Unravelling the Portuguese Coastal and Transitional Waters’ Microbial Resistome as a Biomarker of Differential Anthropogenic Impact
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Environmental Data
2.2. Metagenomic Sequencing and Bioinformatic Processing
3. Results
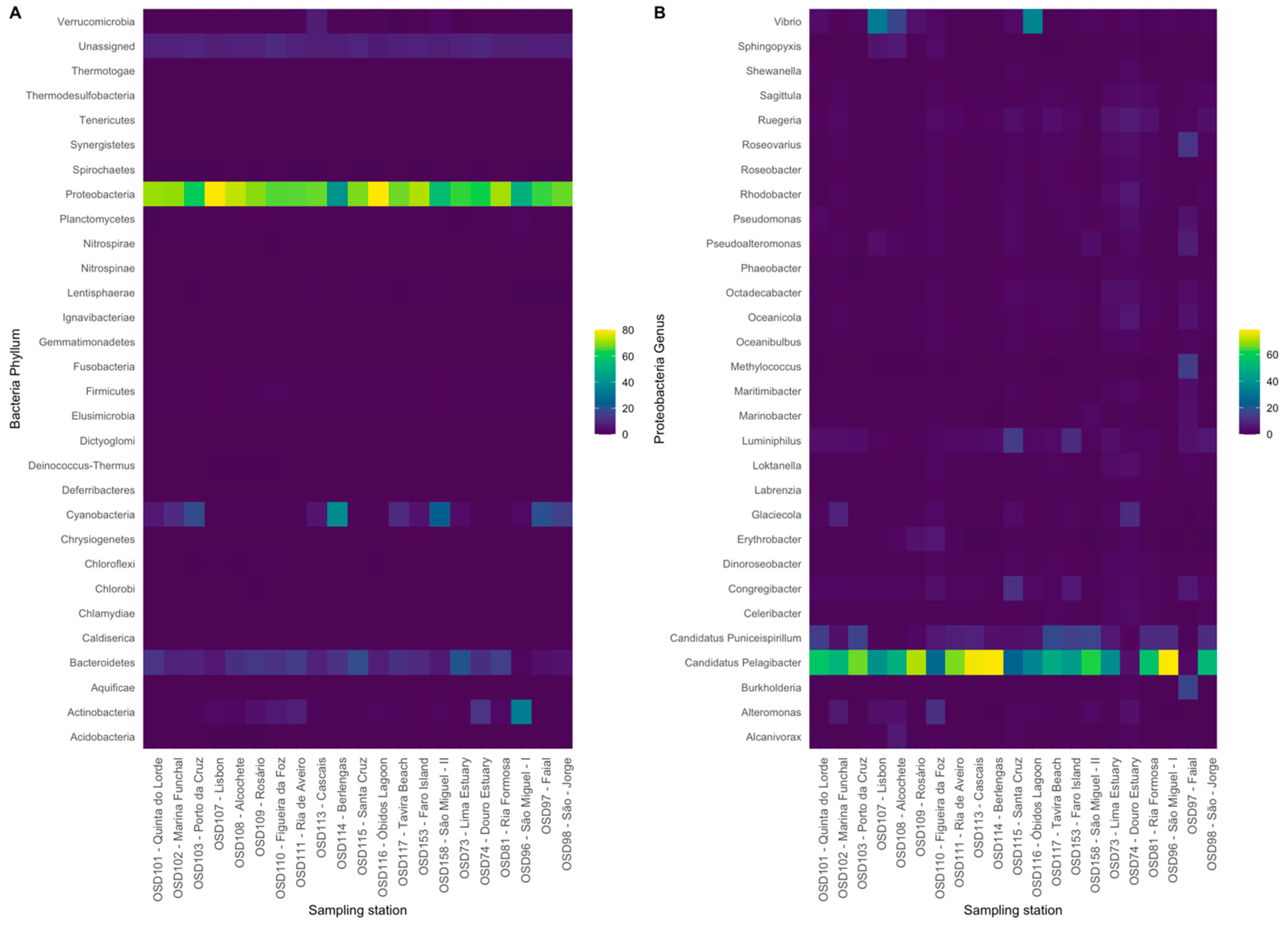
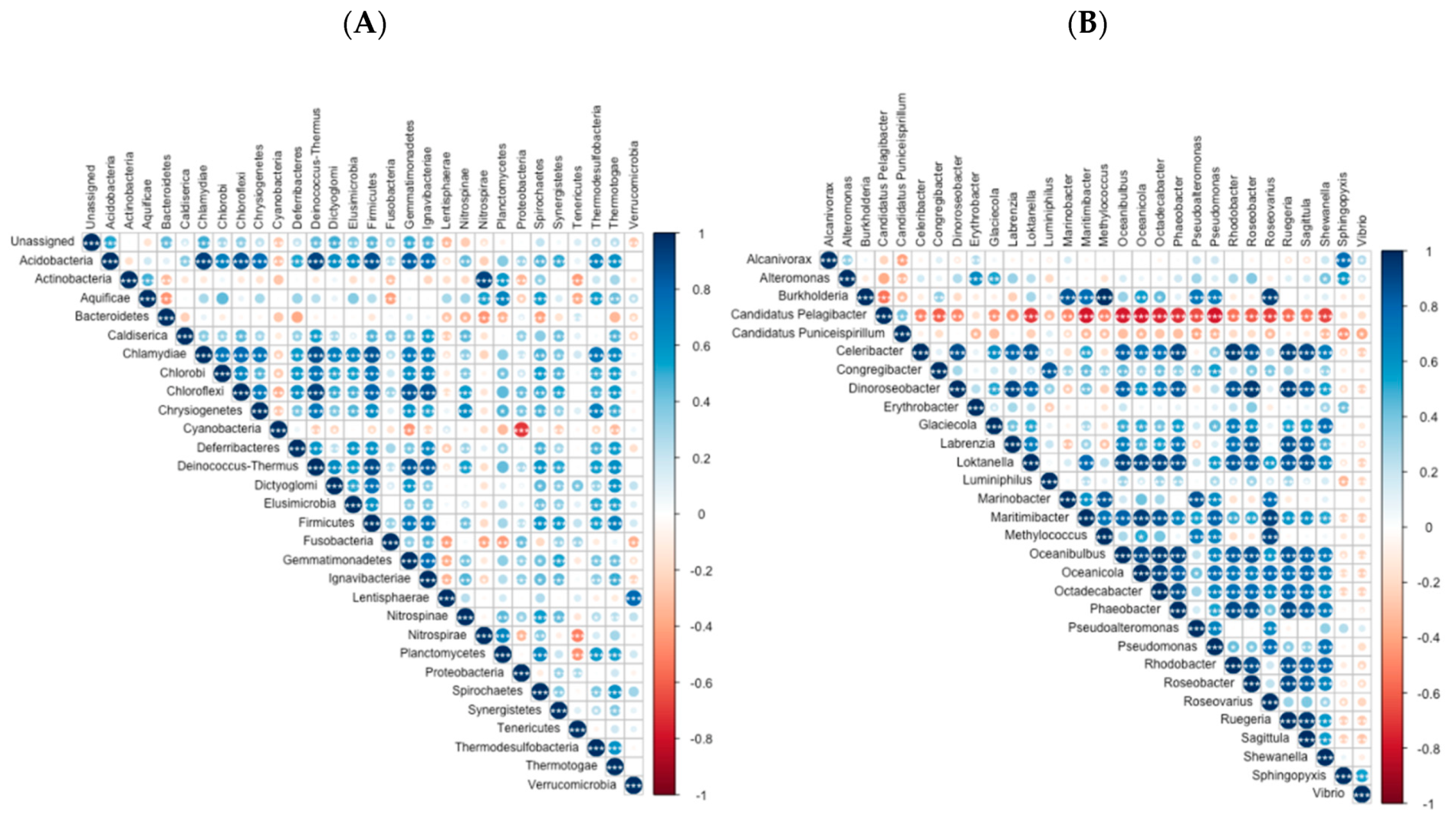
3.1. Coastal Archaea and Bacteria Taxonomic Diversity
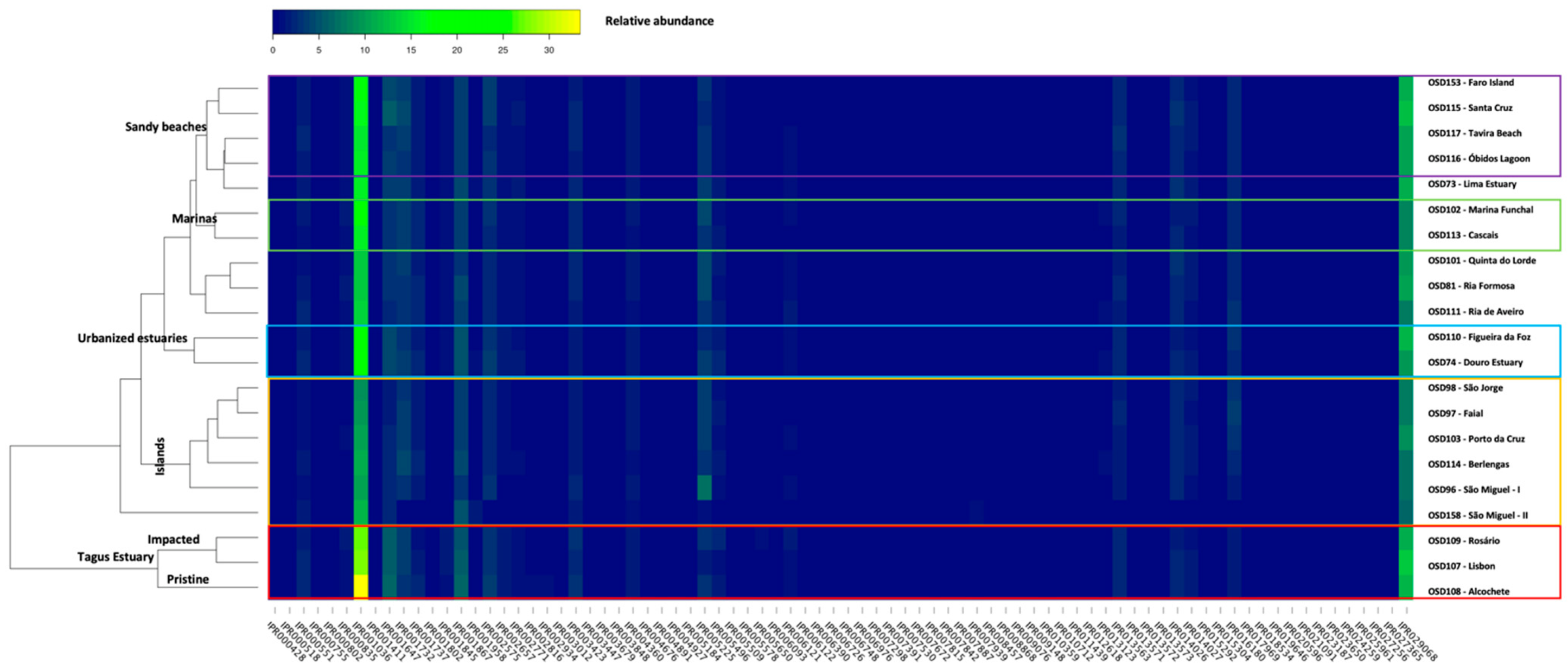
3.2. InterPro Resistome Signatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Concepcion, R.; Dadios, E.; Bandala, A.; Caçador, I.; Fonseca, V.F.; Duarte, B. Applying Limnological Feature-Based Machine Learning Techniques to Chemical State Classification in Marine Transitional Systems. Front. Mar. Sci. 2021, 8, 733. [Google Scholar] [CrossRef]
- Duarte, B.; Caçador, I.; Marques, J.C.; Croudace, I.W. Tagus Estuary Salt Marshes Feedback to Sea Level Rise over a 40-Year Period: Insights from the Application of Geochemical Indices. Ecol. Indic. 2013, 34, 268–276. [Google Scholar] [CrossRef]
- Reis-Santos, P.; Pais, M.; Duarte, B.; Caçador, I.; Freitas, A.; Vila Pouca, A.S.; Barbosa, J.; Leston, S.; Rosa, J.; Ramos, F.; et al. Screening of Human and Veterinary Pharmaceuticals in Estuarine Waters: A Baseline Assessment for the Tejo Estuary. Mar. Pollut. Bull. 2018, 135, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, V.F.; Reis-Santos, P.; Duarte, B.; Cabral, H.N.; Caçador, M.I.; Vaz, N.; Dias, J.M.; Pais, M.P. Roving Pharmacies: Modelling the Dispersion of Pharmaceutical Contamination in Estuaries. Ecol. Indic. 2020, 115, 106437. [Google Scholar] [CrossRef]
- Duarte, B.; Gameiro, C.; Matos, A.R.; Figueiredo, A.; Silva, M.S.; Cordeiro, C.; Caçador, I.; Reis-Santos, P.; Fonseca, V.; Cabrita, M.T. First Screening of Biocides, Persistent Organic Pollutants, Pharmaceutical and Personal Care Products in Antarctic Phytoplankton from Deception Island by FT-ICR-MS. Chemosphere 2021, 274, 129860. [Google Scholar] [CrossRef] [PubMed]
- Vilhena, J.C.E.; Amorim, A.; Ribeiro, L.; Duarte, B.; Pombo, M. Baseline Study of Trace Element Concentrations in Sediments of the Intertidal Zone of Amazonian Oceanic Beaches. Front. Mar. Sci. 2021, 8, 671390. [Google Scholar] [CrossRef]
- Fonseca, V.F.; Duarte, I.A.; Duarte, B.; Freitas, A.; Pouca, A.S.V.; Barbosa, J.; Gillanders, B.M.; Reis-Santos, P. Environmental Risk Assessment and Bioaccumulation of Pharmaceuticals in a Large Urbanized Estuary. Sci. Total Environ. 2021, 783, 147021. [Google Scholar] [CrossRef]
- Duarte, B.; Feijão, E.; Cruz de Carvalho, R.; Duarte, I.A.; Silva, M.; Matos, A.R.; Cabrita, M.T.; Novais, S.C.; Lemos, M.F.L.; Marques, J.C.; et al. Effects of Propranolol on Growth, Lipids and Energy Metabolism and Oxidative Stress Response of Phaeodactylum Tricornutum. Biology 2020, 9, 478. [Google Scholar] [CrossRef]
- Feijão, E.; Cruz de Carvalho, R.; Duarte, I.A.; Matos, A.R.; Cabrita, M.T.; Novais, S.C.; Lemos, M.F.L.; Caçador, I.; Marques, J.C.; Reis-Santos, P.; et al. Fluoxetine Arrests Growth of the Model Diatom Phaeodactylum Tricornutum by Increasing Oxidative Stress and Altering Energetic and Lipid Metabolism. Front. Microbiol. 2020, 11, 1803. [Google Scholar] [CrossRef]
- Cruz de Carvalho, R.; Feijão, E.; Kletschkus, E.; Marques, J.C.; Reis-Santos, P.; Fonseca, V.F.; Papenbrock, J.; Caçador, I.; Duarte, B. Halophyte Bio-Optical Phenotyping: A Multivariate Photochemical Pressure Index (Multi-PPI) to Classify Salt Marsh Anthropogenic Pressures Levels. Ecol. Indic. 2020, 119, 106816. [Google Scholar] [CrossRef]
- Duarte, B.; Santos, D.; Caçador, I. Halophyte Anti-Oxidant Feedback Seasonality in Two Salt Marshes with Different Degrees of Metal Contamination: Search for an Efficient Biomarker. Funct. Plant Biol. 2013, 40, 922. [Google Scholar] [CrossRef]
- Duarte, B.; Carreiras, J.; Pérez-Romero, J.A.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Matos, A.R.; Marques, J.C.; Caçador, I. Halophyte Fatty Acids as Biomarkers of Anthropogenic-Driven Contamination in Mediterranean Marshes: Sentinel Species Survey and Development of an Integrated Biomarker Response (IBR) Index. Ecol. Indic. 2018, 87, 86–96. [Google Scholar] [CrossRef]
- Duarte, B.; Carreiras, J.; Feijão, E.; Reis-Santos, P.; Caçador, I.; Matos, A.R.; Fonseca, V.F. Fatty Acid Profiles of Estuarine Macroalgae Are Biomarkers of Anthropogenic Pressures: Development and Application of a Multivariate Pressure Index. Sci. Total Environ. 2021, 788, 147817. [Google Scholar] [CrossRef]
- Duarte, I.A.; Reis-Santos, P.; França, S.; Cabral, H.; Fonseca, V.F. Biomarker Responses to Environmental Contamination in Estuaries: A Comparative Multi-Taxa Approach. Aquat. Toxicol. 2017, 189, 31–41. [Google Scholar] [CrossRef]
- Duarte, I.A.; Reis-Santos, P.; Novais, S.C.; Rato, L.D.; Lemos, M.F.L.L.; Freitas, A.; Pouca, A.S.V.; Barbosa, J.; Cabral, H.N.; Fonseca, V.F. Depressed, Hypertense and Sore: Long-Term Effects of Fluoxetine, Propranolol and Diclofenac Exposure in a Top Predator Fish. Sci. Total Environ. 2020, 712, 136564. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, Á.; Tangherlini, M.; Dell’Anno, A.; Corinaldesi, C.; Michell, C.T.; Irigoien, X.; Danovaro, R.; Rodríguez-Ezpeleta, N. A Bacterial Community-Based Index to Assess the Ecological Status of Estuarine and Coastal Environments. Mar. Pollut. Bull. 2017, 114, 679–688. [Google Scholar] [CrossRef]
- Goodwin, K.D.K.D.D.; Thompson, L.R.L.R.R.; Duarte, B.; Kahlke, T.; Thompson, A.R.A.R.R.; Marques, J.C.C.J.C.; Caçador, I. DNA Sequencing as a Tool to Monitor Marine Ecological Status. Front. Mar. Sci. 2017, 4, 107. [Google Scholar] [CrossRef]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374. [Google Scholar] [CrossRef]
- Wang, F.; Stedtfeld, R.D.; Kim, O.-S.; Chai, B.; Yang, L. Influence of Soil Characteristics and Proximity to Antarctic Research Stations on Abundance of Antibiotic Resistance Genes in Soils. Environ. Sci. Technol. 2016, 9, 12621–12629. [Google Scholar] [CrossRef]
- Munck, C.; Albertsen, M.; Telke, A.; Ellabaan, M.; Nielsen, P.H.; Sommer, M.O.A. Limited Dissemination of the Wastewater Treatment Plant Core Resistome. Nat. Commun. 2015, 6, 8452. [Google Scholar] [CrossRef]
- Fitzpatrick, D.; Walsh, F. Antibiotic Resistance Genes across a Wide Variety of Metagenomes. FEMS Microbiol. Ecol. 2016, 92, fiv168. [Google Scholar] [CrossRef]
- Poole, K. At the Nexus of Antibiotics and Metals: The Impact of Cu and Zn on Antibiotic Activity and Resistance. Trends Microbiol. 2017, 25, 820–832. [Google Scholar] [CrossRef]
- Poole, K. Mechanisms of Bacterial Biocide and Antibiotic Resistance: Antibiotic and Biocide Resistance. J. Appl. Microbiol. 2002, 92, 55S–64S. [Google Scholar] [CrossRef] [PubMed]
- Wales, A.; Davies, R. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.; Tacão, M.; Henriques, I. Selection of Antibiotic Resistance by Metals in a Riverine Bacterial Community. Chemosphere 2021, 263, 127936. [Google Scholar] [CrossRef] [PubMed]
- Kopf, A.; Bicak, M.; Kottmann, R.; Schnetzer, J.; Kostadinov, I.; Lehmann, K.; Fernandez-Guerra, A.; Jeanthon, C.; Rahav, E.; Ullrich, M.; et al. The Ocean Sampling Day Consortium. GigaSci 2015, 4, 27. [Google Scholar] [CrossRef]
- Sunagawa, S.; Acinas, S.G.; Bork, P.; Bowler, C.; Acinas, S.G.; Babin, M.; Bork, P.; Boss, E.; Bowler, C.; Cochrane, G.; et al. Tara Oceans: Towards Global Ocean Ecosystems Biology. Nat. Rev. Microbiol. 2020, 18, 428–445. [Google Scholar] [CrossRef]
- Halpern, B.S.; Walbridge, S.; Selkoe, K.A.; Kappel, C.V.; Micheli, F.; D’Agrosa, C.; Bruno, J.F.; Casey, K.S.; Ebert, C.; Fox, H.E.; et al. A Global Map of Human Impact on Marine Ecosystems. Science 2008, 319, 948. [Google Scholar] [CrossRef]
- Ocean Sampling Day Consortium, Participants. Registry of Samples and Environmental Context from the Ocean Sampling Day 2014. PANGAEA 2015. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Jansson, J.K.; Knight, R. The Earth Microbiome Project: Successes and Aspirations. BMC Biol. 2014, 12, 69. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Data Compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl Env. Microbiol 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference Database (PR2): A Catalog of Unicellular Eukaryote Small Sub-Unit RRNA Sequences with Curated Taxonomy. Nucleic Acids Res. 2012, 41, D597–D604. [Google Scholar] [CrossRef]
- Tan, G.-M.; Xu, L.; Bu, D.-B.; Feng, S.-Z.; Sun, N.-H. Improvement of Performance of MegaBlast Algorithm for DNA Sequence Alignment. J. Comput. Sci. Technol. 2006, 21, 973–978. [Google Scholar] [CrossRef]
- Apweiler, R. The InterPro Database, an Integrated Documentation Resource for Protein Families, Domains and Functional Sites. Nucleic Acids Res. 2001, 29, 37–40. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat Rev Microbiol 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Centurion, V.B.; Delforno, T.P.; Lacerda-Júnior, G.V.; Duarte, A.W.F.; Silva, L.J.; Bellini, G.B.; Rosa, L.H.; Oliveira, V.M. Unveiling Resistome Profiles in the Sediments of an Antarctic Volcanic Island. Environ. Pollut. 2019, 255, 113240. [Google Scholar] [CrossRef]
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal Homeostasis and Resistance in Bacteria. Nat Rev Microbiol 2017, 15, 338–350. [Google Scholar] [CrossRef]
- DeLong, E.F. Archaea in Coastal Marine Environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef]
- Nimnoi, P. Marine Bacterial Communities in the Upper Gulf of Thailand Assessed by Illumina Next-Generation Sequencing Platform. BMC Microbiol. 2020, 20, 19. [Google Scholar] [CrossRef]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and Function of the Global Ocean Microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Chen, X.-H.; Xie, Z.-X.; Li, D.-X.; Wu, P.-F.; Kong, L.-F.; Lin, L.; Kao, S.-J.; Wang, D.-Z. Bacterial Diversity and Nitrogen Utilization Strategies in the Upper Layer of the Northwestern Pacific Ocean. Front. Microbiol. 2018, 9, 797. [Google Scholar] [CrossRef]
- Cai, L.; Ye, L.; Tong, A.H.Y.; Lok, S.; Zhang, T. Biased Diversity Metrics Revealed by Bacterial 16S Pyrotags Derived from Different Primer Sets. PLoS ONE 2013, 8, e53649. [Google Scholar] [CrossRef]
- Morris, R.M.; Rappé, M.S.; Connon, S.A.; Vergin, K.L.; Siebold, W.A.; Carlson, C.A.; Giovannoni, S.J. SAR11 Clade Dominates Ocean Surface Bacterioplankton Communities. Nature 2002, 420, 806–810. [Google Scholar] [CrossRef]
- Kang, I.; Oh, H.-M.; Kang, D.; Cho, J.-C. Genome of a SAR116 Bacteriophage Shows the Prevalence of This Phage Type in the Oceans. Proc. Natl. Acad. Sci. USA 2013, 110, 12343–12348. [Google Scholar] [CrossRef]
- Kopprio, G.A.; Neogi, S.B.; Rashid, H.; Alonso, C.; Yamasaki, S.; Koch, B.P.; Gärdes, A.; Lara, R.J. Vibrio and Bacterial Communities Across a Pollution Gradient in the Bay of Bengal: Unraveling Their Biogeochemical Drivers. Front. Microbiol. 2020, 11, 594. [Google Scholar] [CrossRef] [PubMed]
- Malhadas, M.S.; Neves, R.J.; Leitão, P.C.; Silva, A. Influence of Tide and Waves on Water Renewal in Óbidos Lagoon, Portugal. Ocean Dyn. 2010, 60, 41–55. [Google Scholar] [CrossRef]
- Duarte, B.; Silva, G.; Costa, J.L.; Medeiros, J.P.; Azeda, C.; Sá, E.; Metelo, I.; Costa, M.J.; Caçador, I. Heavy Metal Distribution and Partitioning in the Vicinity of the Discharge Areas of Lisbon Drainage Basins (Tagus Estuary, Portugal). J. Sea Res. 2014, 93, 101–111. [Google Scholar] [CrossRef]
- Knapp, C.W.; McCluskey, S.M.; Singh, B.K.; Campbell, C.D.; Hudson, G.; Graham, D.W. Antibiotic Resistance Gene Abundances Correlate with Metal and Geochemical Conditions in Archived Scottish Soils. PLoS ONE 2011, 6, e27300. [Google Scholar] [CrossRef]
- Esteban, S.; Moreno-Merino, L.; Matellanes, R.; Catalá, M.; Gorga, M.; Petrovic, M.; López de Alda, M.; Barceló, D.; Durán, J.J.; López-Martínez, J.; et al. Presence of Endocrine Disruptors in Freshwater in the Northern Antarctic Peninsula Region. Environ. Res. 2016, 147, 179–192. [Google Scholar] [CrossRef]
- Verma, H.; Dhingra, G.G.; Sharma, M.; Gupta, V.; Negi, R.K.; Singh, Y.; Lal, R. Comparative Genomics of Sphingopyxis spp. Unravelled Functional Attributes. Genomics 2020, 112, 1956–1969. [Google Scholar] [CrossRef] [PubMed]
- Daury, L.; Orange, F.; Taveau, J.-C.; Verchère, A.; Monlezun, L.; Gounou, C.; Marreddy, R.K.R.; Picard, M.; Broutin, I.; Pos, K.M.; et al. Tripartite Assembly of RND Multidrug Efflux Pumps. Nat. Commun. 2016, 7, 10731. [Google Scholar] [CrossRef]
- Zheng, Q.; Lin, W.; Liu, Y.; Chen, C.; Jiao, N. A Comparison of 14 Erythrobacter Genomes Provides Insights into the Genomic Divergence and Scattered Distribution of Phototrophs. Front. Microbiol. 2016, 7, 984. [Google Scholar] [CrossRef][Green Version]
- Sanz-Sáez, I.; Pereira García, C.; Bravo, A.G.; Trujillo, L.; Pla i Ferriol, M.; Capilla, M.; Sánchez, P.; Rodríguez Martín-Doimeadios, R.D.C.; Acinas, S.G.; Sánchez, O. Prevalence of Heterotrophic Methylmercury Detoxifying Bacteria across Oceanic Regions. Environ. Sci. Technol. 2022, 56, 3452–3461. [Google Scholar] [CrossRef]
- Kovac Virsek, M.; Hubad, B.; Lapanje, A. Mercury Induced Community Tolerance in Microbial Biofilms Is Related to Pollution Gradients in a Long-Term Polluted River. Aquat. Toxicol. 2013, 144–145, 208–217. [Google Scholar] [CrossRef]
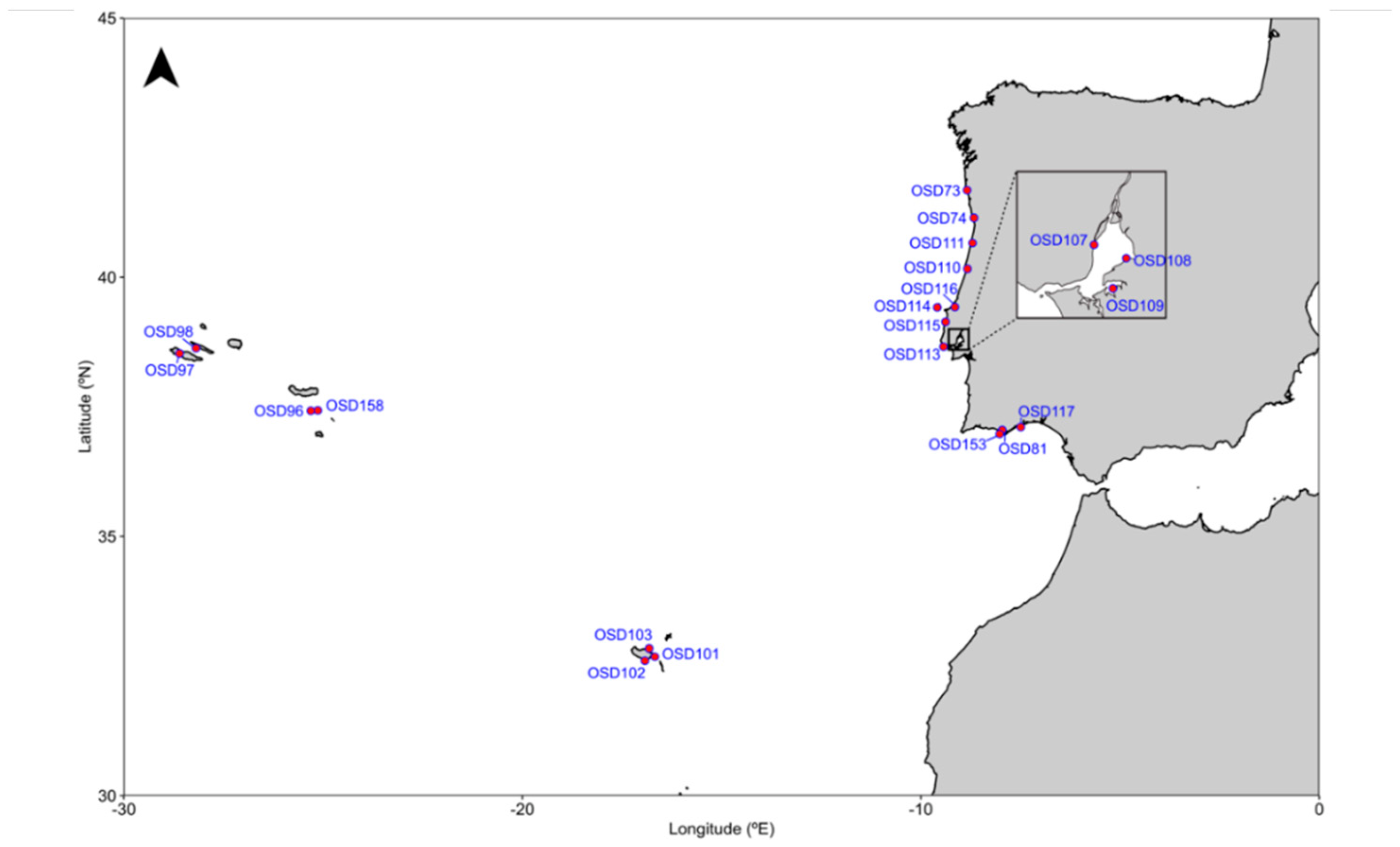



| Site | Number of Sequences | Temperature (°C) | Salinity (PSU) | Distance to Coast (Km) | Inorganic Pressure (a.u.) | Pollution (a.u.) | Plume Influence (a.u.) | Shipping Influence (a.u.) | Nearby Population (a.u.) |
|---|---|---|---|---|---|---|---|---|---|
| OSD101—Quinta do Lorde | 107,432 | 20.50 | 37.00 | 0.47 | 0.46 | 0.40 | 0.51 | 59.13 | 959.381 |
| OSD102—Marina Funchal | 67,004 | 20.80 | 36.00 | 0.40 | 1.89 | 0.5 | 0.81 | 20.61 | 4686.554 |
| OSD103—Porto da Cruz | 73,592 | 20.20 | 37.00 | 1.69 | 0.35 | 0.20 | 0.41 | 38.53 | 1960.618 |
| OSD107—Lisbon | 44,340 | 20.20 | 30.00 | 0.05 | 13.49 | 0.21 | 7.26 | 1043.46 | 8346.504 |
| OSD108—Alcochete | 47,730 | 20.10 | 30.00 | 0.36 | 2.86 | 0.21 | 2.32 | 1163.10 | 500.485 |
| OSD109—Rosário | 42,947 | 20.50 | 30.00 | 3.82 | 26.38 | 0.37 | 23.72 | 746.34 | 5045.171 |
| OSD110—Figueira da Foz | 37,659 | 23.00 | 22.50 | 1.13 | 22.23 | 0.52 | 8.36 | 2217.20 | 1444.238 |
| OSD111—Ria de Aveiro | 89,743 | 25.20 | 30.00 | 0.65 | 16.19 | 1.02 | 6.18 | 8763.42 | 941.3195 |
| OSD113—Cascais | 112,246 | 20.20 | 30.00 | 0.05 | 23.35 | 1.16 | 8.30 | 3875.05 | 3864.33 |
| OSD114—Berlengas | 107,131 | 18.50 | 33.57 | 9.22 | 0.00 | 0.50 | 0.00 | 226.06 | 0.00 |
| OSD115—Santa Cruz | 88,923 | 20.30 | 40.00 | 0.10 | 2.94 | 0.22 | 2.38 | 1161.79 | 488.97 |
| OSD116—Óbidos Lagoon | 43,828 | 24.70 | 22.50 | 0.74 | 1.11 | 0.09 | 0.86 | 1239.22 | 208.93 |
| OSD117—Tavira Beach | 81,348 | 23.64 | 37.93 | 0.75 | 80.71 | 0.72 | 206.89 | 1727.71 | 1173.97 |
| OSD153—Faro Island | 129,914 | 21.10 | 34.40 | 0.51 | 4.25 | 0.51 | 2.54 | 2168.11 | 360.03 |
| OSD158—São Miguel—II | 152,437 | 19.20 | 35.70 | 33.87 | 0.00 | 0.34 | 0.00 | 43.57 | 0.00 |
| OSD73—Lima Estuary | 116,182 | 18.40 | 32.30 | 0.58 | 14.57 | 1.34 | 6.15 | 13,681.47 | 1176.63 |
| OSD74—Douro Estuary | 121,027 | 20.20 | 13.75 | 0.70 | 10.16 | 1.52 | 4.01 | 15,309.65 | 438.40 |
| OSD81—Ria Formosa | 115,610 | 22.20 | 34.30 | 0.08 | 4.217 | 0.49 | 2.53 | 2277.45 | 388.36 |
| OSD96—São Miguel—I | 61,474 | 18.50 | 35.00 | 32.81 | 0.00 | 0.33 | 0.00 | 42.70 | 0.00 |
| OSD97—Faial | 90,089 | 16.90 | 35.60 | 2.13 | 0.22 | 0.43 | 0.99 | 57.91 | 432.38 |
| OSD98—São—Jorge | 91,817 | 18.70 | 35.60 | 0.86 | 0.07 | 0.30 | 0.74 | 84.24 | 106.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, B.; Figueiredo, A.; Ramalhosa, P.; Canning-Clode, J.; Caçador, I.; Fonseca, V.F. Unravelling the Portuguese Coastal and Transitional Waters’ Microbial Resistome as a Biomarker of Differential Anthropogenic Impact. Toxics 2022, 10, 613. https://doi.org/10.3390/toxics10100613
Duarte B, Figueiredo A, Ramalhosa P, Canning-Clode J, Caçador I, Fonseca VF. Unravelling the Portuguese Coastal and Transitional Waters’ Microbial Resistome as a Biomarker of Differential Anthropogenic Impact. Toxics. 2022; 10(10):613. https://doi.org/10.3390/toxics10100613
Chicago/Turabian StyleDuarte, Bernardo, Andreia Figueiredo, Patrício Ramalhosa, João Canning-Clode, Isabel Caçador, and Vanessa F. Fonseca. 2022. "Unravelling the Portuguese Coastal and Transitional Waters’ Microbial Resistome as a Biomarker of Differential Anthropogenic Impact" Toxics 10, no. 10: 613. https://doi.org/10.3390/toxics10100613
APA StyleDuarte, B., Figueiredo, A., Ramalhosa, P., Canning-Clode, J., Caçador, I., & Fonseca, V. F. (2022). Unravelling the Portuguese Coastal and Transitional Waters’ Microbial Resistome as a Biomarker of Differential Anthropogenic Impact. Toxics, 10(10), 613. https://doi.org/10.3390/toxics10100613












