Evaluation of Metabolism of a Defined Pesticide Mixture through Multiple In Vitro Liver Models
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Cell Culture Reagents and Materials
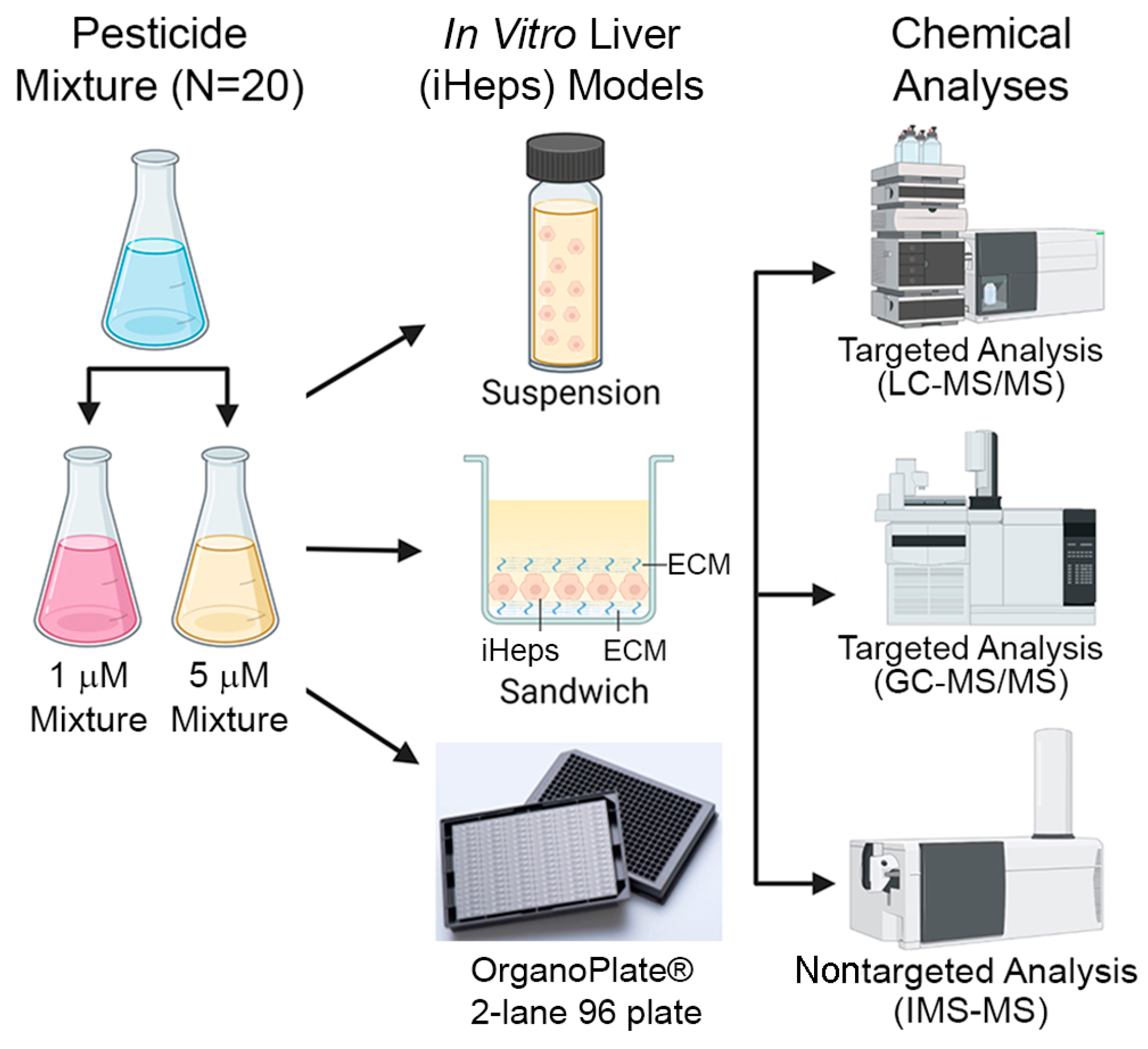
2.3. Preparation of Chemical Mixtures
2.4. iHep Suspension Assays
2.5. iHep Culture in OrganoPlate® 2-Lane 96, 384-Well Plates, and Chemical Treatments
2.6. iHep 2D Sandwich Culture and Chemical Treatments
2.7. Functional Assays
2.8. LC-MS/MS Analyses
2.9. GC-MS/MS Analyses
2.10. IMS-MS Analyses
2.11. Determination of Intrinsic Clearance
2.12. Statistical Analyses
3. Results
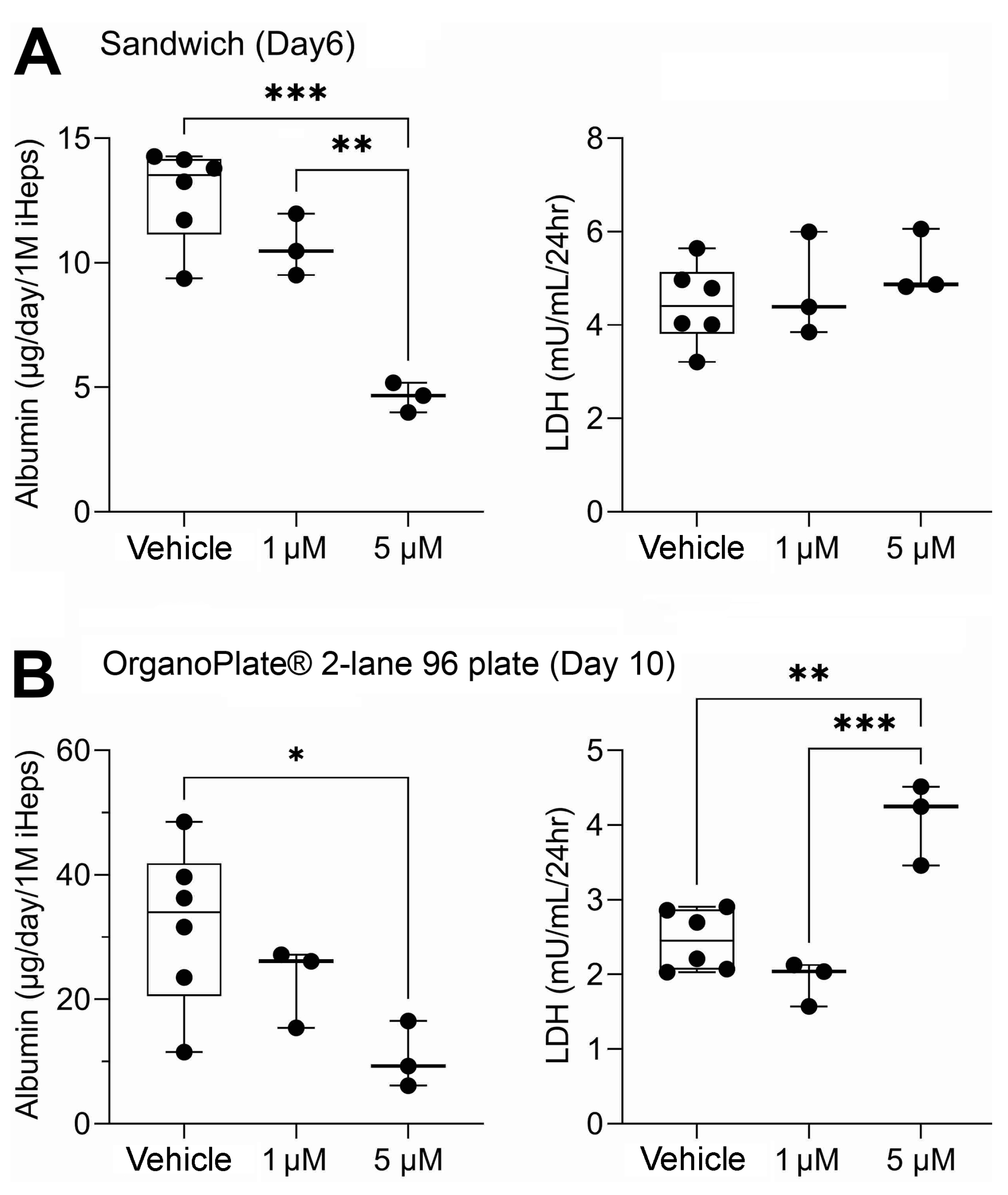
3.1. Liver Function Comparison between 2D Sandwich and OrganoPlate® 2-Lane 96
3.2. Liver Metabolism Assessment through Targeted Mass Spectrometry Analyses
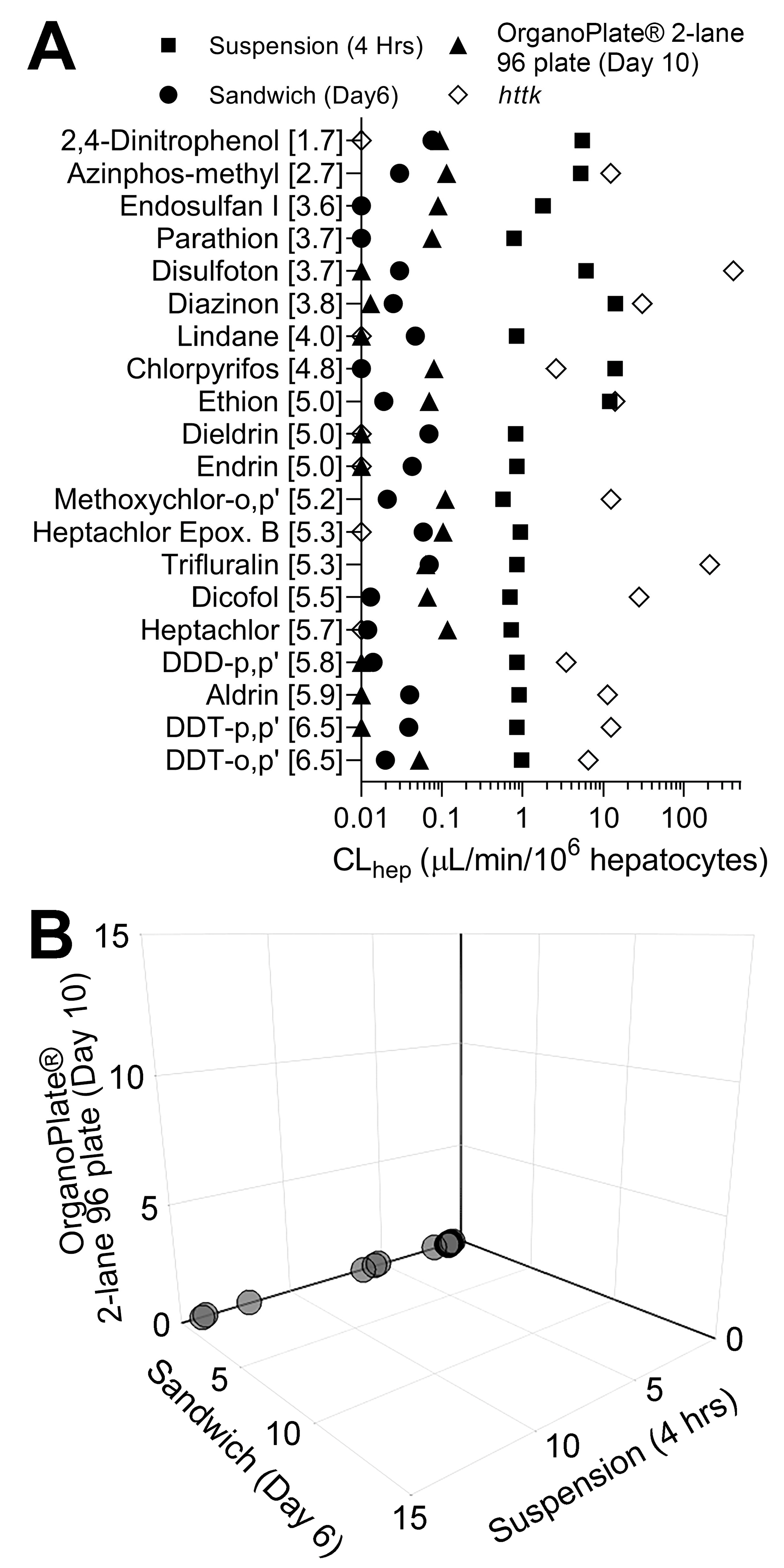
3.3. Comparison of In Vitro Hepatocyte Clearance Values Obtained from Targeted and Nontargeted Analyses
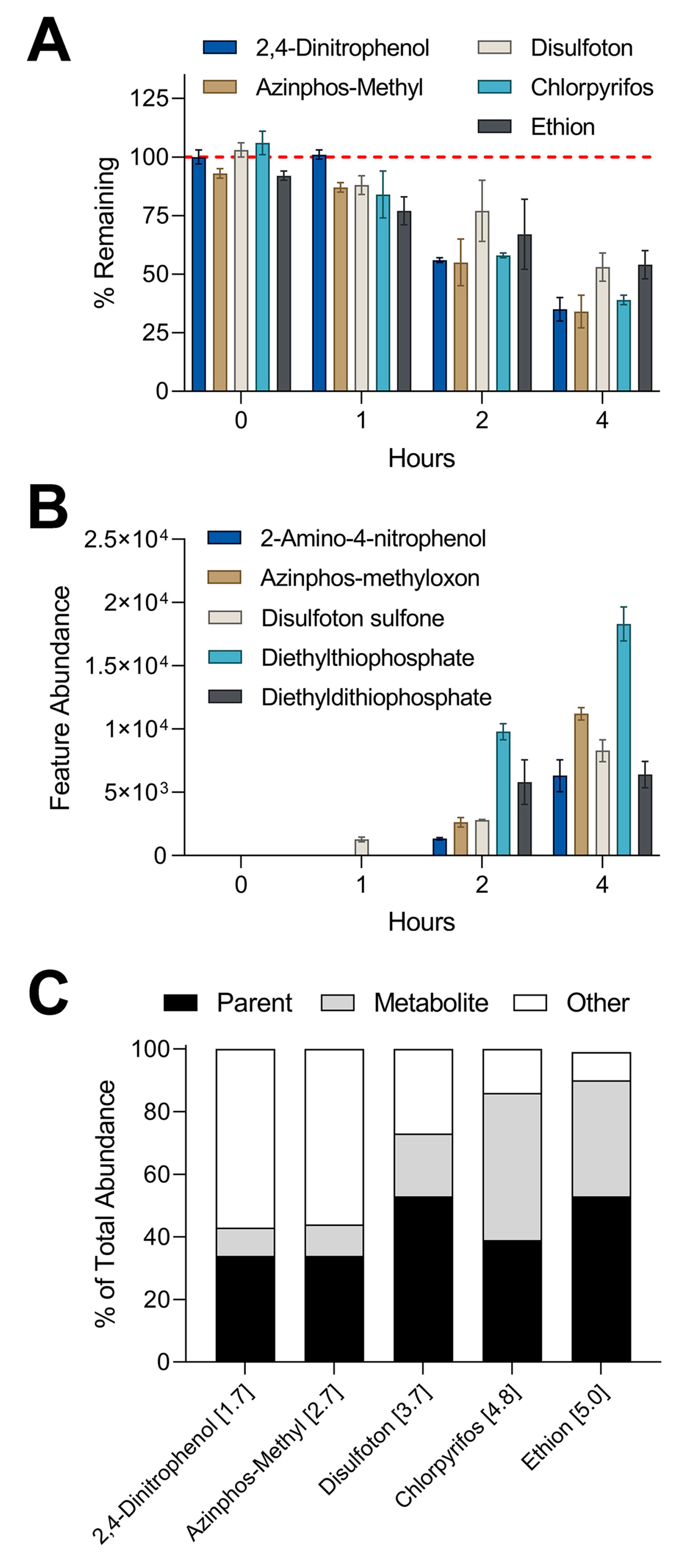
3.4. Metabolite Detection Using IMS-MS Nontargeted Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Omiecinski, C.J.; Vanden Heuvel, J.P.; Perdew, G.H.; Peters, J.M. Xenobiotic metabolism, disposition, and regulation by receptors: From biochemical phenomenon to predictors of major toxicities. Toxicol. Sci. 2011, 120 (Suppl. 1), S49–S75. [Google Scholar] [CrossRef]
- Meyer, U.A. Overview of enzymes of drug metabolism. J. Pharmacokinet. Biopharm. 1996, 24, 449–459. [Google Scholar] [CrossRef]
- Wrighton, S.A.; Vandenbranden, M.; Ring, B.J. The human drug metabolizing cytochromes p450. J. Pharmacokinet. Biopharm. 1996, 24, 461–473. [Google Scholar] [CrossRef]
- Esteves, F.; Rueff, J.; Kranendonk, M. The central role of cytochrome p450 in xenobiotic metabolism-a brief review on a fascinating enzyme family. J. Xenobiotics 2021, 11, 94–114. [Google Scholar] [CrossRef]
- Graham, S.E.; Peterson, J.A. How similar are p450s and what can their differences teach us? Arch. Biochem. Biophys. 1999, 369, 24–29. [Google Scholar] [CrossRef]
- Williams, D.P.; Kitteringham, N.R.; Naisbitt, D.J.; Pirmohamed, M.; Smith, D.A.; Park, B.K. Are chemically reactive metabolites responsible for adverse reactions to drugs? Curr. Drug Metab. 2002, 3, 351–366. [Google Scholar] [CrossRef]
- Hodges, R.E.; Minich, D.M. Modulation of metabolic detoxification pathways using foods and food-derived components: A scientific review with clinical application. J. Nutr. Metab. 2015, 2015, 760689. [Google Scholar] [CrossRef]
- Gómez-Lechón, M.J.; Castell, J.V.; Donato, M.T. Hepatocytes--the choice to investigate drug metabolism and toxicity in man: In vitro variability as a reflection of in vivo. Chem. Biol. Interact. 2007, 168, 30–50. [Google Scholar] [CrossRef]
- Park, K.; Williams, D.P.; Naisbitt, D.J.; Kitteringham, N.R.; Pirmohamed, M. Investigation of toxic metabolites during drug development. Toxicol. Appl. Pharmacol. 2005, 207, 425–434. [Google Scholar] [CrossRef]
- Luo, Y.S.; Chen, Z.; Hsieh, N.H.; Lin, T.E. Chemical and biological assessments of environmental mixtures: A review of current trends, advances, and future perspectives. J. Hazard. Mater. 2022, 432, 128658. [Google Scholar] [CrossRef]
- Hsieh, N.H.; Chen, Z.; Rusyn, I.; Chiu, W.A. Risk characterization and probabilistic concentration-response modeling of complex environmental mixtures using new approach methodologies (nams) data from organotypic in vitro human stem cell assays. Environ. Health Perspect. 2021, 129, 17004. [Google Scholar] [CrossRef]
- Hernández, A.F.; Tsatsakis, A.M. Human exposure to chemical mixtures: Challenges for the integration of toxicology with epidemiology data in risk assessment. Food Chem. Toxicol. 2017, 103, 188–193. [Google Scholar] [CrossRef]
- Tsatsakis, A.M.; Docea, A.O.; Tsitsimpikou, C. New challenges in risk assessment of chemicals when simulating real exposure scenarios; simultaneous multi-chemicals’ low dose exposure. Food Chem. Toxicol. 2016, 96, 174–176. [Google Scholar] [CrossRef]
- Krewski, D.; Acosta, D., Jr.; Andersen, M.; Anderson, H.; Bailar, J.C., 3rd; Boekelheide, K.; Brent, R.; Charnley, G.; Cheung, V.G.; Green, S., Jr.; et al. Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 2010, 13, 51–138. [Google Scholar] [CrossRef]
- Krewski, D.; Andersen, M.E.; Tyshenko, M.G.; Krishnan, K.; Hartung, T.; Boekelheide, K.; Wambaugh, J.F.; Jones, D.; Whelan, M.; Thomas, R.; et al. Toxicity testing in the 21st century: Progress in the past decade and future perspectives. Arch. Toxicol. 2020, 94, 1–58. [Google Scholar] [CrossRef]
- Andersen, M.E.; Krewski, D. Toxicity testing in the 21st century: Bringing the vision to life. Toxicol. Sci. 2009, 107, 324–330. [Google Scholar] [CrossRef]
- Griffin, S.J.; Houston, J.B. Prediction of in vitro intrinsic clearance from hepatocytes: Comparison of suspensions and monolayer cultures. Drug Metab. Dispos. 2005, 33, 115–120. [Google Scholar] [CrossRef]
- Soldatow, V.Y.; Lecluyse, E.L.; Griffith, L.G.; Rusyn, I. In vitro models for liver toxicity testing. Toxicol. Res. 2013, 2, 23–39. [Google Scholar] [CrossRef]
- Brown, H.S.; Griffin, M.; Houston, J.B. Evaluation of cryopreserved human hepatocytes as an alternative in vitro system to microsomes for the prediction of metabolic clearance. Drug Metab. Dispos. 2007, 35, 293–301. [Google Scholar] [CrossRef]
- Rusyn, I.; Sakolish, C.; Kato, Y.; Stephan, C.; Vergara, L.; Hewitt, P.; Bhaskaran, V.; Davis, M.; Hardwick, R.N.; Ferguson, S.S.; et al. Microphysiological systems evaluation: Experience of tex-val tissue chip testing consortium. Toxicol. Sci. 2022, 188, 143–152. [Google Scholar] [CrossRef]
- Baudy, A.R.; Otieno, M.A.; Hewitt, P.; Gan, J.; Roth, A.; Keller, D.; Sura, R.; Van Vleet, T.R.; Proctor, W.R. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip 2020, 20, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Pelkonen, O.; Bennekou, S.H.; Crivellente, F.; Terron, A.; Hernandez, A.F. Integration of epidemiological findings with mechanistic evidence in regulatory pesticide risk assessment: Efsa experiences. Arch. Toxicol. 2019, 93, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.; Vernetti, L.A.; Gough, A. A 3d microfluidic liver model for high throughput compound toxicity screening in the organoplate(r). Toxicology 2021, 450, 152667. [Google Scholar] [CrossRef] [PubMed]
- Sakolish, C.; Reese, C.E.; Luo, Y.S.; Valdiviezo, A.; Schurdak, M.E.; Gough, A.; Taylor, D.L.; Chiu, W.A.; Vernetti, L.A.; Rusyn, I. Analysis of reproducibility and robustness of a human microfluidic four-cell liver acinus microphysiology system (lamps). Toxicology 2021, 448, 152651. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Lim, A.Y.; Sakolish, C.; Valdiviezo, A.; Moyer, H.L.; Hewitt, P.; Bajaj, P.; Han, G.; Rusyn, I. Analysis of reproducibility and robustness of organoplate(r) 2-lane 96, a liver microphysiological system for studies of pharmacokinetics and toxicological assessment of drugs. Toxicol. Vitr. 2022, 85, 105464. [Google Scholar] [CrossRef] [PubMed]
- Lash, L.H.; Qian, W.; Putt, D.A.; Jacobs, K.; Elfarra, A.A.; Krause, R.J.; Parker, J.C. Glutathione conjugation of trichloroethylene in rats and mice: Sex-, species-, and tissue-dependent differences. Drug Metab. Dispos. 1998, 26, 12–19. [Google Scholar] [PubMed]
- Rubiano, A.; Indapurkar, A.; Yokosawa, R.; Miedzik, A.; Rosenzweig, B.; Arefin, A.; Moulin, C.M.; Dame, K.; Hartman, N.; Volpe, D.A.; et al. Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism, and accumulation. Clin. Transl. Sci. 2021, 14, 1049–1061. [Google Scholar] [CrossRef]
- Plattard, N.; Venisse, N.; Carato, P.; Dupuis, A.; Haddad, S. Hepatic metabolism of chlorinated derivatives of bisphenol a (cl(x)bpa) and interspecies differences between rats and humans. Arch. Toxicol. 2022, 96, 783–792. [Google Scholar] [CrossRef]
- Sobus, J.R.; Wambaugh, J.F.; Isaacs, K.K.; Williams, A.J.; McEachran, A.D.; Richard, A.M.; Grulke, C.M.; Ulrich, E.M.; Rager, J.E.; Strynar, M.J.; et al. Integrating tools for non-targeted analysis research and chemical safety evaluations at the us epa. J. Expo. Sci. Environ. Epidemiol. 2018, 28, 411–426. [Google Scholar] [CrossRef]
- Valdiviezo, A.; Aly, N.A.; Luo, Y.S.; Cordova, A.; Casillas, G.; Foster, M.; Baker, E.S.; Rusyn, I. Analysis of per- and polyfluoroalkyl substances in houston ship channel and galveston bay following a large-scale industrial fire using ion-mobility-spectrometry-mass spectrometry. J. Environ. Sci. 2022, 115, 350–362. [Google Scholar] [CrossRef]
- Mairinger, T.; Causon, T.J.; Hann, S. The potential of ion mobility-mass spectrometry for non-targeted metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Mesa, M.; Monteau, F.; Le Bizec, B.; Dervilly-Pinel, G. Potential of ion mobility-mass spectrometry for both targeted and non-targeted analysis of phase ii steroid metabolites in urine. Anal. Chim. Acta X 2019, 1, 100006. [Google Scholar] [CrossRef] [PubMed]
- ATSDR Atsdr’s Substance Priority List. Available online: http://www.atsdr.cdc.gov/SPL/index.html (accessed on 8 April 2022).
- Valdiviezo, A.; Luo, Y.S.; Chen, Z.; Chiu, W.A.; Rusyn, I. Quantitative in vitro-to-in vivo extrapolation for mixtures: A case study of superfund priority list pesticides. Toxicol. Sci. 2021, 183, 60–69. [Google Scholar] [CrossRef]
- Lee-Montiel, F.T.; George, S.M.; Gough, A.H.; Sharma, A.D.; Wu, J.; DeBiasio, R.; Vernetti, L.A.; Taylor, D.L. Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp. Biol. Med. 2017, 242, 1617–1632. [Google Scholar] [CrossRef]
- Dodds, J.N.; May, J.C.; McLean, J.A. Correlating resolving power, resolution, and collision cross section: Unifying cross-platform assessment of separation efficiency in ion mobility spectrometry. Anal. Chem. 2017, 89, 12176–12184. [Google Scholar] [CrossRef]
- Smith, C.M.; Nolan, C.K.; Edwards, M.A.; Hatfield, J.B.; Stewart, T.W.; Ferguson, S.S.; Lecluyse, E.L.; Sahi, J. A comprehensive evaluation of metabolic activity and intrinsic clearance in suspensions and monolayer cultures of cryopreserved primary human hepatocytes. J. Pharm. Sci. 2012, 101, 3989–4002. [Google Scholar] [CrossRef] [PubMed]
- Wetmore, B.A. Quantitative in vitro-to-in vivo extrapolation in a high-throughput environment. Toxicology 2015, 332, 94–101. [Google Scholar] [CrossRef]
- Vega, L.; Valverde, M.; Elizondo, G.; Leyva, J.F.; Rojas, E. Diethylthiophosphate and diethyldithiophosphate induce genotoxicity in hepatic cell lines when activated by further biotransformation via cytochrome p450. Mutat. Res. 2009, 679, 39–43. [Google Scholar] [CrossRef]
- Heudorf, U.; Angerer, J. Metabolites of organophosphorous insecticides in urine specimens from inhabitants of a residential area. Environ. Res. 2001, 86, 80–87. [Google Scholar] [CrossRef]
- Luo, Y.S.; Long, T.Y.; Chiang, S.Y.; Wu, K.Y. Characterization of primary glutathione conjugates with acrylamide and glycidamide: Toxicokinetic studies in sprague dawley rats treated with acrylamide. Chem. Biol. Interact. 2021, 350, 109701. [Google Scholar] [CrossRef]
- Luo, Y.S.; Furuya, S.; Soldatov, V.Y.; Kosyk, O.; Yoo, H.S.; Fukushima, H.; Lewis, L.; Iwata, Y.; Rusyn, I. Metabolism and toxicity of trichloroethylene and tetrachloroethylene in cytochrome p450 2e1 knockout and humanized transgenic mice. Toxicol. Sci. 2018, 164, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Balls, M. Replacement of animal procedures: Alternatives in research, education and testing. Lab. Anim. 1994, 28, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Balls, M. Alternatives to laboratory animals: Trends in replacement and the three rs. Altern. Lab. Anim. 2022, 50, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Berger, D.R.; Ballinger, K.R.; Davidson, M.D.; Lin, C.; Ware, B.R. Microengineered liver tissues for drug testing. J. Lab. Autom. 2015, 20, 216–250. [Google Scholar] [CrossRef]
- March, S.; Ramanan, V.; Trehan, K.; Ng, S.; Galstian, A.; Gural, N.; Scull, M.A.; Shlomai, A.; Mota, M.M.; Fleming, H.E.; et al. Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat. Protoc. 2015, 10, 2027–2053. [Google Scholar] [CrossRef]
- Wang, W.W.; Khetani, S.R.; Krzyzewski, S.; Duignan, D.B.; Obach, R.S. Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab. Dispos. 2010, 38, 1900–1905. [Google Scholar] [CrossRef]
- LeCluyse, E.L.; Audus, K.L.; Hochman, J.H. Formation of extensive canalicular networks by rat hepatocytes cultured in collagen-sandwich configuration. Am. J. Physiol. 1994, 266, C1764–C1774. [Google Scholar] [CrossRef]
- Guillouzo, A. Liver cell models in in vitro toxicology. Environ. Health Perspect. 1998, 106 (Suppl. 2), 511–532. [Google Scholar]
- Hewitt, N.J.; Lechon, M.J.; Houston, J.B.; Hallifax, D.; Brown, H.S.; Maurel, P.; Kenna, J.G.; Gustavsson, L.; Lohmann, C.; Skonberg, C.; et al. Primary hepatocytes: Current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab. Rev. 2007, 39, 159–234. [Google Scholar] [CrossRef]
- LeCluyse, E.L. Human hepatocyte culture systems for the in vitro evaluation of cytochrome p450 expression and regulation. Eur. J. Pharm. Sci. 2001, 13, 343–368. [Google Scholar] [CrossRef]
- LeCluyse, E.L.; Ahlgren-Beckendorf, J.A.; Carroll, K.; Parkinson, A.; Johnson, J. Regulation of glutathione s-transferase enzymes in primary cultures of rat hepatocytes maintained under various matrix configurations. Toxicol. Vitr. 2000, 14, 101–115. [Google Scholar] [CrossRef]
- Liu, X.; Chism, J.P.; LeCluyse, E.L.; Brouwer, K.R.; Brouwer, K.L. Correlation of biliary excretion in sandwich-cultured rat hepatocytes and in vivo in rats. Drug Metab. Dispos. 1999, 27, 637–644. [Google Scholar] [PubMed]
- Dunn, J.C.; Yarmush, M.L.; Koebe, H.G.; Tompkins, R.G. Hepatocyte function and extracellular matrix geometry: Long-term culture in a sandwich configuration. FASEB J. 1989, 3, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Mathijs, K.; Kienhuis, A.S.; Brauers, K.J.; Jennen, D.G.; Lahoz, A.; Kleinjans, J.C.; van Delft, J.H. Assessing the metabolic competence of sandwich-cultured mouse primary hepatocytes. Drug Metab. Dispos. 2009, 37, 1305–1311. [Google Scholar] [CrossRef]
- Ehrlich, A.; Duche, D.; Ouedraogo, G.; Nahmias, Y. Challenges and opportunities in the design of liver-on-chip microdevices. Annu. Rev. Biomed. Eng. 2019, 21, 219–239. [Google Scholar] [CrossRef] [PubMed]
- Underhill, G.H.; Khetani, S.R. Bioengineered liver models for drug testing and cell differentiation studies. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 426–439.e421. [Google Scholar] [CrossRef] [PubMed]
- Telles-Silva, K.A.; Pacheco, L.; Komatsu, S.; Chianca, F.; Caires-Júnior, L.C.; Araujo, B.H.S.; Goulart, E.; Zatz, M. Applied hepatic bioengineering: Modeling the human liver using organoid and liver-on-a-chip technologies. Front. Bioeng. Biotechnol. 2022, 10, 845360. [Google Scholar] [CrossRef]
- Vernetti, L.A.; Senutovitch, N.; Boltz, R.; DeBiasio, R.; Shun, T.Y.; Gough, A.; Taylor, D.L. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 2016, 241, 101–114. [Google Scholar] [CrossRef]
- Eckstrum, K.; Striz, A.; Ferguson, M.; Zhao, Y.; Welch, B.; Solomotis, N.; Olejnik, N.; Sprando, R. Utilization of a model hepatotoxic compound, diglycolic acid, to evaluate liver organ-chip performance and in vitro to in vivo concordance. Food Chem. Toxicol. 2020, 146, 111850. [Google Scholar] [CrossRef]
- Usta, O.B.; McCarty, W.J.; Bale, S.; Hegde, M.; Jindal, R.; Bhushan, A.; Golberg, I.; Yarmush, M.L. Microengineered cell and tissue systems for drug screening and toxicology applications: Evolution of in-vitro liver technologies. Technology 2015, 3, 1–26. [Google Scholar] [CrossRef]
- Messelmani, T.; Morisseau, L.; Sakai, Y.; Legallais, C.; Le Goff, A.; Leclerc, E.; Jellali, R. Liver organ-on-chip models for toxicity studies and risk assessment. Lab Chip 2022, 22, 2423–2450. [Google Scholar] [CrossRef] [PubMed]
- Docci, L.; Milani, N.; Ramp, T.; Romeo, A.A.; Godoy, P.; Franyuti, D.O.; Krähenbühl, S.; Gertz, M.; Galetin, A.; Parrott, N.; et al. Exploration and application of a liver-on-a-chip device in combination with modelling and simulation for quantitative drug metabolism studies. Lab Chip 2022, 22, 1187–1205. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, K.C.; Luo, Y.S.; Rusyn, I.; Chiu, W.A. Comparative analysis of rapid equilibrium dialysis (red) and solid phase micro-extraction (spme) methods for in vitro-in vivo extrapolation of environmental chemicals. Toxicol. Vitr. 2019, 60, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lloyd, D.; Zhou, Y.H.; Chiu, W.A.; Wright, F.A.; Rusyn, I. Risk characterization of environmental samples using in vitro bioactivity and polycyclic aromatic hydrocarbon concentrations data. Toxicol. Sci. 2021, 179, 108–120. [Google Scholar] [CrossRef]
- Gearhart-Serna, L.M.; Davis, J.B.; Jolly, M.K.; Jayasundara, N.; Sauer, S.J.; Di Giulio, R.T.; Devi, G.R. A polycyclic aromatic hydrocarbon-enriched environmental chemical mixture enhances ahr, antiapoptotic signaling and a proliferative phenotype in breast cancer cells. Carcinogenesis 2020, 41, 1648–1659. [Google Scholar] [CrossRef]
- Vignati, L.; Turlizzi, E.; Monaci, S.; Grossi, P.; Kanter, R.; Monshouwer, M. An in vitro approach to detect metabolite toxicity due to cyp3a4-dependent bioactivation of xenobiotics. Toxicology 2005, 216, 154–167. [Google Scholar] [CrossRef]
- Kaabia, Z.; Laparre, J.; Cesbron, N.; Le Bizec, B.; Dervilly-Pinel, G. Comprehensive steroid profiling by liquid chromatography coupled to high resolution mass spectrometry. J. Steroid. Biochem. Mol. Biol. 2018, 183, 106–115. [Google Scholar] [CrossRef]
- Zheng, X.; Dupuis, K.T.; Aly, N.A.; Zhou, Y.; Smith, F.B.; Tang, K.; Smith, R.D.; Baker, E.S. Utilizing ion mobility spectrometry and mass spectrometry for the analysis of polycyclic aromatic hydrocarbons, polychlorinated biphenyls, polybrominated diphenyl ethers and their metabolites. Anal. Chim. Acta 2018, 1037, 265–273. [Google Scholar] [CrossRef]
- Foster, M.; Rainey, M.; Watson, C.; Dodds, J.N.; Kirkwood, K.I.; Fernandez, F.M.; Baker, E.S. Uncovering pfas and other xenobiotics in the dark metabolome using ion mobility spectrometry, mass defect analysis, and machine learning. Environ. Sci. Technol. 2022, 56, 9133–9143. [Google Scholar] [CrossRef]
- ATSDR. Toxicological Profile for Dinitrophenols; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 2021.
- Hernández, A.F.; Lozano-Paniagua, D.; González-Alzaga, B.; Kavvalakis, M.P.; Tzatzarakis, M.N.; López-Flores, I.; Aguilar-Garduño, C.; Caparros-Gonzalez, R.A.; Tsatsakis, A.M.; Lacasaña, M. Biomonitoring of common organophosphate metabolites in hair and urine of children from an agricultural community. Environ. Int. 2019, 131, 104997. [Google Scholar] [CrossRef]
- Hollingworth, R.M. Comparative metabolism and selectivity of organophosphate and carbamate insecticides. Bull. World Health Organ. 1971, 44, 155–170. [Google Scholar] [PubMed]
- ATSDR. Toxicological Profile for Chlorpyrifos; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 1997.
- Zhou, J.; Yin, Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Bennett, B.D.; Rabinowitz, J.D. Analytical strategies for lc-ms-based targeted metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 871, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Sabatine, M.S.; Liu, E.; Morrow, D.A.; Heller, E.; McCarroll, R.; Wiegand, R.; Berriz, G.F.; Roth, F.P.; Gerszten, R.E. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation 2005, 112, 3868–3875. [Google Scholar] [CrossRef] [PubMed]
- Suman, T.Y.; Kim, S.Y.; Yeom, D.H.; Jeon, J. Transformation products of emerging pollutants explored using non-target screening: Perspective in the transformation pathway and toxicity mechanism-a review. Toxics 2022, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Williams, J.P.; Menikarachchi, L.; Thompson, J.W.; Tyldesley-Worster, R.; Halldórsson, S.; Rolfsson, O.; Moseley, A.; Grant, D.; Langridge, J.; et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. 2014, 86, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Sherrod, S.D.; McLean, J.A. Improving the discovery of secondary metabolite natural products using ion mobility-mass spectrometry. Curr. Opin. Chem. Biol. 2018, 42, 160–166. [Google Scholar] [CrossRef]
- Amara, A.; Frainay, C.; Jourdan, F.; Naake, T.; Neumann, S.; Novoa-Del-Toro, E.M.; Salek, R.M.; Salzer, L.; Scharfenberg, S.; Witting, M. Networks and graphs discovery in metabolomics data analysis and interpretation. Front. Mol. Biosci. 2022, 9, 841373. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. Metlin: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. Massbank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Pearce, R.G.; Setzer, R.W.; Strope, C.L.; Wambaugh, J.F.; Sipes, N.S. Httk: R package for high-throughput toxicokinetics. J. Stat. Softw. 2017, 79, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Matsuzaki, J.; Katsuda, T.; Saito, Y.; Saito, H.; Ochiya, T. Generation of functional human hepatocytes in vitro: Current status and future prospects. Inflamm. Regen. 2019, 39, 13. [Google Scholar] [CrossRef] [PubMed]
- Sirenko, O.; Hesley, J.; Rusyn, I.; Cromwell, E.F. High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev. Technol. 2014, 12, 43–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Chemical | CASRN | Vendor | Purity | Catalog No. |
|---|---|---|---|---|
| Test chemicals (Parent compounds) | ||||
| Aldrin | 309-00-2 | Chem Service | 97.9% | N-11049 |
| DDD-p,p’ | 72-54-8 | Sigma-Aldrich | ≥98% | 35486 |
| DDT-o,p’ | 789-02-6 | Chem Service | 99.5% | N-12708 |
| DDT-p,p’ | 50-29-3 | Sigma-Aldrich | ≥98% | 31041 |
| Dicofol | 115-32-2 | Sigma-Aldrich | ≥98% | 36677 |
| Dieldrin | 60-57-1 | Sigma-Aldrich | ≥95% | 33491 |
| Endosulfan I | 115-29-7 | Sigma-Aldrich | ≥98% | 32015 |
| Endrin | 72-20-8 | Sigma-Aldrich | ≥98% | 32014 |
| Heptachlor epoxide B | 1024-57-3 | Chem Service | 99.5% | N-12148 |
| Heptachlor | 76-44-8 | Chem Service | 98.6% | N-12147 |
| Lindane | 58-89-9 | Sigma-Aldrich | ≥96.5% | 233390 |
| Methoxychlor-o,p’ | 72-43-5 | Sigma-Aldrich | ≥98% | 36161 |
| Parathion | 56-38-2 | Chem Service | 98.4% | N-12819 |
| Trifluralin | 1582-09-8 | Sigma-Aldrich | ≥98% | 32061 |
| 2,4-Dinitrophenol | 51-28-5 | Sigma-Aldrich | ≥98% | 34334 |
| Azinphos-methyl | 86-50-0 | Sigma-Aldrich | ≥95% | 45333 |
| Chlorpyrifos | 2921-88-2 | Sigma-Aldrich | ≥98% | 45395 |
| Diazinon | 333-41-5 | Sigma-Aldrich | ≥98% | 45428 |
| Disulfoton | 298-04-4 | Sigma-Aldrich | ≥98% | 45460 |
| Ethion | 563-12-2 | Sigma-Aldrich | ≥95% | 45477 |
| Metabolites | ||||
| 2-Amino-4-Nitrophenol | 99-57-0 | Sigma-Aldrich | 96% | A70402 |
| 4-Amino-2-Nitrophenol | 119-34-6 | Sigma-Aldrich | ≥95% | 45946 |
| Azinphos-methyl oxon | 961-22-8 | TRC | ≥95% | G855650 |
| DDA-p,p’ | 5359-38-6 | Sigma-Aldrich | 98% | 100870 |
| DDE-p,p’ | 72-55-9 | Chem Service | 99.3% | N-10875 |
| Diazoxon | 962-58-3 | TRC | ≥95% | D416890 |
| Diethylthiophosphate | 5871-17-0 | Sigma-Aldrich | 98% | 445177 |
| Diethyldithiophosphate | 298-06-6 | Sigma-Aldrich | 90% | D93600 |
| Dimethylthiophosphate | 1112-38-5 | TRC | ≥95% | D495418 |
| Disulfoton sulfone | 2497-06-5 | Sigma-Aldrich | ≥95% | 45871 |
| Internal Standards | ||||
| Atrazine | 1912-24-9 | Sigma-Aldrich | ≥98% | 45330 |
| Benzo[a]anthracene | 56-55-3 | Sigma-Aldrich | ≥98.5% | B2209 |
| Terbutryn | 886-50-0 | Sigma-Aldrich | ≥98% | 45677 |
| Mifepristone | 84371-65-3 | Selleck Chem | >99% | S2606 |
| Troglitazone | 97322-87-7 | Sigma-Aldrich | ≥98% | T2573 |
| Chemical | Parent Compound(s) | m/z | CCS |
|---|---|---|---|
| 2,4-Dinitrophenol | - | 183.01 | 127.81 |
| Azinphos-methyl | - | 339.99 | 169.57 |
| Disulfoton | - | 297.02 | 165.2 |
| Chlorpyrifos | - | 371.91 | 172.35 |
| Ethion | - | 406.98 | 180.81 |
| Heptachlor epoxide B | - | 386.82 | 177.49 |
| Trifluralin | - | 336.12 | 161.69 |
| Diazinon | - | 327.09 | 174.89 |
| Endosulfan I | - | 404.82 | 175.19 |
| Dieldrin | - | 378.88 | 160.76 |
| Aldrin | - | 361.88 | 157.56 |
| DDD-p,p’ | - | 316.95 | 170.62 |
| Metabolites | |||
| 2-Amino-4-nitrophenol | 2,4-Dinitrophenol | 153.03 | 116.54 |
| 4-Amino-2-nitrophenol | 2,4-dinitrophenol | 153.03 | 117.87 |
| Azinphos-methyl oxon | Azinphos-methyl | 324.02 | 177.67 |
| DDA-p,p’ | DDD-p,p’ | 278.98 | 152.46 |
| Diethylthiophosphate | Diazinon, Chlorpyrifos, Ethion | 171.02 | 129.33 |
| Diethyldithiophosphate | Ethion | 187.00 | 135.08 |
| Dimethylthiophosphate | Azinphos-methyl | 142.99 | 113.21 |
| Disulfoton sulfone | Disulfoton | 307.03 | 156.79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdiviezo, A.; Kato, Y.; Baker, E.S.; Chiu, W.A.; Rusyn, I. Evaluation of Metabolism of a Defined Pesticide Mixture through Multiple In Vitro Liver Models. Toxics 2022, 10, 566. https://doi.org/10.3390/toxics10100566
Valdiviezo A, Kato Y, Baker ES, Chiu WA, Rusyn I. Evaluation of Metabolism of a Defined Pesticide Mixture through Multiple In Vitro Liver Models. Toxics. 2022; 10(10):566. https://doi.org/10.3390/toxics10100566
Chicago/Turabian StyleValdiviezo, Alan, Yuki Kato, Erin S. Baker, Weihsueh A. Chiu, and Ivan Rusyn. 2022. "Evaluation of Metabolism of a Defined Pesticide Mixture through Multiple In Vitro Liver Models" Toxics 10, no. 10: 566. https://doi.org/10.3390/toxics10100566
APA StyleValdiviezo, A., Kato, Y., Baker, E. S., Chiu, W. A., & Rusyn, I. (2022). Evaluation of Metabolism of a Defined Pesticide Mixture through Multiple In Vitro Liver Models. Toxics, 10(10), 566. https://doi.org/10.3390/toxics10100566









