain Reaction (qPCR)
2.8. Protein Preparation
2.9. Immunoblotting Analysis
2.10. MDA Assay
2.11. Dual-Labeled Immunofluorescence Analysis
2.12. Statistical Analysis
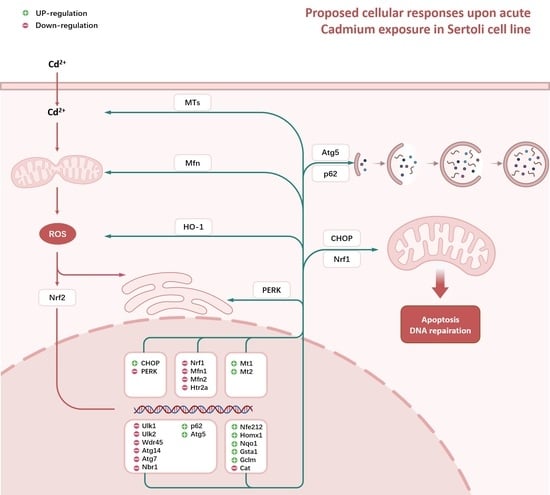
3. Results
3.1. Subsection
3.1.1. The Viability of TM4 Cells upon CdCl2 Treatment
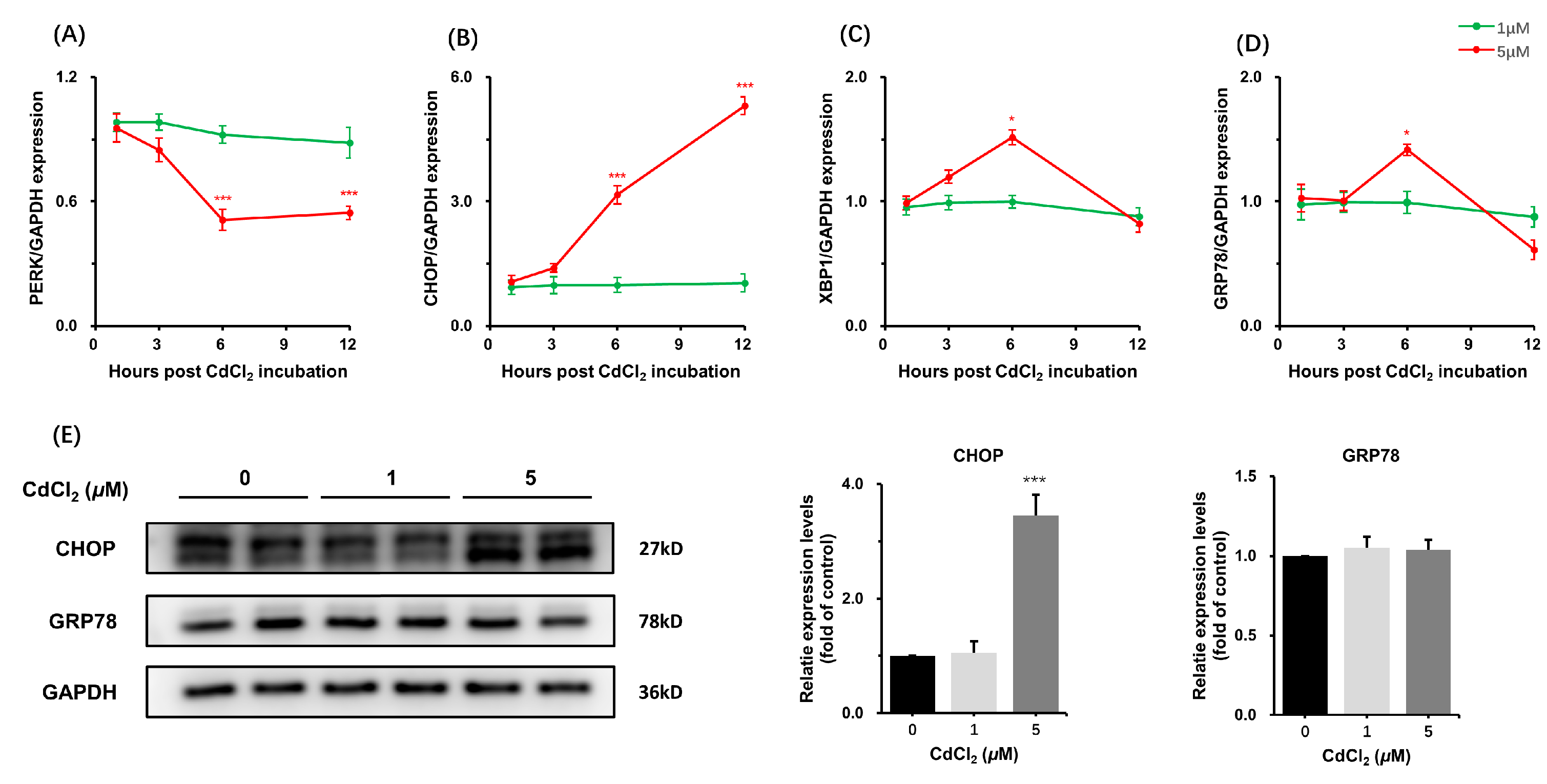
3.1.2. Effects of CdCl2 Treatment on Endoplasmic Reticulum Stress
3.1.3. Effects of CdCl2 Treatment on Mitochondrial Unfolded Protein Response (UPRmt) and Mitochondrial Dynamics
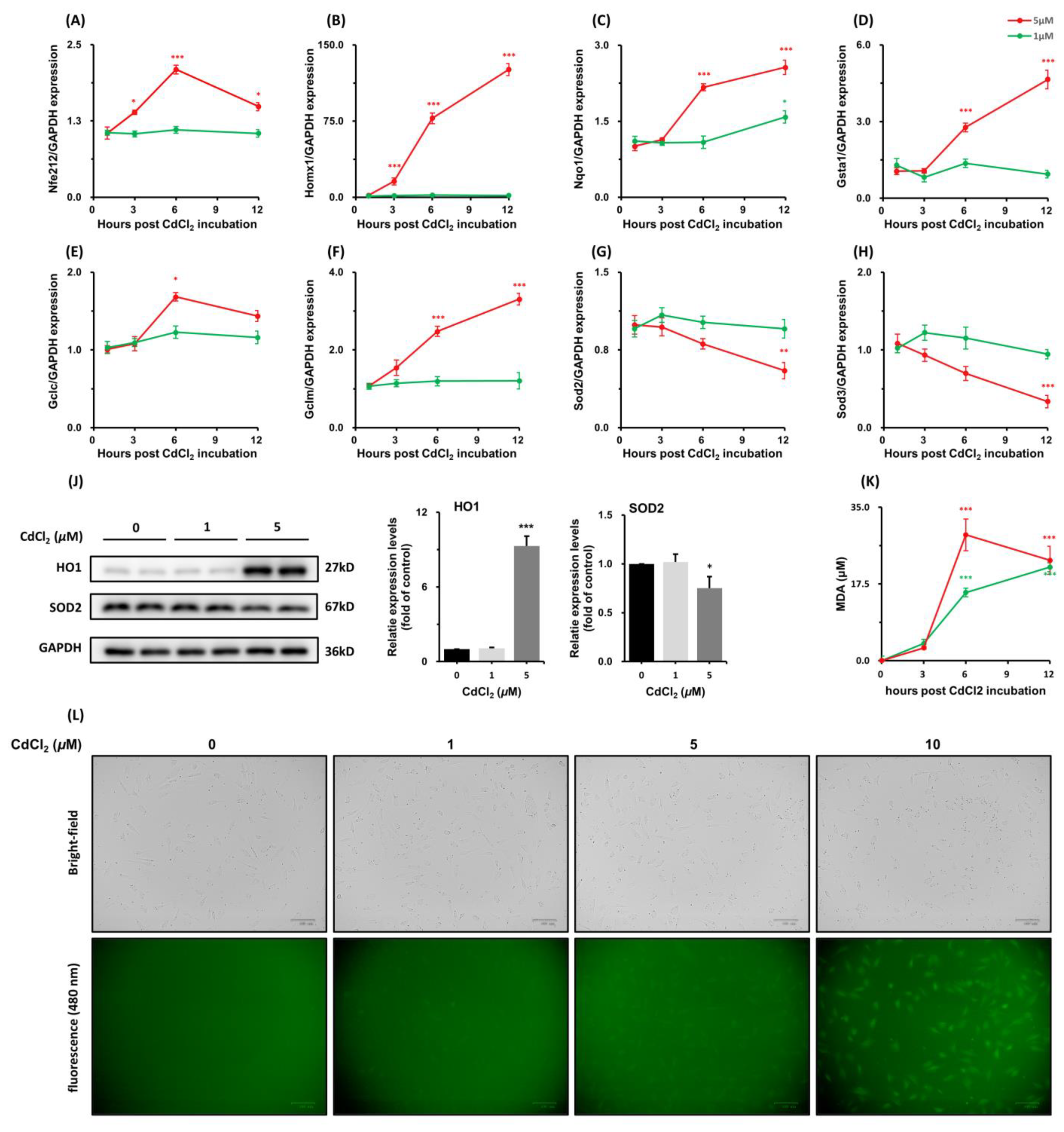
3.1.4. Effects of CdCl2 Treatment on Nrf2 Antioxidative Response-Related Genes
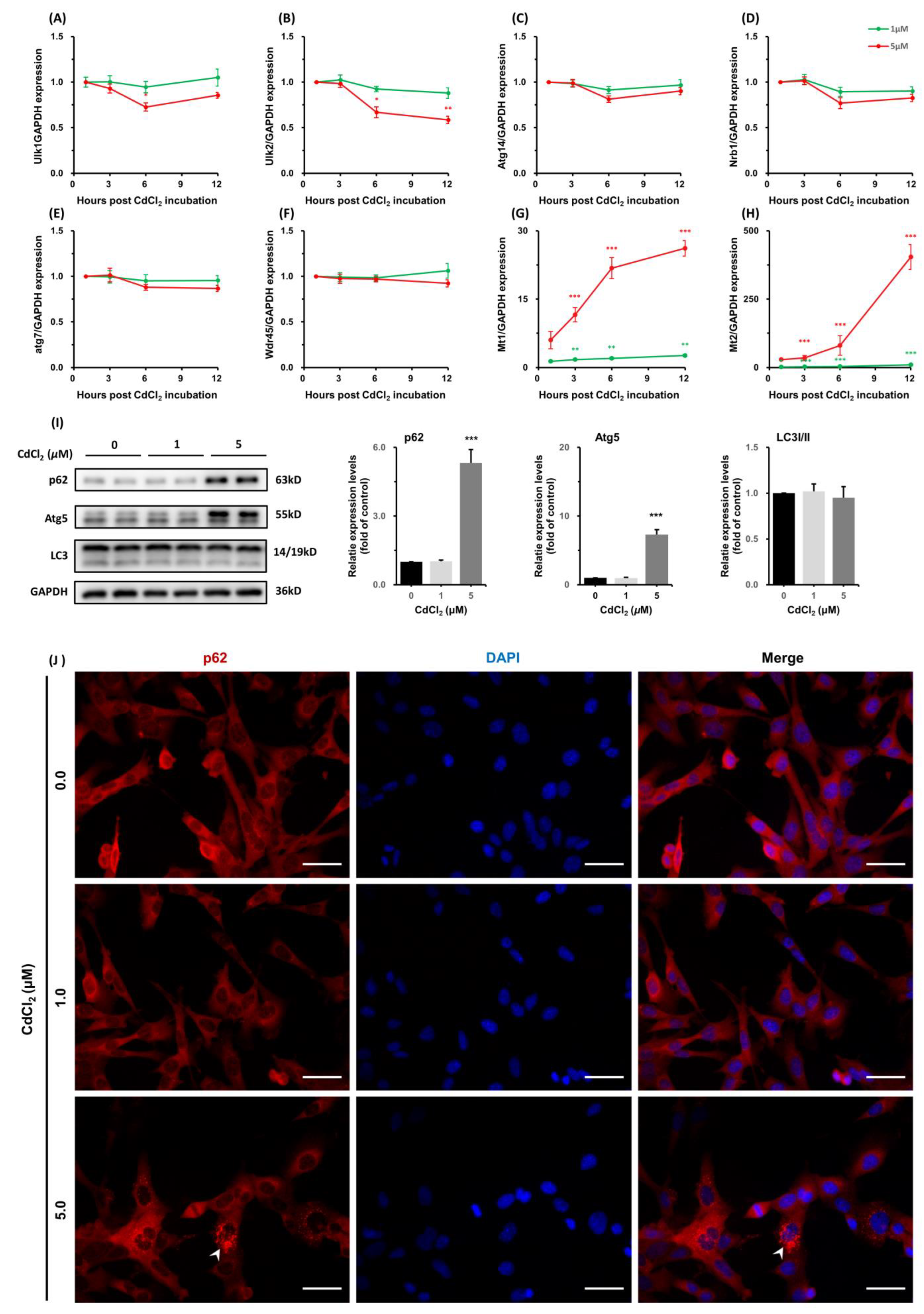
3.1.5. CdCl2 Caused Interferences in the Expression of Autophagy-Related Genes and Increases in Metallothioneins (MTs) Transcription
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anetor, G.O.; Nwobi, N.L.; Igharo, G.O.; Sonuga, O.O.; Anetor, J.I. Environmental Pollutants and Oxidative Stress in Terrestrial and Aquatic Organisms: Examination of the Total Picture and Implications for Human Health. Front. Physiol. 2022, 13, 931386. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef] [PubMed]
- Satarug, S.; Garrett, S.H.; Sens, M.A.; Sens, D.A. Cadmium, environmental exposure, and health outcomes. Environ. Health Perspect. 2010, 118, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, B.; Barregard, L. Review of cadmium exposure and smoking-independent effects on atherosclerotic cardiovascular disease in the general population. J. Intern. Med. 2021, 290, 1153–1179. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, Z.; Song, W.; Hong, D.; Huang, L.; Li, Y. A review on Cadmium Exposure in the Population and Intervention Strategies Against Cadmium Toxicity. Bull. Environ. Contam. Toxicol. 2021, 106, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhou, B.; Liu, H.; Cai, L. Comprehensive Review of Cadmium Toxicity Mechanisms in Male Reproduction and Therapeutic Strategies. Rev. Environ. Contam. Toxicol. 2021, 258, 151–193. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-L.; Wang, J.-Y.; Gong, L.-L.; Gan, S.; Gu, C.-M.; Wang, S.-S. Association between cadmium exposure and urolithiasis risk: A systematic review and meta-analysis. Medicine 2018, 97, e9460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-X.; Zhu, H.-L.; Shi, X.-T.; Nan, Y.; Liu, W.-B.; Dai, L.-M.; Xiong, Y.-W.; Yi, S.-J.; Cao, X.-L.; Xu, D.-X.; et al. Autophagy in Sertoli cell protects against environmental cadmium-induced germ cell apoptosis in mouse testes. Environ. Pollut. 2021, 270, 116241. [Google Scholar] [CrossRef]
- Ramos-Treviño, J.; Bassol-Mayagoitia, S.; Hernández-Ibarra, J.A.; Ruiz-Flores, P.; Nava-Hernández, M.P. Toxic Effect of Cadmium, Lead, and Arsenic on the Sertoli Cell: Mechanisms of Damage Involved. DNA Cell Biol. 2018, 37, 600–608. [Google Scholar] [CrossRef]
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef]
- Yokonishi, T.; McKey, J.; Ide, S.; Capel, B. Sertoli cell ablation and replacement of the spermatogonial niche in mouse. Nat. Commun. 2020, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-R.; Liu, Y.-X. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 2015, 149, R159–R167. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Matsumura, T.; Akiyama, H.; Kanai, Y.; Ogawa, T.; Sato, T. Sertoli cell replacement in explanted mouse testis tissue supporting host spermatogenesis. Biol. Reprod. 2021, 105, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Thévenod, F.; Fels, J.; Lee, W.K.; Zarbock, R. Channels, transporters and receptors for cadmium and cadmium complexes in eukaryotic cells: Myths and facts. Biometals 2019, 32, 469–489. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Yu, D.; He, Z.; Bao, L.; Feng, L.; Chen, L.; Liu, Z.; Hu, X.; Zhang, N.; Wang, T.; et al. Endoplasmic reticulum stress-mediated autophagy activation is involved in cadmium-induced ferroptosis of renal tubular epithelial cells. Free Radic. Biol. Med. 2021, 175, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-R.; Ou, Y.-C.; Wu, C.-C.; Wang, J.-D.; Lin, S.-Y.; Wang, Y.-Y.; Chen, W.-Y.; Liao, S.-L.; Chen, C.-J. Endoplasmic reticulum stress and autophagy contributed to cadmium nephrotoxicity in HK-2 cells and Sprague-Dawley rats. Food Chem. Toxicol. 2020, 146, 111828. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, M.; Hiramatsu, N. The oxidative stress: Endoplasmic reticulum stress axis in cadmium toxicity. Biometals 2010, 23, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Pi, H.; Zhang, L.; Zhang, N.; Li, Y.; Zhang, H.; Tang, J.; Li, H.; Feng, M.; Deng, P.; et al. Melatonin prevents abnormal mitochondrial dynamics resulting from the neurotoxicity of cadmium by blocking calcium-dependent translocation of Drp1 to the mitochondria. J. Pineal Res. 2016, 60, 291–302. [Google Scholar] [CrossRef]
- Tang, J.; Duan, W.; Deng, P.; Li, H.; Liu, C.; Duan, Y.; Feng, M.; Xu, S. Cadmium disrupts mitochondrial distribution and activates excessive mitochondrial fission by elevating cytosolic calcium independent of MCU-mediated mitochondrial calcium uptake in its neurotoxicity. Toxicology 2021, 453, 152726. [Google Scholar] [CrossRef]
- Zheng, F.; Gonçalves, F.M.; Abiko, Y.; Li, H.; Kumagai, Y.; Aschner, M. Redox toxicology of environmental chemicals causing oxidative stress. Redox Biol. 2020, 34, 101475. [Google Scholar] [CrossRef]
- Miura, N.; Koizumi, S. Heavy metal responses of the human metallothionein isoform genes. Yakugaku Zasshi 2007, 127, 665–673. [Google Scholar] [CrossRef]
- Ma, W.; Li, X.; Wang, Q.; Ren, Z.; Crabbe, M.J.C.; Wang, L. Tandem oligomeric expression of metallothionein enhance heavy metal tolerance and bioaccumulation in Escherichia coli. Ecotoxicol. Environ. Saf. 2019, 181, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Vinken, M.; Blaauboer, B.J. In vitro testing of basal cytotoxicity: Establishment of an adverse outcome pathway from chemical insult to cell death. Toxicol. In Vitro 2017, 39, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Qiao, S.; Xiang, Y.; Cui, M.; Yao, X.; Lin, R.; Zhang, X. Endoplasmic reticulum stress: Multiple regulatory roles in hepatocellular carcinoma. Biomed. Pharmacother. 2021, 142, 112005. [Google Scholar] [CrossRef]
- Che, L.; Yang, C.-L.; Chen, Y.; Wu, Z.-L.; Du, Z.-B.; Wu, J.-S.; Gan, C.-L.; Yan, S.-P.; Huang, J.; Guo, N.-J.; et al. Mitochondrial redox-driven mitofusin 2 S-glutathionylation promotes neuronal necroptosis via disrupting ER-mitochondria crosstalk in cadmium-induced neurotoxicity. Chemosphere 2021, 262, 127878. [Google Scholar] [CrossRef]
- Zhang, C.; Ge, J.; Lv, M.; Zhang, Q.; Talukder, M.; Li, J.-L. Selenium prevent cadmium-induced hepatotoxicity through modulation of endoplasmic reticulum-resident selenoproteins and attenuation of endoplasmic reticulum stress. Environ. Pollut. 2020, 260, 113873. [Google Scholar] [CrossRef]
- Hu, H.; Tian, M.; Ding, C.; Yu, S. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front. Immunol. 2018, 9, 3083. [Google Scholar] [CrossRef]
- Tang, Q.; Ren, L.; Liu, J.; Li, W.; Zheng, X.; Wang, J.; Du, G. Withaferin A triggers G2/M arrest and intrinsic apoptosis in glioblastoma cells via ATF4-ATF3-CHOP axis. Cell Prolif. 2020, 53, e12706. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020, 15, 235–259. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.-M.; Williams, J.A.; Ding, W.-X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015, 4, 6–13. [Google Scholar] [CrossRef]
- Dong, F.; Xiao, P.; Li, X.; Chang, P.; Zhang, W.; Wang, L. Cadmium triggers oxidative stress and mitochondrial injury mediated apoptosis in human extravillous trophoblast HTR-8/SVneo cells. Reprod. Toxicol. 2021, 101, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Smyrnias, I. The mitochondrial unfolded protein response and its diverse roles in cellular stress. Int. J. Biochem. Cell Biol. 2021, 133, 105934. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Zhang, X.; Xin, Z.; Xu, B.; Jin, Z.; Wu, J.; Hu, W.; Yang, Y. Does perturbation in the mitochondrial protein folding pave the way for neurodegeneration diseases? Ageing Res. Rev. 2020, 57, 100997. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Zhang, C.; Sun, Y.-C.; Zhang, Q.; Lv, M.-W.; Guo, K.; Li, J.-L. Cadmium exposure triggers mitochondrial dysfunction and oxidative stress in chicken (Gallus gallus) kidney via mitochondrial UPR inhibition and Nrf2-mediated antioxidant defense activation. Sci. Total Environ. 2019, 689, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yin, F.; Xu, J.; Zhang, T.; Wang, G.; Mao, M.; Wang, Z.; Sun, W.; Han, J.; Yang, M.; et al. CYT997 (Lexibulin) induces apoptosis and autophagy through the activation of mutually reinforced ER stress and ROS in osteosarcoma. J. Exp. Clin. Cancer Res. 2019, 38, 44. [Google Scholar] [CrossRef] [PubMed]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef]
- Jiang, T.; Harder, B.; La Rojo de Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015, 88, 199–204. [Google Scholar] [CrossRef]
- Singh, P.; Chandrasekaran, V.; Hardy, B.; Wilmes, A.; Jennings, P.; Exner, T.E. Temporal transcriptomic alterations of cadmium exposed human iPSC-derived renal proximal tubule-like cells. Toxicol. In Vitro 2021, 76, 105229. [Google Scholar] [CrossRef]
- Ding, H.; Li, Z.; Li, X.; Yang, X.; Zhao, J.; Guo, J.; Lu, W.; Liu, H.; Wang, J. FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells. Int. J. Mol. Sci. 2022, 23, 4948. [Google Scholar] [CrossRef]
- Kamiya, T.; Izumi, M.; Hara, H.; Adachi, T. Propolis suppresses CdCl₂-induced cytotoxicity of COS7 cells through the prevention of intracellular reactive oxygen species accumulation. Biol. Pharm. Bull. 2012, 35, 1126–1131. [Google Scholar] [CrossRef]
- Waza, A.A.; Hamid, Z.; Ali, S.; Bhat, S.A.; Bhat, M.A. A review on heme oxygenase-1 induction: Is it a necessary evil. Inflamm. Res. 2018, 67, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, G.; Zhang, K.; Tan, Y.; Zou, H.; Yuan, Y.; Gu, J.; Song, R.; Zhu, J.; Liu, Z. Cadmium exposure induces rat proximal tubular cells injury via p62-dependent Nrf2 nucleus translocation mediated activation of AMPK/AKT/mTOR pathway. Ecotoxicol. Environ. Saf. 2021, 214, 112058. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; DesMarais, T.; Costa, M. Metals and Mechanisms of Carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 537–554. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Tan, J.; Miao, Y.; Zhang, Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J. Cell. Physiol. 2018, 233, 3867–3874. [Google Scholar] [CrossRef] [PubMed]
- Fukase, Y.; Tsugami, H.; Nakamura, Y.; Ohba, K.; Ohta, H. The role of metallothionein and metal transporter on cadmium transport from mother to fetus. Yakugaku Zasshi 2014, 134, 801–804. [Google Scholar] [CrossRef]
- Kheradmand, F.; Nourmohammadi, I.; Ahmadi-Faghih, M.A.; Firoozrai, M.; Modarressi, M.H. Zinc and low-dose of cadmium protect sertoli cells against toxic-dose of cadmium: The role of metallothionein. Iran. J. Reprod. Med. 2013, 11, 487–494. [Google Scholar]



| Antibody | Host Species | Vendor | Catalog Number | Applications (s)/Dilutions (s) |
|---|---|---|---|---|
| SQSTM1/p62 | Rabbit | CST | 39,749 | IB (1:1000), IF (1:100) |
| Atg5 | Rabbit | CST | 12,994 | IB (1:1000) |
| Atg7 | Rabbit | CST | 8558 | IB (1:1000) |
| LC3 | Rabbit | ProteinTech | 14,600-1-AP | IB (1:2500) |
| GAPDH | Mouse | ProteinTech | 60,004-1-Ig | IB (1:50,000) |
| CHOP | Rabbit | ProteinTech | 15,204-1-AP | IB (1:1000) |
| GRP78/BIP | Rabbit | ProteinTech | 11,587 | IB (1:3000) |
| HO-1 | Rabbit | ProteinTech | 10,701-1-AP | IB (1:3000) |
| Nrf1 | Rabbit | ProteinTech | 12482-1-AP | IB (1:2000) |
| Mfn1 | Rabbit | ProteinTech | 13798-1-AP | IB (1:1900) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Man, Y.; Liu, Y.; Xiong, C.; Zhang, Y.; Zhang, L. Non-Lethal Concentrations of CdCl2 Cause Marked Alternations in Cellular Stress Responses within Exposed Sertoli Cell Line. Toxics 2023, 11, 167. https://doi.org/10.3390/toxics11020167
Man Y, Liu Y, Xiong C, Zhang Y, Zhang L. Non-Lethal Concentrations of CdCl2 Cause Marked Alternations in Cellular Stress Responses within Exposed Sertoli Cell Line. Toxics. 2023; 11(2):167. https://doi.org/10.3390/toxics11020167
Chicago/Turabian StyleMan, Yonghong, Yunhao Liu, Chuanzhen Xiong, Yang Zhang, and Ling Zhang. 2023. "Non-Lethal Concentrations of CdCl2 Cause Marked Alternations in Cellular Stress Responses within Exposed Sertoli Cell Line" Toxics 11, no. 2: 167. https://doi.org/10.3390/toxics11020167
APA StyleMan, Y., Liu, Y., Xiong, C., Zhang, Y., & Zhang, L. (2023). Non-Lethal Concentrations of CdCl2 Cause Marked Alternations in Cellular Stress Responses within Exposed Sertoli Cell Line. Toxics, 11(2), 167. https://doi.org/10.3390/toxics11020167