Do Your Kids Get What You Paid for? Evaluation of Commercially Available Probiotic Products Intended for Children in the Republic of the Philippines and the Republic of Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Acquisition of Commercially Available Probiotics
2.2. Evaluation of the Cell Viability
2.2.1. Sample Processing
2.2.2. Enumeration of Lactobacillus Species
2.2.3. Enumeration of Bifidobacterium Species
2.2.4. Enumeration of Streptococcus thermophilus
2.2.5. Enumeration of Enterococcus Species
2.2.6. Detection of Staphylococcus spp. as Contaminants
2.3. Isolation and Purification of Microorganisms from Probiotic Products
2.4. Culture-Based Molecular Identification of Microorganisms from Probiotic Products
Phylogenetic Analysis of Isolated Microorganisms
2.5. Culture-Independent Metagenomic Analysis of Microorganisms from Probiotic Products
2.5.1. DNA Extraction
2.5.2. Metagenomic Analysis of Probiotic Products
2.6. Safety Assessment of Isolated Strains
2.6.1. Hemolytic Activity of the Isolates
2.6.2. Biogenic Amine Production
2.6.3. Antibiotic Resistance Test
2.7. In Vitro Survival of Isolates to Simulated Stomach Duodenum-Passage
2.8. Investigation of Antimicrobial Properties of Isolates
2.9. Statistical Analyses
3. Results and Discussion
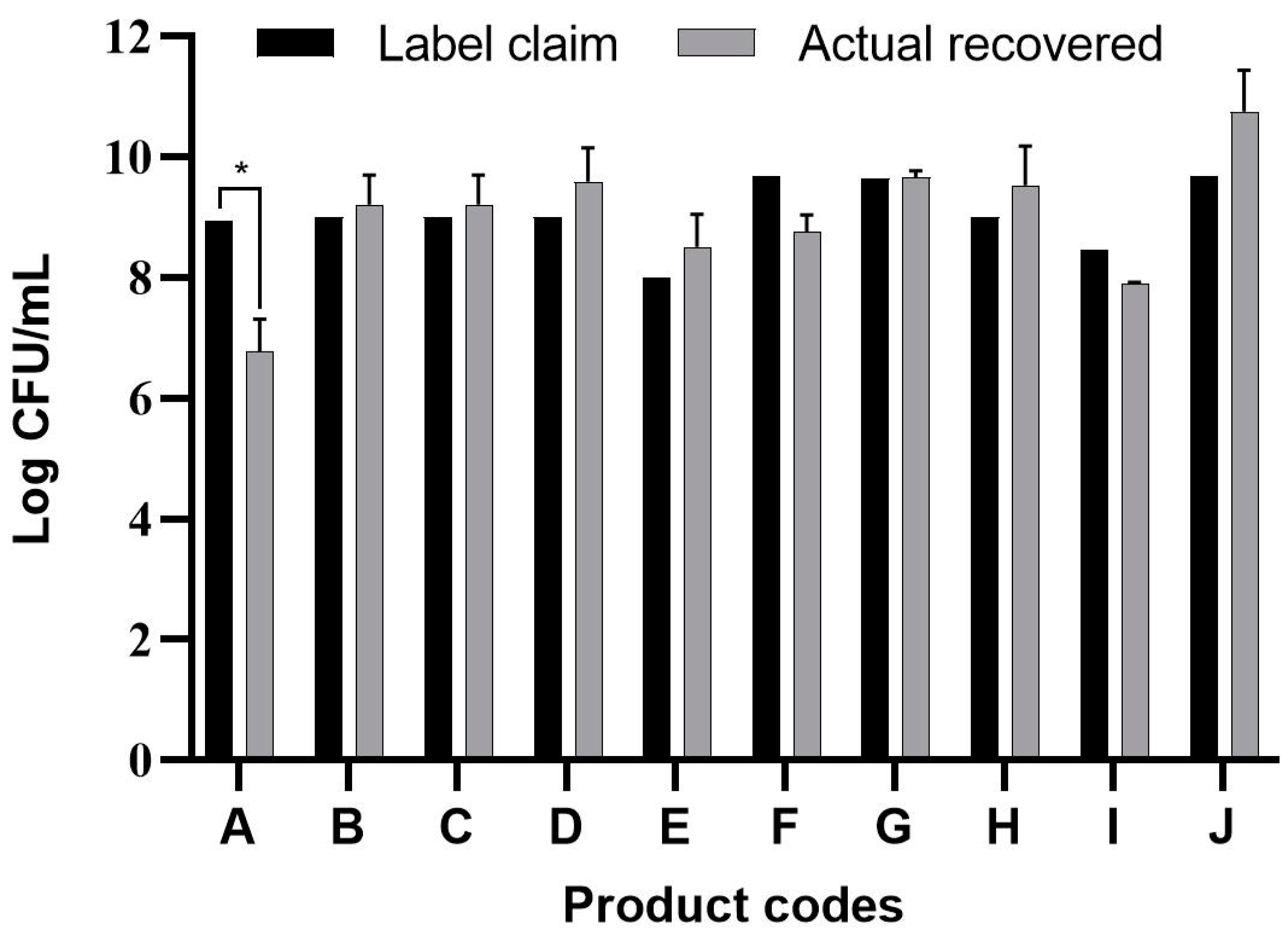
3.1. Viable Count Assay
3.2. Culture-Dependent Molecular Identification
3.3. Metagenomic Analyses of Probiotic Products by Next Generation Sequencing
3.4. Safety Evaluation of the Isolates
3.4.1. Hemolysis Activity of the Isolates
3.4.2. Detection of Biogenic Amines Produced by the Isolates
3.4.3. Antibiotic Resistance of the Isolates
3.5. Simulated Stomach-Duodenum Passage
3.6. Antagonistic Activity of the Isolates Against Common GIT Pathogens
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hyams, J.S.; Di Lorenzo, C.; Saps, M.; Shulman, R.J.; Staiano, A.; Van Tilburg, M. Childhood Functional Gastrointestinal Disorders: Child/Adolescent. Gastroenterology 2016, 150, 1456–1468. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, A.; Farver, M.; Smilowitz, J.T. Article Commentary: The Influence of Early Infant-Feeding Practices on the Intestinal Microbiome and Body Composition in Infants. Nutr. Metab. Insights 2015, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.; Illi, M.; Lötscher, M.; Wehrli, M.; Von Gunten, S. Isolation of Antibodies from Human Plasma, Saliva, Breast Milk, and Gastrointestinal Fluid. In Natural Antibodies; Kaveri, S.V., Bayry, J., Eds.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1643, pp. 23–31. ISBN 978-1-4939-7179-4. [Google Scholar]
- Canani, R.B.; Cirillo, P.; Terrin, G.; Cesarano, L.; Spagnuolo, M.I.; Vincenzo, A.D.; Albano, F.; Passariello, A.; Marco, G.D.; Manguso, F.; et al. Probiotics for treatment of acute diarrhoea in children: Randomised clinical trial of five different preparations. BMJ 2007, 335, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, S.J.; Martinez, E.G.; Gregorio, G.V.; Dans, L.F. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst. Rev. 2010, 11, 1–96. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO. FAO/WHO Joint Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food (30 April 2002 and 1 May 2002); Scientific Research Publishing: London, ON, Canada, 2002. [Google Scholar]
- Szajewska, H.; Mrukowicz, J.Z. Probiotics in the Treatment and Prevention of Acute Infectious Diarrhea in Infants and Children: A Systematic Review of Published Randomized, Double-Blind, Placebo-Controlled Trials. J. Pediatr. Gastroenterol. Nutr. 2001, 33, S17–S25. [Google Scholar] [CrossRef] [Green Version]
- Szajewska, H.; Guarino, A.; Hojsak, I.; Indrio, F.; Kolacek, S.; Shamir, R.; Vandenplas, Y.; Weizman, Z. Use of Probiotics for Management of Acute Gastroenteritis: A Position Paper by the ESPGHAN Working Group for Probiotics and Prebiotics. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Perceval, C.; Szajewska, H.; Indrio, F.; Weizman, Z.; Vandenplas, Y. Prophylactic use of probiotics for gastrointestinal disorders in children. Lancet Child Adolesc. Health 2019, 3, 655–662. [Google Scholar] [CrossRef]
- Schnadower, D.; Finkelstein, Y.; Freedman, S.B. Ondansetron and probiotics in the management of pediatric acute gastroenteritis in developed countries. Curr. Opin. Gastroenterol. 2015, 31, 1–6. [Google Scholar] [CrossRef]
- Bernaola Aponte, G.; Bada Mancilla, C.A.; Carreazo Pariasca, N.Y.; Rojas Galarza, R.A. Probiotics for Treating Persistent Diarrhoea in Children. In Cochrane Database of Systematic Reviews 2013, Issue 8; The Cochrane Collaboration, Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2010. [Google Scholar] [CrossRef] [Green Version]
- Sanders, M.E. Probiotics: Considerations for Human Health. Nutr. Rev. 2003, 61, 91–99. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Arnold, J.W.; Simpson, J.B.; Roach, J.; Kwintkiewicz, J.; Azcarate-Peril, M.A. Intra-species Genomic and Physiological Variability Impact Stress Resistance in Strains of Probiotic Potential. Front. Microbiol. 2018, 9, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huys, G.; Botteldoorn, N.; Delvigne, F.; De Vuyst, L.; Heyndrickx, M.; Pot, B.; Dubois, J.J.; Daube, G. Microbial characterization of probiotics-Advisory report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol. Nutr. Food Res. 2013, 57, 1479–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, L.C.; Sanders, M.E. Probiotics and Prebiotics in Dietetics Practice. J. Acad. Nutr. Diet. 2008, 108, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Reports and Data. Probiotics Market to Reach USD 78.3 Billion By 2026. Available online: http://www.globenewswire.com/news-release/2020/02/06/1981205/0/en/Probiotics-Market-To-Reach-USD-78-3-Billion-By-2026-Reports-And-Data.html (accessed on 3 August 2020).
- Lux Research. Probiotic Infant Formula to Claim 76% Share of $22.9 Billion Market in 2024. Available online: http://www.globenewswire.com/news-release/2015/04/21/920815/0/en/Probiotic-Infant-Formula-to-Claim-76-Share-of-22-9-Billion-Market-in-2024.html (accessed on 3 August 2020).
- Ullah, M.; Raza, A.; Ye, L.; Yu, Z. Viability and Composition Validation of Commercial Probiotic Products by Selective Culturing Combined with Next-Generation Sequencing. Microorganisms 2019, 7, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fasoli, S.; Marzotto, M.; Rizzotti, L.; Rossi, F.; Dellaglio, F.; Torriani, S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int. J. Food Microbiol. 2003, 82, 59–70. [Google Scholar] [CrossRef]
- Lugli, G.A.; Mangifesta, M.; Mancabelli, L.; Milani, C.; Turroni, F.; Viappiani, A.; van Sinderen, D.; Ventura, M. Compositional assessment of bacterial communities in probiotic supplements by means of metagenomic techniques. Int. J. Food Microbiol. 2019, 294, 1–9. [Google Scholar] [CrossRef]
- Terzaghi, B.E. Morphologics and host sensitives of lactic streptococcal phages from cheese factories. N.Z.J. Dairy Sci. Technol. 1976, 11, 155–163. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Han, Y.; Park, H.; Choi, B.R.; Ji, Y.; Kwon, E.Y.; Choi, M.S. Alteration of Microbiome Profile by D-Allulose in Amelioration of High-Fat-Diet-Induced Obesity in Mice. Nutrients 2020, 12, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Kim, H.; Park, H.; Lee, J.; Lee, H.; Shin, H.; Kim, B.; Franz, C.M.A.P.; Holzapfel, W.H. Functionality and safety of lactic bacterial strains from Korean kimchi. Food Control. 2013, 31, 467–473. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, CLSI Supplement M100, 29th ed.; Clinical and Laboratory Science Institute: Wayne, NJ, USA, 2019; pp. 1–281. [Google Scholar]
- Klare, I.; Konstabel, C.; Müller-Bertling, S.; Reissbrodt, R.; Huys, G.; Vancanneyt, M.; Swings, J.; Goossens, H.; Witte, W. Evaluation of New Broth Media for Microdilution Antibiotic Susceptibility Testing of Lactobacilli, Pediococci, Lactococci, and Bifidobacteria. Appl. Environ. Microbiol. 2005, 71, 8982–8986. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing: EUCAST. Available online: https://eucast.org/ (accessed on 3 August 2020).
- Guidance on EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 1–10. [Google Scholar] [CrossRef]
- Mathara, J.; Schillinger, U.; Guigas, C.; Franz, C.; Kutima, P.; Mbugua, S.; Shin, H.; Holzapfel, W. Functional characteristics of Lactobacillus spp. from traditional Maasai fermented milk products in Kenya. Int. J. Food Microbiol. 2008, 126, 57–64. [Google Scholar] [CrossRef]
- Toscano, M.; Vecchi, E.; Rodighiero, V.; Drago, L. Microbiological and genetic identification of some probiotics proposed for medical use in 2011. J. Chemother. 2013, 25, 156–161. [Google Scholar] [CrossRef]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to Qualify Microorganisms as “Probiotic” in Foods and Dietary Supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef]
- Hill, D.; Sugrue, I.; Tobin, C.; Hill, C.; Stanton, C.; Ross, R.P. The Lactobacillus casei Group: History and Health Related Applications. Front. Microbiol. 2018, 9, 2107. [Google Scholar] [CrossRef] [Green Version]
- Salvetti, E.; Torriani, S.; Felis, G.E. The Genus Lactobacillus: A Taxonomic Update. Probiotics Antimicrob. Proteins 2012, 4, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Pot, B.; Felis, G.E.; Bruyne, K.D.; Tsakalidou, E.; Papadimitriou, K.; Leisner, J.; Vandamme, P. The Genus Lactobacillus. In Lactic Acid Bacteria; Holzapfel, W.H., Wood, B.J.B., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 249–353. ISBN 978-1-118-65525-2. [Google Scholar]
- Huang, C.H.; Li, S.W.; Huang, L.; Watanabe, K. Identification and Classification for the Lactobacillus casei Group. Front. Microbiol. 2018, 9, 1974. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Sun, Z.; Liu, W.; Bao, Q.; Zhang, J.; Zhang, H. Phylogenetic study of Lactobacillus acidophilus group, L. casei group and L. plantarum group based on partial hsp60, pheS and tuf gene sequences. Eur. Food Res. Technol. 2012, 234, 927–934. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masco, L.; Huys, G.; De Brandt, E.; Temmerman, R.; Swings, J. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 2005, 102, 221–230. [Google Scholar] [CrossRef]
- Lewis, Z.T.; Shani, G.; Masarweh, C.F.; Popovic, M.; Frese, S.A.; Sela, D.A.; Underwood, M.A.; Mills, D.A. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr. Res. 2016, 79, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Breed, R.S.; Murray, E.G.D.; Smith, N.R. Bergey’s Manual of Determinative Bacteriology, 7th ed.; The Williams & Wilkins Company: Baltimore, MD, USA, 1972; p. 1094. [Google Scholar]
- Hill, C.; Scott, K.; Klaenhammer, T.R.; Quigley, E.; Sanders, M.E. Probiotic nomenclature matters. Gut Microbes 2016, 7, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Vitali, B.; Candela, M.; Matteuzzi, D.; Brigidi, P. Quantitative Detection of Probiotic Bifidobacterium Strains in Bacterial Mixtures by Using Real-time PCR. Syst. Appl. Microbiol. 2003, 26, 269–276. [Google Scholar] [CrossRef]
- Holzapfel, W.; Arini, A.; Aeschbacher, M.; Coppolecchia, R.; Pot, B. Enterococcus faecium SF68 as a model for efficacy and safety evaluation of pharmaceutical probiotics. Benef. Microbes 2018, 9, 375–388. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S. Microbiologic evaluation of commercial probiotics. J. Am. Vet. Med. Assoc. 2002, 220, 794–797. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcus epidermidis—The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Sharon, I.; Morowitz, M.J.; Thomas, B.C.; Costello, E.K.; Relman, D.A.; Banfield, J.F. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013, 23, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Dong, D.; Ni, Q.; Wang, C.; Zhang, L.; Li, Z.; Jiang, C.; Enqiang, M.; Peng, Y. Effects of intestinal colonization by Clostridium difficile and Staphylococcus aureus on microbiota diversity in healthy individuals in China. BMC Infect. Dis. 2018, 18, 207. [Google Scholar] [CrossRef]
- Theunissen, J.; Britz, T.; Torriani, S.; Witthuhn, R. Identification of probiotic microorganisms in South African products using PCR-based DGGE analysis. Int. J. Food Microbiol. 2005, 98, 11–21. [Google Scholar] [CrossRef]
- Patro, J.N.; Ramachandran, P.; Barnaba, T.; Mammel, M.K.; Lewis, J.L.; Elkins, C.A. Culture-Independent Metagenomic Surveillance of Commercially Available Probiotics with High-Throughput Next-Generation Sequencing. MSphere 2016, 1, e00057-16. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Flint, S.; Yu, P.L. Enterocins in food preservation. Int. J. Food Microbiol. 2010, 141, 1–10. [Google Scholar] [CrossRef]
- Noguchi, N.; Nakaminami, H.; Nakase, K.; Sasatsu, M. Characterization of Enterococcus Strains Contained in Probiotic Products. Biol. Pharm. Bull. 2011, 34, 1469–1473. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.; Goin, C.; O’Flaherty, S.; Altermann, E.; Hutkins, R. Specialized adaptation of a lactic acid bacterium to the milk environment: The comparative genomics of Streptococcus thermophilus LMD-9. Microb. Cell Fact. 2011, 10, S22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdichon, F.; Alper, I.; Bibiloni, R.; Dubois, A.; Laulund, S.; Miks, M.; Morelli, L.; Zuliani, V.; Yao, S. Inventory of Microbial Food Cultures with Safety Demonstration in Fermented Food Products. Update of the Bulletin of the IDF 455–2012. Bull. Int. Dairy Federat. 2018, 495, 5–71. [Google Scholar]
- Vesterlund, S.; Vankerckhoven, V.; Saxelin, M.; Goossens, H.; Salminen, S.; Ouwehand, A.C. Safety assessment of Lactobacillus strains: Presence of putative risk factors in faecal, blood and probiotic isolates. Int. J. Food Microbiol. 2007, 116, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Özogul, Y.; Özogul, F. Chapter 1: Biogenic Amines Formation, Toxicity, Regulations in Food. In Biogenic Amines in Food: Analysis, Occurrence and Toxicity; Royal Society of Chemistry: London, UK, 2019; pp. 1–17. [Google Scholar]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Inf. Microbio. 2012, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, C.; Teuber, S.; Gershwin, M.E. Histamine (Scombroid) Fish Poisoning: A Comprehensive Review. Clin. Rev. Allerg. Immunol. 2016, 50, 64–69. [Google Scholar] [CrossRef]
- Shalaby, A.R. Significance of biogenic amines to food safety and human health. Food Res. Int. 1996, 29, 675–690. [Google Scholar] [CrossRef]
- Linares, D.M.; Del Rio, B.; Redruello, B.; Ladero, V.; Martin, M.C.; Fernandez, M.; Ruas-Madiedo, P.; Alvarez, M.A. Comparative analysis of the in vitro cytotoxicity of the dietary biogenic amines tyramine and histamine. Food Chem. 2016, 197, 658–663. [Google Scholar] [CrossRef] [Green Version]
- Bargossi, E.; Gardini, F.; Gatto, V.; Montanari, C.; Torriani, S.; Tabanelli, G. The Capability of Tyramine Production and Correlation between Phenotypic and Genetic Characteristics of Enterococcus faecium and Enterococcus faecalis Strains. Front. Microbiol. 2015, 6, 1371. [Google Scholar] [CrossRef] [Green Version]
- Ladero, V.; Fernández, M.; Calles-Enríquez, M.; Sánchez-Llana, E.; Cañedo, E.; Martín, M.C.; Alvarez, M.A. Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 2012, 30, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Coton, E.; Mulder, N.; Coton, M.; Pochet, S.; Trip, H.; Lolkema, J.S. Origin of the Putrescine-Producing Ability of the Coagulase-Negative Bacterium Staphylococcus epidermidis 2015B. Appl. Environ. Microbiol. 2010, 76, 5570–5576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, H.; Park, J.H.; Choi, A.; Hwang, H.J.; Mah, J.H. Validation of an HPLC Analytical Method for Determination of Biogenic Amines in Agricultural Products and Monitoring of Biogenic Amines in Korean Fermented Agricultural Products. Toxicol. Res. 2015, 31, 299–305. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific opinion on risk-based control of biogenic amine formation in fermented foods. EFSA J. 2011, 10, 1–93. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Tomar, S.K.; Sangwan, V.; Goswami, P.; Singh, R. Antibiotic Resistance of Lactobacillus sp. Isolated from Commercial Probiotic Preparations: Antibiotic-Resistant Lactobacillus. J. Food Saf. 2016, 36, 38–51. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef]
- Mater, D.D.G.; Langella, P.; Corthier, G.; Flores, M.J. A Probiotic Lactobacillus Strain Can Acquire Vancomycin Resistance during Digestive Transit in Mice. J. Mol. Microbiol. Biotechnol. 2008, 14, 123–127. [Google Scholar] [CrossRef]
- Devirgiliis, C.; Zinno, P.; Perozzi, G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol. 2013, 4, 301. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.; Ngu, D.Y.S.; Dan, L.A.; Ooi, A.; Lim, R.L.H. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015, 14, 95. [Google Scholar] [CrossRef]
- Schillinger, U.; Guigas, C.; Holzapfel, W.H. In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 2005, 15, 1289–1297. [Google Scholar] [CrossRef]
- İspirli, H.; Demirbaş, F.; Dertli, E. Characterization of functional properties of Enterococcus faecium strains isolated from human gut. Can. J. Microl. 2015, 61, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.M.; Li, N.; Zheng, K.; Hao, J.F. Enhancing acid tolerance of the probiotic bacterium Lactobacillus acidophilus NCFM with trehalose. FEMS Microbiol. Lett. 2018, 365, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress Physiology of Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [Green Version]
- Bezkorovainy, A. Probiotics: Determinants of survival and growth in the gut. Am. J. Clin. Nutr. 2001, 73, 399s–405s. [Google Scholar] [CrossRef] [PubMed]
- Mathipa, M.G.; Thantsha, M.S. Probiotic engineering: Towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog. 2017, 9, 28. [Google Scholar] [CrossRef]
- Todorov, S.D.; De Melo Franco, B.D.G.; Tagg, J.R. Bacteriocins of Gram-positive bacteria having activity spectra extending beyond closely-related species. Benef. Microbes 2019, 10, 315–328. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Bacteriocin production by Lactobacillus pentosus ST712BZ isolated from boza. Braz. J. Microbiol. 2007, 38, 166–172. [Google Scholar] [CrossRef]
- De Kwaadsteniet, M.; Todorov, S.D.; Knoetze, H.; Dicks, L.M.T. Characterization of a 3944 Da bacteriocin, produced by Enterococcus mundtii ST15, with activity against Gram-positive and Gram-negative bacteria. Int. J. Food Microbiol. 2005, 105, 433–444. [Google Scholar] [CrossRef]
- Browne, P.D.; Van Der Waal, M.B.; Claassen, E. The ‘healthy’ gut microbiota. In Microbiota in Health and Disease: From Pregnancy to Childhood, 1st ed.; Browne, P.D., Claassen, E., Cabana, M.D., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2017; pp. 17–35. [Google Scholar]
- Vandenplas, Y.; Huysentruyt, K. Probiotic Interventions to Optimize the Infant and Child Microbiota. In Microbiota in Health and Disease: From Pregnancy to Childhood, 1st ed.; Browne, P.D., Claassen, E., Cabana, M.D., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2017; pp. 299–311. [Google Scholar]
- Van Den Nieuwboer, M.; Browne, P.D.; Claassen, E. Safety of Probiotics in Infants and Children. In Microbiota in Health and Disease: From Pregnancy to Childhood, 1st ed.; Browne, P.D., Claassen, E., Cabana, M.D., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2017; pp. 313–340. [Google Scholar]
- FAO/WHO Codex Alimentarius Commission. Discussion Paper on Harmonized Probiotic Guidelines for Use in Foods and Dietary Supplements. Agenda Item 11; CX/NFSDU 19/41/11; FAO/WHO Codex Alimentarius Commission: Rome, Italy, 2019; Available online: http://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FMeetings%252FCX-720-41%252FWD%252Fnf41_11e.pdf (accessed on 3 August 2020).
- Degnan, F.H. The US Food and Drug Administration and probiotics: Regulatory categorization. Clin. Infect. Dis. 2008, 46, S133–S136. [Google Scholar] [CrossRef]
- Saldanha, L.G. US Food and Drug Administration regulations governing label claims for food products, including probiotics. Clin. Infect. Dis. 2008, 46, S119–S151. [Google Scholar] [CrossRef]
- Indian Council of Medical Research Task Force. Co-ordinating Unit ICMR.; Co-ordinating Unit DBT. ICMR-DBT guidelines for evaluation of probiotics in food. Indian J. Med. Res. 2011, 134, 22–25. [Google Scholar]
| Product | Label Claim | Species Identified Using 16S rRNA Sequencing | ||
|---|---|---|---|---|
| Bacterial Species Declared | Identified Microorganisms from Probiotic Products | Assigned Strain Number | Phylogenetic Tree Showing Relationship of Microorganisms Isolated from Each Probiotic Product | |
| A | Lactobacillus plantarum | Lactobacillus plantarum# | HEM C163 |  Phylogeny of isolated microorganisms from product A based on 1466 nucleotides of the partial 16S rRNA gene using the neighbor-joining method. The sum of branch length = 0.17651297 is shown. Scale bar represents one nucleotide substitution for every 100 nucleotides. † |
| Bifidobacterium breve subsp. breve | Bacillus coagulans | HEM C18 | ||
| Bifidobacterium infantis subsp. infantis | HEM C19 | |||
| Bifidobacterium longum | HEM C41 | |||
| Enterococcus faecalis | HEM C42 | |||
| Lactobacillus acidophilus | HEM C43 | |||
| Lactobacillus brevis | HEM C140 | |||
| Lactobacillus bulgaricus | Enterococcus faecium | HEM C143 | ||
| Lactobacillus casei | ||||
| Lactobacillus fermentum | ||||
| Lactobacillus helveticus subsp. jugurti | ||||
| Streptococcus thermophilus | ||||
| B | Streptococcus thermophilus | Streptococcus thermophilus# | HEM C52 |  Phylogeny of isolated microorganisms from product B based on 1445 nucleotides of the partial 16S rRNA gene using the neighbor-joining method. The sum of branch length = 0.22020732 is shown. Scale bar represents two nucleotide substitutions for every 200 nucleotides. † |
| Lactobacillus acidophilus | Lactobacillus acidophilus# | HEM C16 | ||
| Lactobacillus rhamnosus | Lactobacillus rhamnosus# | HEM C14 | ||
| Lactobacillus casei | HEM C147 | |||
| Lactobacillus bulgaricus | HEM C151 | |||
| Bifidobacterium breve | HEM C153 | |||
| Bifidobacterium infantis | ||||
| Lactobacillus paracasei | HEM C39 | |||
| Lactobacillus gallinarum | HEM C10 | |||
| HEM C11 | ||||
| HEM C12 | ||||
| HEM C13 | ||||
| HEM C146 | ||||
| HEM C52 | ||||
| C | Lactobacillus reuteri | Lactobacillus reuteri# | HEM C1 | |
| Enterococcus durans | HEM C121 * | |||
| D | Lactobacillus rhamnosus | Lactobacillus rhamnosus# | HEM C3 | |
| E | Lactobacillus acidophilus | Lactobacillus acidophilus# | HEM C21 | |
| Bifidobacterium longum | Enterococcus faecalis | HEM C48 | ||
| F | Lactobacillus plantarum | Lactobacillus plantarum# | HEM C5 | |
| G | Enterococcus faecium | Enterococcus faecium# | HEM C22 |  Phylogeny of isolated microorganisms from product G based on 1390 nucleotides of the partial 16S rRNA gene using the neighbor-joining method. The sum of branch length = 0.26710396 is shown. Scale bar represents two nucleotide substitutions for every 200 nucleotides. † |
| Bifidobacterium longum | HEM C23 | |||
| Bifidobacterium bifidum | HEM C27 | |||
| Lactobacillus acidophilus | HEM C51 | |||
| HEM C57 | ||||
| Bifidobacterium breve | HEM C28 | |||
| Lactobacillus plantarum | HEM C25 | |||
| H | Streptococcus thermophilus | Streptococcus thermophilus# | HEM C31 |  Phylogeny of isolated microorganisms from product H based on 1376 nucleotides of the partial 16S rRNA gene using the neighbor-joining method. The sum of branch length = 0.39666877 is shown. Scale bar represents two nucleotide substitutions for every 200 nucleotides. † |
| Bifidobacterium breve | Bifidobacterium breve# | HEM C30 | ||
| Lactobacillus plantarum | Lactobacillus plantarum# | HEM C29 | ||
| Bifidobacterium longum | HEM C33 | |||
| HEM C138 | ||||
| Enterococcus faecium | HEM C54 | |||
| Staphylococcus epidermidis * | HEM C56 | |||
| I | Lactobacillus sporogenes | Bacillus coagulans# | HEM C136 |  Phylogeny of isolated microorganisms from Product I based on 1371 nucleotides of the partial 16S rRNA gene using the neighbor-joining method. The sum of branch length = 0.23418728 is shown. Scale bar represents one nucleotide substitutions for every 100 nucleotides. † |
| Lactobacillus johnsonii | Lactobacillus sakei | HEM C61 | ||
| Lactobacillus rhamnosus | Lactobacillus reuteri | HEM C79 | ||
| Bifidobacterium lactis | Enterococcus faecium | HEM C103 | ||
| HEM C128 | ||||
| Lactobacillus plantarum | HEM C127 | |||
| J | Lactobacillus reuteri | Lactobacillus reuteri# | HEM C65 |  Phylogeny of isolated microorganisms from product I based on 1385 nucleotides of the partial 16S rRNA gene using the neighbor-joining method. The sum of branch length = 0.13083425 is shown. Scale bar represents one nucleotide substitutions for every 100 nucleotides. † |
| Lactobacillus acidophilus | HEM C74 | |||
| Bifidobacterium bifidum | HEM C130 | |||
| Bifidobacterium animalis subsp. lactis | HEM C132 | |||
| Streptococcus thermophilus | HEM C148 | |||
| Enterococcus faecium | HEM C149 | |||
| Enterococcus faecium# | HEM C82 | |||
| HEM C93 | ||||
| HEM C98 | ||||
| HEM C99 | ||||
| HEM C100 | ||||
| HEM C115 | ||||
| Enterococcus durans | HEM C64 | |||
| HEM C134 | ||||
| Lactobacillus sakei | HEM C81 |
| Culture-Independent (Metagenomic Analysis) | ||||
|---|---|---|---|---|
| Product | Phylum | Relative Abundance | Genera | Relative Abundance |
| A | Proteobacteria | 15.4% | Unidentified genus | 10.3% |
| Delftia | 5.1% | |||
| Firmicutes | 56.4% | Lactobacillus | 7.7% | |
| Enterococcus | 43.6% | |||
| Bacillus | 5.1% | |||
| Actinobacteria | 28.2% | Bifidobacterium | 28.2% | |
| G | Firmicutes | 30.4% | Enterococcus | 30.4% |
| Actinobacteria | 69.6% | Bifidobacterium | 69.6% | |
| Minimum Inhibitory Concentration (mg/L) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Product | Isolate | Strain | Hemo-lysis | BA Produced | AMP | CHL | CLI | ERY | GEN | KAN | STR | TET | VAN | |
| A | Bac. coagulans | HEM C18 | γ | Tyr, His | n.r. | 1 | ≤0.25 | ≤0.25 | 1 | 8 | 4 | ≤1 | 16 * | |
| E. faecium | HEM C143 | γ | None | 2 | 32 * | >16 * | 16 * | 64 * | >1024 * | 128 | ≤1 | 2 | ||
| L. plantarum# | HEM C163 | γ | None | 2 | 16 * | 8 * | 2* | 32 * | >128 * | n.r. | >128 * | n.r. | ||
| B | L. rhamnosus# | HEM C14 | γ | None | 16 * | 1 | ≤0.50 | 0.5 | 16 | >128 * | >128 * | 2 | n.r. | |
| L. acidophilus# | HEM C16 | γ | None | ≤0.25 | 8* | 0.5 | ≤0.25 | ≤8 | 32 | ≤8 | 4 | 1 | ||
| L. paracasei | HEM C39 | γ | None | 1 | 2 | ≤0.5 | ≤0.25 | 32 | >128 * | 64 | 4 | n.r | ||
| L. gallinarum | HEM C9 | γ | None | ≤0.25 | 1 | ≤0.25 | ≤0.25 | 1 | 32 | ≤2 | ≤1 | 1 | ||
| Strep. thermophilus# | HEM C52 | γ | None | ≤0.50 | 4 | ≤0.50 | ≤0.50 | 2 | 32 | ≤2 | ≤1 | ≤1 | ||
| C | L. reuteri# | HEM C1 | γ | None | >8 * | 2 | ≤0.25 | ≤0.25 | 1 | 32 | 8 | >64 * | n.r. | |
| E. durans | HEM C121 | γ | None | 1 | 64 * | >16 * | 8 * | 64 * | >1024 * | 128 | ≤1 | 2 | ||
| D | L. rhamnosus# | HEM C3 | γ | None | 1 | 1 | ≤0.50 | ≤0.25 | 8 | 64 | ≤32 | 2 | n.r | |
| E | L. acidophilus# | HEM C21 | γ | None | 0.5 | 1 | ≤0.25 | ≤0.25 | 4 | 64 | 4 | 4 | ≤0.50 | |
| E. faecalis | HEM C48 | γ | Tyr | ≤0.25 | 1 | ≤0.25 | ≤0.25 | 4 | 64 | 4 | 2 | 2 | ||
| F | L. plantarum# | HEM C5 | γ | None | 1 | 1 | ≤0.50 | ≤0.25 | 8 | 128 * | n.r. | 64 * | n.r | |
| G | L. plantarum | HEM C25 | γ | None | 1 | 8 | 0.5 | ≤0.25 | ≤2 | ≤16 | n.r. | 32 | n.r | |
| E. faecium# | HEM C22 | γ | None | 1 | ≤4 | ≤1 | 16 * | 32 | 512 | 128 | ≤1 | ≤1 | ||
| Bif. breve | HEM C28 | γ | None | 16 * | 8 | >8 * | >8 * | 64 | n.r. | 64 | 1 | >16 * | ||
| H | Strep. thermophilus# | HEM C31 | γ | None | ≤0.50 | 2 | ≤0.50 | ≤0.50 | ≤1 | 8 | ≤2 | ≤1 | ≤1 | |
| L. plantarum | HEM C33 | γ | None | 1 | 8 | 0.5 | ≤0.25 | ≤2 | ≤16 | n.r. | 32 | n.r | ||
| E. faecium | HEM C54 | γ | None | 1 | ≤4 | ≤1 | 8* | 16 | >1024 * | >128 * | ≤1 | ≤1 | ||
| Bif. breve# | HEM C30 | γ | None | 2 | 8 | >8 * | 8 | 128* | n.r. | >128 * | 2 | >16 * | ||
| Staph. epidermidis | HEM C56 | β | Put | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| I | Bac. coagulans# | HEM C136 | γ | None | n.r. | 4 | 4 | ≤0.50 | 2 | 8 | 32 * | >32 * | ≤1 | |
| L. sakei | HEM C61 | γ | None | 1 | 16 * | ≤0.5 | ≤0.25 | 4 | 16 | 32 | 4 | n.r. | ||
| L. reuteri | HEM C79 | γ | None | 1 | 16* | ≤0.5 | ≤0.25 | ≤2 | 64 | 32 | 8 | n.r. | ||
| E. faecium | HEM C128 | γ | None | 4 * | 32 * | >16 * | 16 * | 64 * | >1024 * | 128 | ≤1 | 4 | ||
| L. plantarum | HEM C127 | γ | None | 1 | 8 | ≤0.25 | ≤0.25 | ≤2 | 64 | n.r. | 16 | n.r. | ||
| J | L. reuteri# | HEM C148 | γ | None | 1 | 4 | ≤0.25 | ≤0.25 | ≤2 | 16 | ≤8 | ≤2 | n.r. | |
| L. sakei | HEM C81 | γ | None | ≤1 | 8 * | ≤0.25 | ≤0.25 | 8 | 64 | 32 | >32 * | n.r. | ||
| E. faecium# | HEM C100 | γ | None | 1 | 8 | >16 * | 8 * | 64 * | 1024 | 128 | ≤1 | 2 | ||
| E. durans | HEM C64 | γ | None | 1 | 8 | >16 * | ≤1 | 64 * | >1024 * | 128 | ≤1 | >16 * | ||
| Controls | Bacillus cereus | ATCC 27348 | β | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| Escherichia coli | ATCC 25922 | α | His, Put, Tyr, Cad | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| Lactobacillus plantarum | 299V | γ | None | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| EFSA Cut-off Values | Bacillus spp. | n.r | 8 | 4 | 4 | 4 | 8 | 8 | 8 | 4 | ||||
| Strep. thermophilus | 2 | 4 | 2 | 2 | 32 | 64 | 64 | 4 | 4 | |||||
| Bifidobacterium spp. | 2 | 4 | 1 | 1 | 64 | n.r | 128 | 8 | 2 | |||||
| E. faecium | 2 | 16 | 4 | 4 | 32 | 1024 | 128 | 4 | 4 | |||||
| L. plantarum | 2 | 8 | 2 | 1 | 16 | 64 | n.r | 32 | n.r | |||||
| L. acidophilus/L. gallinarum | 1 | 4 | 1 | 1 | 16 | 64 | 16 | 4 | 2 | |||||
| L. reuteri | 2 | 4 | 1 | 1 | 8 | 64 | 64 | 16 | n.r | |||||
| L. rhamnosus | 4 | 4 | 1 | 1 | 16 | 64 | 32 | 8 | n.r | |||||
| L. sakei | 4 | 4 | 1 | 1 | 16 | 64 | 64 | 8 | n.r | |||||
| Product | Strains Isolated from Probiotic Products | Initial Counts | After 1 h (Stomach) | After 3h (Duodenum) | ||
|---|---|---|---|---|---|---|
| (Log CFU/mL) | (Log CFU/mL) | Survival % | (Log CFU/mL) | Survival % | ||
| A | E. faecium HEM C143 * | 8.77 ± 0.04 | 8.62 ± 0.08 | 72.22 | 7.43 ± 0.04 | 45.79 |
| L. plantarum HEM C163 | 9.41 ± 0.07 | 9.33 ± 0.04 | 84.48 | 8.77 ± 0.07 | 67.97 | |
| B | L. rhamnosus HEM C14 | 8.67 ± 0.01 | 8.02 ± 0.03 | 22.60 | 3.15 ± 0.25 | 0.002 |
| L. acidophilus HEM C16 | 9.29 ± 0.05 | 9.29 ± 0.12 | 100.05 | 3.88 ± 0.62 | 0.00077 | |
| L. paracasei HEM C39 * | 9.42 ± 0.07 | 9.16 ± 0.27 | 59.43 | 3.09 ± 0.30 | 0.000056 | |
| Strep. thermophilus HEM C52 | 9.98 ± 0.03 | 9.69 ± 0.04 | 51.72 | 8.31 ± 0.01 | 2.17 | |
| C | L. reuteri HEM C1 | 9.34 ± 0.20 | 9.34 ± 0.15 | 101.53 | 5.53 ± 0.21 | 0.016 |
| D | L. rhamnosus HEM C3 | 9.20 ± 0.18 | 8.70 ± 0.08 | 35.08 | <3.49 | <0.001 |
| E | L. acidophilus HEM C21 | 8.74 ± 0.06 | 8.83 ± 0.18 | 131.67 | 5.21 ± 0.17 | 0.0031 |
| F | L. plantarum HEM C5 | 9.53 ± 0.07 | 9.49 ± 0.08 | 91.61 | 8.31 ± 0.02 | 25.81 |
| G | E. faecium HEM C22 | 8.83 ± 0.17 | 8.58 ± 0.15 | 64.98 | 5.88 ± 0.06 | 0.011 |
| L. plantarum HEM C25 | 9.04 ± 0.37 | 8.58 ± 0.15 | 10.58 | 7.34 ± 0.22 | 1.011 | |
| Bif. breve HEM C28 * | 9.15 ± 0.00 | <5.00 | <0.10 | 4.21 ± 0.17 | 0.000122 | |
| H | L. plantarum HEM C33 | 9.36 ± 0.00 | 9.16± 0.11 | 77.81 | 7.49 ± 0.21 | 1.96 |
| Bif. breve HEM C30 | 8.81 ± 0.05 | 6.59 ± 0.16 | 0.61 | 5.60 ± 0.05 | 0.063 | |
| Strep. thermophilus HEM C31 | 9.78 ± 0.04 | 9.17 ± 0.00 | 26.01 | 7.47 ± 0.08 | 0.0495 | |
| I | L. reuteri HEM C79 * | 9.30 ± 0.08 | 8.30 ± 0.09 | 77.81 | 5.73 ± 0.05 | 0.022 |
| L. plantarum HEM C127 * | 9.48 ± 0.03 | 9.35 ± 0.17 | 78.29 | 6.37 ± 0.10 | 0.080 | |
| J | E. faecium HEM C99 | 9.01 ± 0.11 | 8.65 ± 0.11 | 44.98 | 6.71 ± 0.04 | 0.513 |
| L. reuteri HEM C148 | 9.38 ± 0.08 | 9.30 ± 0.09 | 87.50 | 6.73 ± 0.05 | 0.226 | |
| Test Pathogen Strains | ||||||
|---|---|---|---|---|---|---|
| Product | Isolates from Probiotic Products | Staphylococcus aureus subsp. aureus ATCC 6538 | Bacillus cereus ATCC 11778 | Escherichia coli ATCC 8739 | Listeria innocua ATCC 33090 | Salmonella enterica subsp. enterica var. TyphimuriumATCC 14028 |
| A | E. faecium HEM C143 * | - | - | - | - | - |
| L. plantarum HEM C163 | - | - | - | - | - | |
| B | L. rhamnosus HEM C14 | - | - | - | - | - |
| L. acidophilus HEM C16 | + | - | - | - | + | |
| L. paracasei HEM C39 * | - | - | - | - | - | |
| Strep. thermophilus HEM C52 | - | - | - | - | - | |
| C | L. reuteri HEM C1 | - | - | - | - | - |
| D | L. rhamnosus HEM C3 | - | + | - | - | + |
| E | L. acidophilus HEM C21 | - | - | - | - | - |
| F | L. rhamnosus HEM C5 | - | - | - | - | - |
| G | E. faecium HEM C22 | +++ | - | - | +++ | - |
| L. plantarum HEM C25 * | +++ | - | - | +++ | ++ | |
| Bif. breve HEM C30 * | - | - | - | - | - | |
| H | L. plantarum HEM C33 | - | - | - | - | - |
| Bif. breve HEM C30 | - | - | - | - | - | |
| Strep. thermophilus HEM C31 | - | - | - | - | - | |
| I | L. sakei HEM C61 * | +++ | ++ | - | - | ++ |
| L. reuteri HEM C79 * | - | - | - | - | - | |
| J | E. faecium HEM C99 | - | - | - | - | - |
| L. reuteri HEM C148 | - | - | - | - | - | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dioso, C.M.; Vital, P.; Arellano, K.; Park, H.; Todorov, S.D.; Ji, Y.; Holzapfel, W. Do Your Kids Get What You Paid for? Evaluation of Commercially Available Probiotic Products Intended for Children in the Republic of the Philippines and the Republic of Korea. Foods 2020, 9, 1229. https://doi.org/10.3390/foods9091229
Dioso CM, Vital P, Arellano K, Park H, Todorov SD, Ji Y, Holzapfel W. Do Your Kids Get What You Paid for? Evaluation of Commercially Available Probiotic Products Intended for Children in the Republic of the Philippines and the Republic of Korea. Foods. 2020; 9(9):1229. https://doi.org/10.3390/foods9091229
Chicago/Turabian StyleDioso, Clarizza May, Pierangeli Vital, Karina Arellano, Haryung Park, Svetoslav Dimitrov Todorov, Yosep Ji, and Wilhelm Holzapfel. 2020. "Do Your Kids Get What You Paid for? Evaluation of Commercially Available Probiotic Products Intended for Children in the Republic of the Philippines and the Republic of Korea" Foods 9, no. 9: 1229. https://doi.org/10.3390/foods9091229
APA StyleDioso, C. M., Vital, P., Arellano, K., Park, H., Todorov, S. D., Ji, Y., & Holzapfel, W. (2020). Do Your Kids Get What You Paid for? Evaluation of Commercially Available Probiotic Products Intended for Children in the Republic of the Philippines and the Republic of Korea. Foods, 9(9), 1229. https://doi.org/10.3390/foods9091229





