Impact of Simulated Gastrointestinal Digestion on the Biological Activity of an Alcalase Hydrolysate of Orange Seed (Siavaraze, Citrus sinensis) by-Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Productions of Orange Seed Proteins Concentrate
2.3. Optimization of Enzymatic Hydrolysis
2.4. Determination of Hydrolysis Degree
2.5. Assay of DPPH Radical Scavenging Activity Measurement
2.6. Assay of Ferric-Reducing Antioxidant Power Measurement
2.7. Assay of the ACE-Inhibitory Activity Measurement
2.8. Assay of α-Amylase-Inhibitory Activity
2.9. Assay of α-Glucosidase-Inhibitory Activity
2.10. SEC Separation of Hydrolyzed Protein
2.11. Isolation of Most Active Fractions Using RP-HPLC
2.12. Simulated in Vitro Gastrointestinal Digestion
2.13. Free Amino Acid Analysis
2.14. Identification of Peptides, Using Mass Spectrometry in Tandem
2.15. Statistical Analysis
3. Results and Discussion
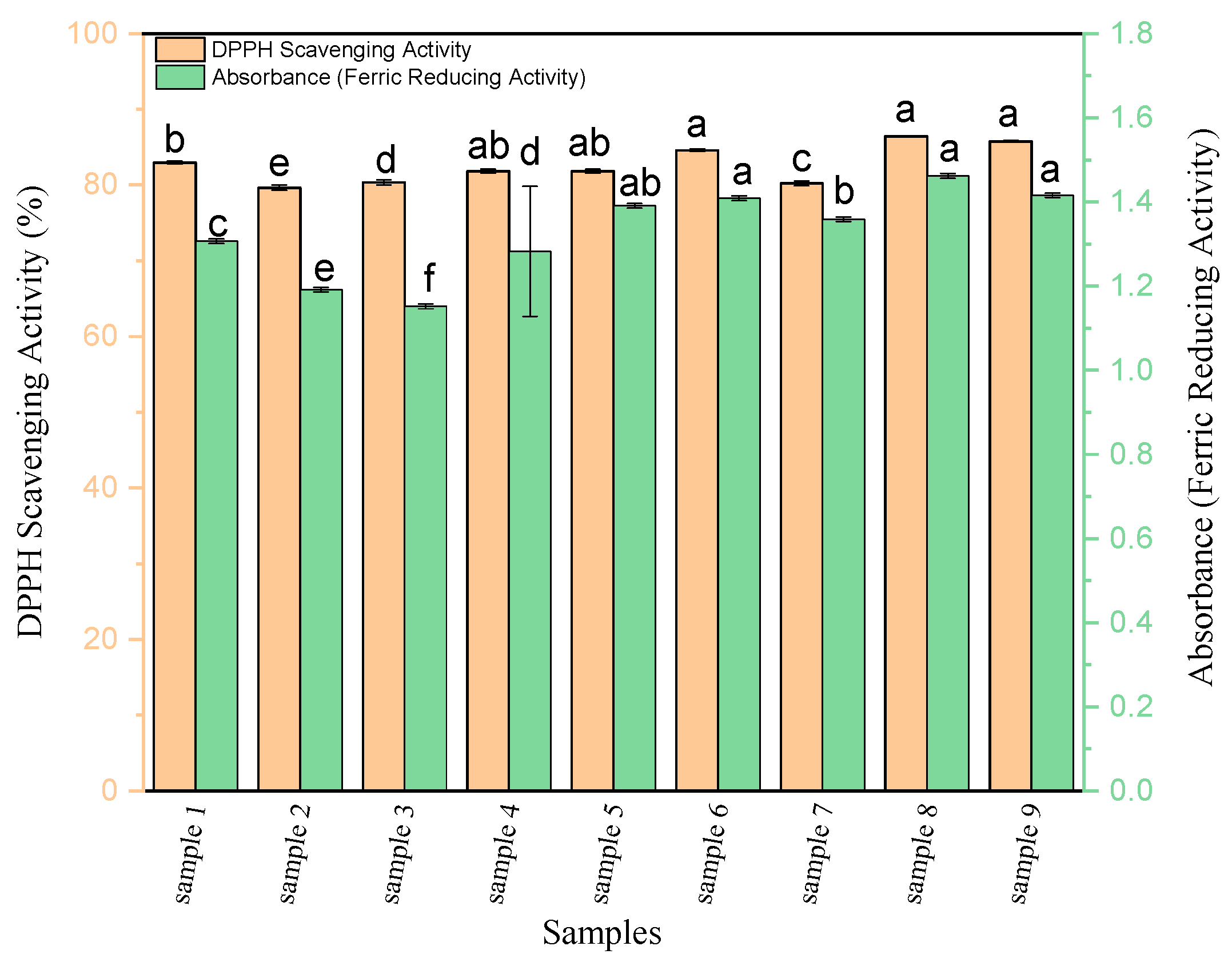
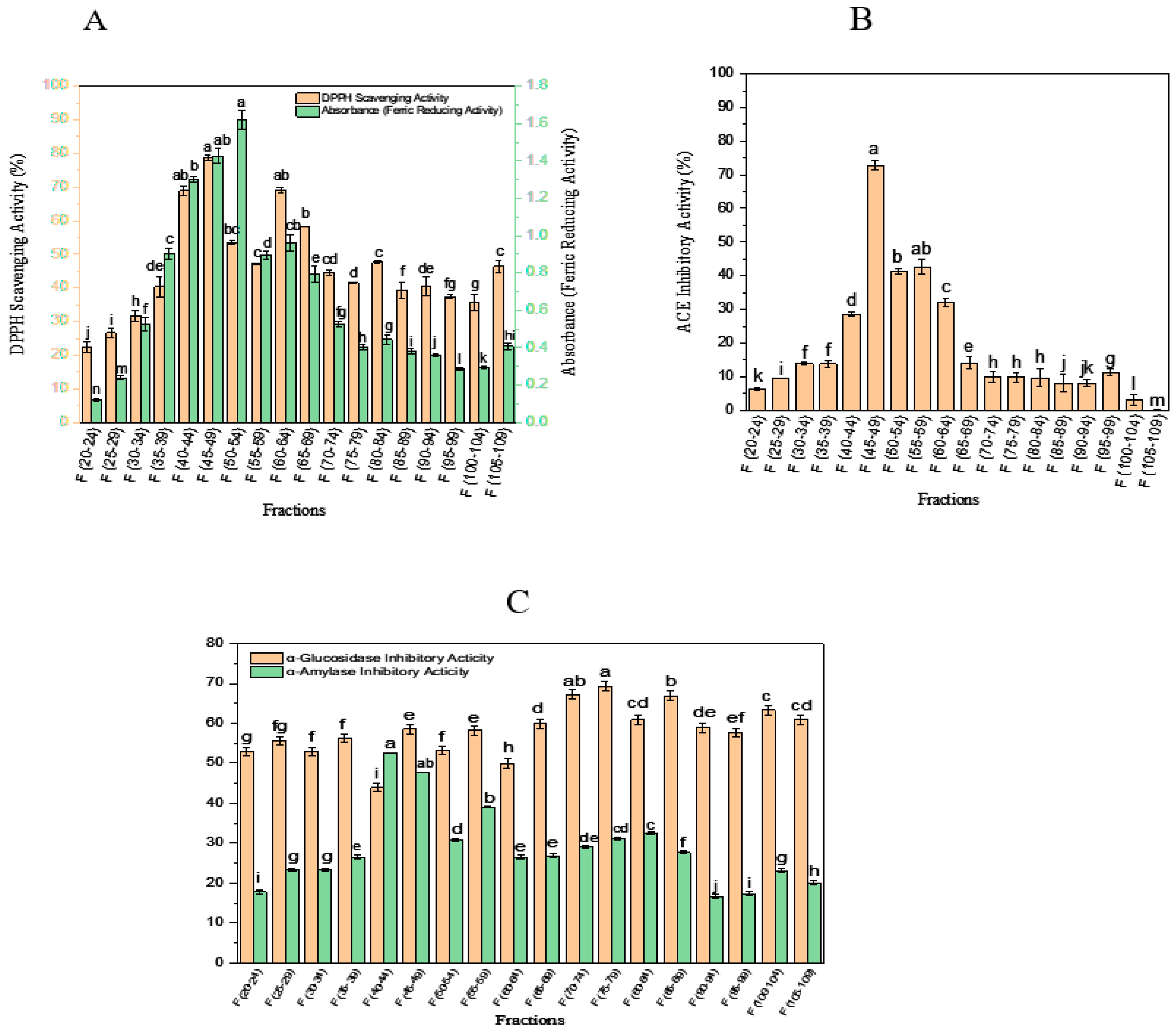
3.1. Antioxidant Activity
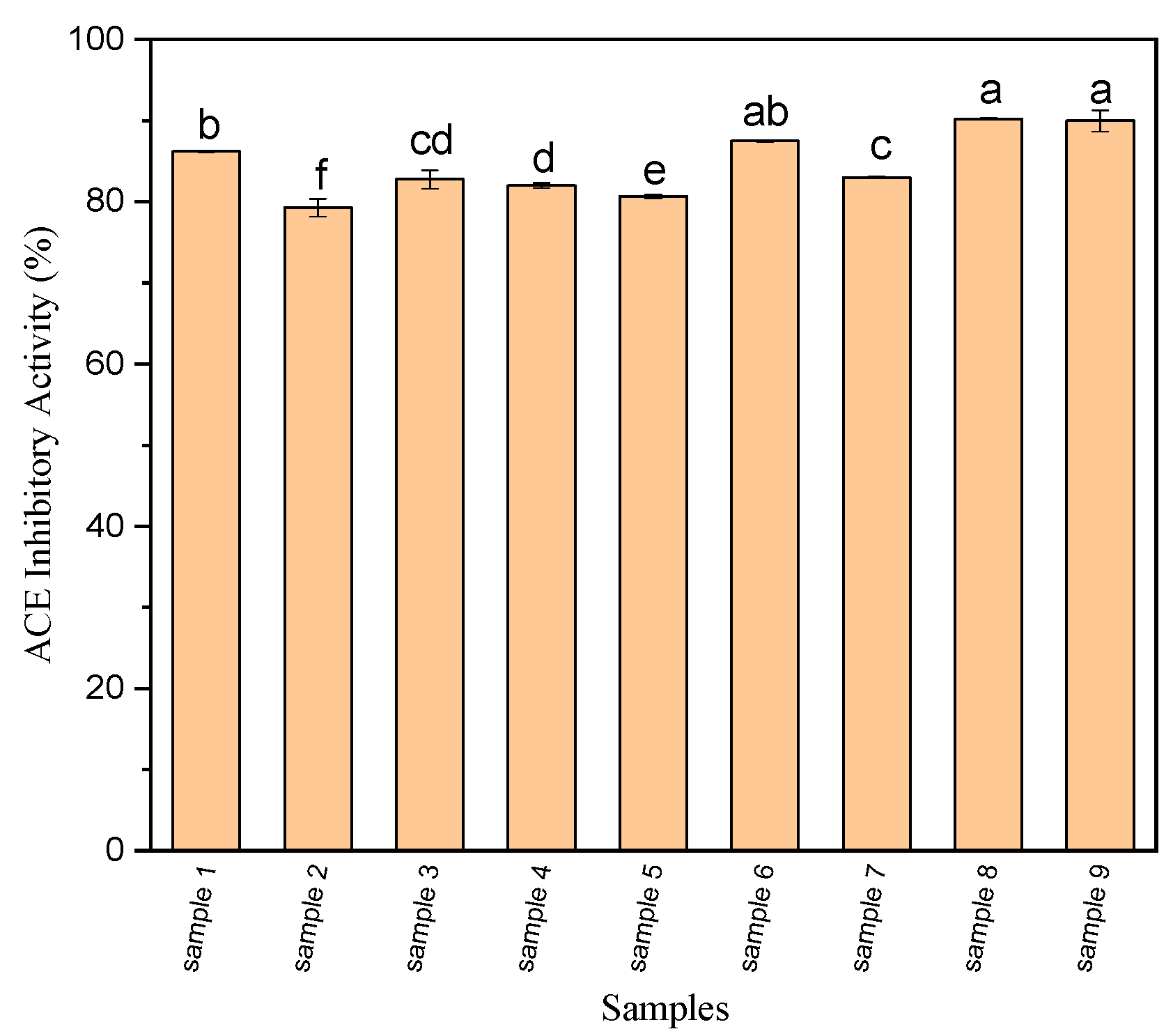
3.2. ACE-Inhibitory Activity
3.3. α-Amylase- and α-Glucosidase-Inhibitory Activity
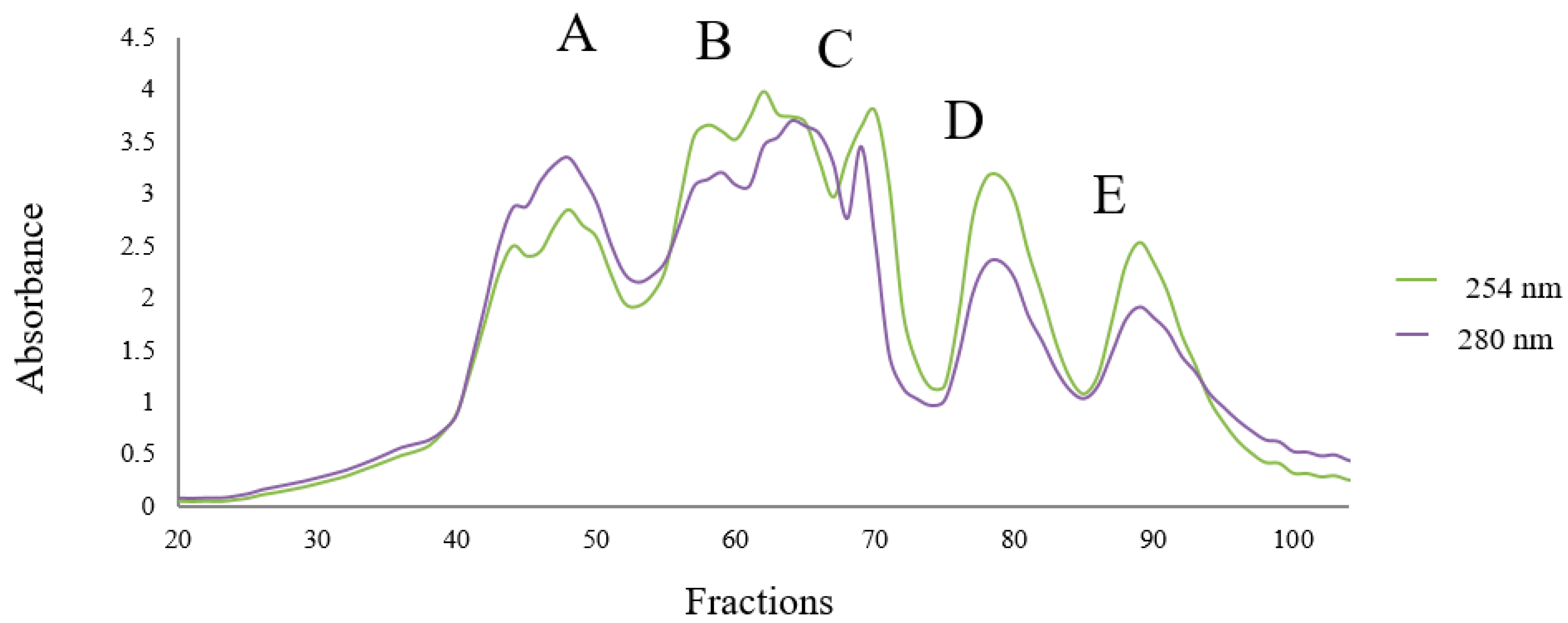
3.4. Fractionation of OSPH by SEC
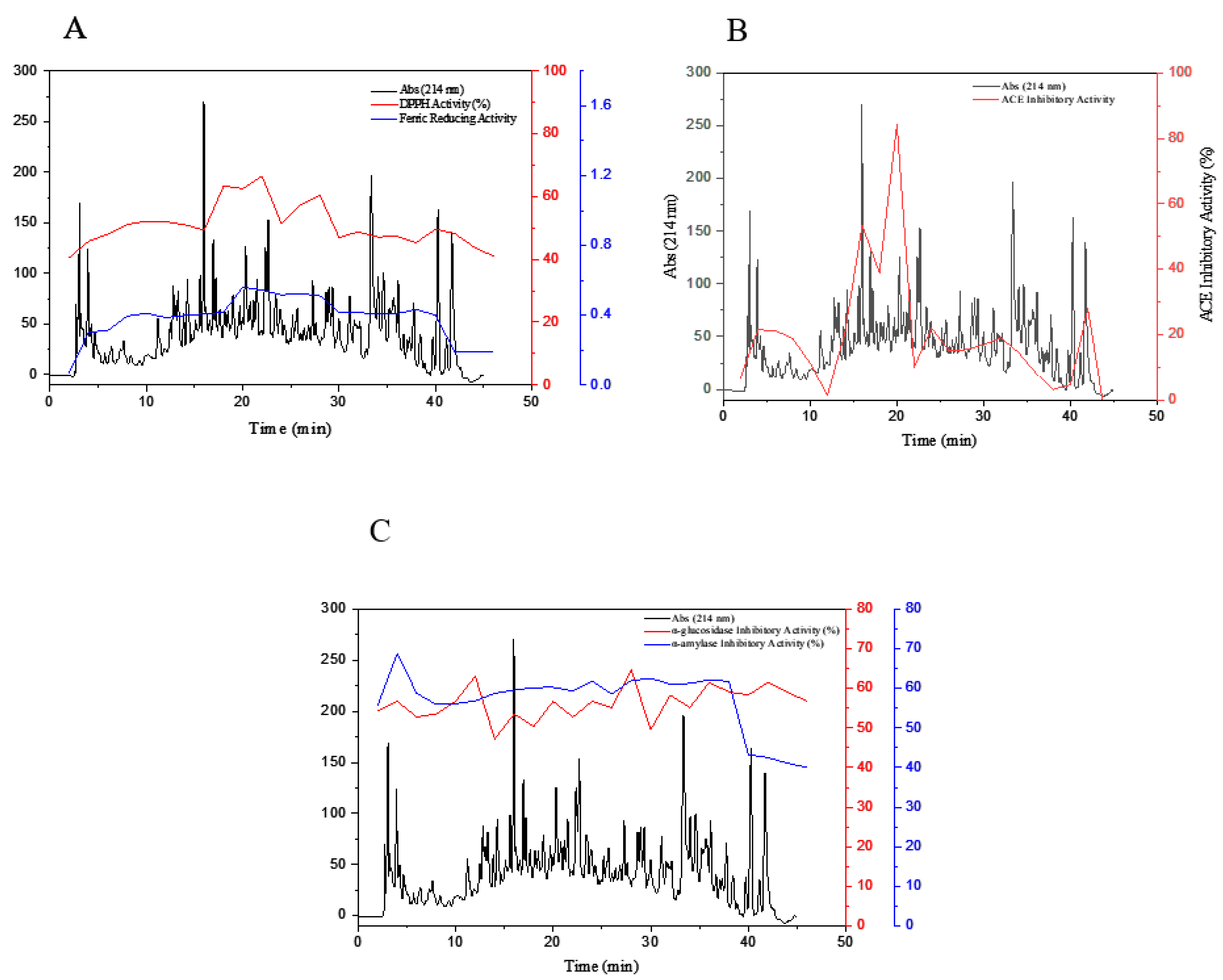
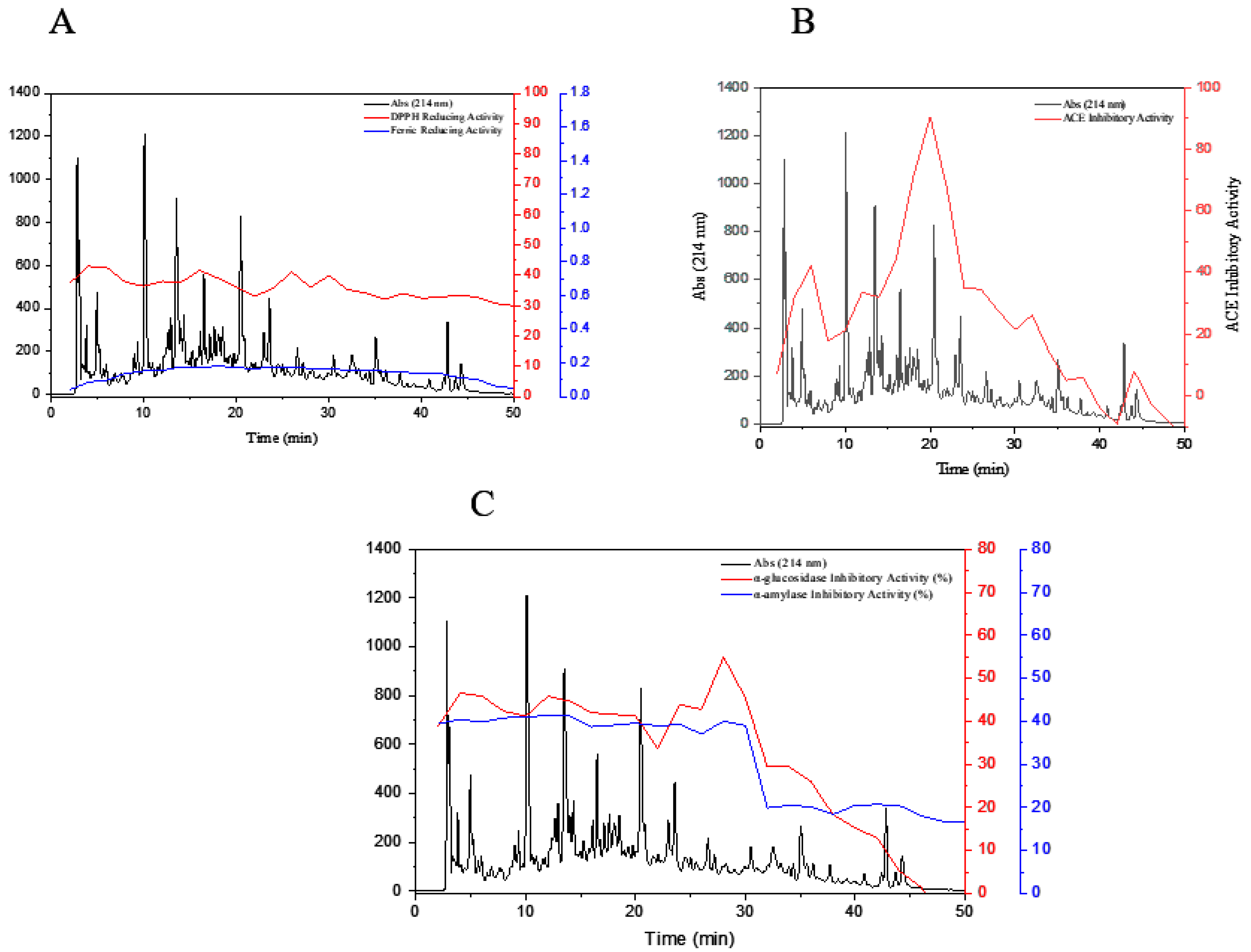
3.5. Purification of Peptide Fractions by RP-HPLC
3.6. Free Amino Acid Composition
3.7. Influence of Simulated In Vitro Gastrointestinal Digestion on the Bioactivity of Peptides
3.8. Identification of Peptide in Most Active HPLC Fractions by MS/MS
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Danquah, M.K.; Agyei, D. Pharmaceutical applications of bioactive peptides. OA Biotechnol. 2012, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Garcia, R.A.; Pyle, D.J.; Piazza, G.J.; Wen, Z. Hydrolysis of animal protein meals for improved utility in non-feed applications. ASABE 2011, 27, 269–275. [Google Scholar]
- Zambrowicz, A.; Timmer, M.; Polanowski, A.; Lubec, G.; Trziszka, T. Manufacturing of peptides exhibiting biological activity. Amino Acids 2013, 44, 315–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taha, S.F.; Mohamed, S.S.; Wagdy, M.S.; Mohamed, F.G. Antioxidant and antimicrobial activities of enzymatic hydrolysis products from sunflower protein isolate. World Appl. Sci. J. 2013, 21, 651–658. [Google Scholar]
- Cheng, X.; Tang, X.; Wang, Q.; Mao, X.Y. Antibacterial effect and hydrophobicity of yak κ-casein hydrolysate and its fractions. Int. Dairy J. 2013, 31, 16–111. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-derived bioactive peptides on inflammation and oxidative stress. Biomed. Res. Int. 2014, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Szwajkowska, M.; Wolanciuk, A.; Barłowska, J.; Król, J.; Litwinczuk, Z. Bovine milk proteins as the source of bioactive peptides influencing the consumers’ immune system—A review. Food Sci. Tech. 2011, 7, 120–125. [Google Scholar]
- Wiriyaphan, C.; Chitsomboon, B.; Yongsawadigul, J. Antioxidant activity of protein hydrolysates derived from threadfin bream surimi byproducts. Food Chem. 2012, 132, 104–111. [Google Scholar] [CrossRef]
- Wouters, A.G.B.; Rombouts, I.; Fierens, E.; Brijs, K.; Delcour, J.A. Relevance of the functional properties of enzymatic plant protein hydrolysates in food systems. CRFSFS 2016, 15, 786–800. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Singh, R.; Rana, S. Bioactive Peptides: A Review. Int. J. Bioautom. 2011, 15, 223–250. [Google Scholar]
- Weiguang, S.; Xiangzhen, K.; Yufei, H.; Xingfei, L.; Caimeng, Z.; Yeming, C. Antioxidant and antibacterial activity and in vitro digestion stability of cottonseed protein hydrolysates. LWT Food Sci. Technol. 2020, 118, 10–18. [Google Scholar]
- Delgado, M.C.O.; Tironi, V.A.; Añón, M.C. Antioxidant activity of amaranth protein or their hydrolysates under simulated gastrointestinal digestion. LWT Food Sci. Technol. 2011, 44, 1752–1760. [Google Scholar] [CrossRef]
- Gul Akıllıoglu, H.G.; Karakaya, S. Effects of heat treatment and in vitro digestion on the Angiotensin converting enzyme inhibitory activity of some legume species. Eur. Food Res. Technol. 2009, 229, 915–921. [Google Scholar] [CrossRef]
- Mazloomi, S.N.; Mahoonak, A.S.; Ghorbani, M.; Houshmand, G. Physicochemical properties of chitosan-coated nanoliposome loaded with orange seed protein hydrolysate. J. Food Eng. 2020, 280, 109976. [Google Scholar] [CrossRef]
- Mohamed, B.; El-Shenawi, M. Functional properties and In-vitro digestibility of bitter orange (Citrus aurantium) seed flour. MRJASSS 2013, 1, 042–047. [Google Scholar]
- Horax, R.; Hettiarachchy, N.; Over, K.; Chen, P.; Gbur, E. Extraction, fractionation and characterization of Bitter Melon seed proteins. J. Agric. Food Chem. 2010, 58, 1892–1897. [Google Scholar] [CrossRef]
- Matsuoka, T.; Kawashima, T.; Nakamura, T.; Kanamaru, Y.; Yabe, T. Isolation and characterization of proteases that hydrolyze royal jelly proteins from queen bee larvae of the honeybee, Apis mellifera. Apidologie 2012, 43, 685–697. [Google Scholar] [CrossRef] [Green Version]
- Kaewka, K.; Therakulkait, C.R.; Cadwallader, K. Effect of preparation conditions on composition and sensory aroma characteristics of acid hydrolyzed rice bran protein concentrate. J. Cereal Sci. 2009, 50, 56–60. [Google Scholar] [CrossRef]
- Bersuder, P.; Hole, M.; Smith, G. Antioxidants from a heated histidine-glucose model system. I: Investigation of the antioxidant role of histidine and isolation of antioxidants by high-performance liquid chromatography. J. Am. Oil Chem. Soc. 1998, 75, 181–187. [Google Scholar] [CrossRef]
- Bougatef, A.; Hajji, M.; Balti, R. Antioxidant and free radical—Scavenging activities of smoth hound muscle protein hydrolysates obtained by gastro intestinal proteases. Food Chem. 2010, 15, 1198–1255. [Google Scholar]
- Sentandreu, M.; Toldra, F. A Fluorescence-based protocol for quantifying angiotensin-converting enzyme activity. Nat. Protoc. 2006, 1, 2423–2427. [Google Scholar] [CrossRef] [PubMed]
- Lassoued, I.; Mora, L.; Barkia, A.; Aristoy, M.C.; Nasri, M.; Toldra, F. Bioactive peptides identified in thornback ray skin’s gelatin hydrolysates by proteases from Bacillus subtilis and Bacillus amyloliquefaciens. J. Proteom. 2015, 128, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Jemil, I.; Abdelhedi, O.; Mora, L.; Nasri, R.; Aristoy, M.-C.; Jridi, M.; Hajji, M.; Toldrá, F.; Nasri, M. Peptidomic analysis of bioactive peptides in zebra blenny (Salaria basilisca) muscle protein hydrolysate exhibiting antimicrobial activity obtained by fermentation with Bacillus mojavensis A21. Process Biochem. 2016, 51, 2186–2197. [Google Scholar] [CrossRef] [Green Version]
- Moayedi, A.; Mora, L.; Aristoy, C.M.; Hashemi, M.; Safari, M.; Toldrá, M. ACE inhibitory and antioxidant activities of peptide fragments obtained from tomato processing by-products fermented usingbacillus subtilis: Effect of amino acid composition and peptides molecular mass distribution. Appl. Biochem. Biotechnol. 2016, 16, 18–35. [Google Scholar]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Brodkorb, A. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Flores, M.; Aristoy, M.C.; Spanier, A.M.; Toldrá, F. Non-volatile components effects on quality of “Serrano” dry-cured ham as related to processing time. Food Sci. 1997, 62, 1235–1239. [Google Scholar] [CrossRef]
- Mora, L.; Escudero, E.; Aristoy, M.C.; Toldrá, F. A peptidomic approach to study the contribution of added casein proteins to the peptide profile in Spanish dryfermented sausages. Int. J. Food Microbiol. 2015, 212, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Mora, L.; Aristoy, M.C.; Toldra, F. Bioactive peptides in foods. Encycl. Food Health 2016, 35, 395–400. [Google Scholar]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Guerar, F.; Guimas, L.; Binet, A. Production of tuna waste hydrolysates by a commercial neutral protease preparation. J. Mol. Catal. 2002, 19, 489–498. [Google Scholar] [CrossRef]
- Jamdar, S.N.; Rajalakshmi, V.; Pednekar, M.D.; Juan, F.; Yardi, V.; Sharma, A. Influence of degree of hydrolysis on functional properties, antioxidant activity and ACE inhibitory activity of peanut protein hydrolysate. Food Chem. 2010, 121, 178–184. [Google Scholar] [CrossRef]
- Je, J.Y.; Lee, M.H.; Lee, K.H.; Ahn, C.B. Antioxidant and hypertensive protein hydrolysates produced from tuna liver by enzymatic hydrolysis. Food Res. 2009, 42, 1266–1272. [Google Scholar] [CrossRef]
- Ren, J.; Zhao, M.; Shi, J.; Wang, J.; Jiang, Y.; Cui, C.; Xue, S.J. Purification and identification of antioxidant peptides from grass carp muscle hydrolysates by consecutive chromatography and electrospray ionization-mass spectrometry. Food Chem. 2008, 108, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Power, O.; Jakeman, P.; FitzGerald, R. Antioxidative peptides: Enzymatic production, in vitro and in vivo antioxidant activity and potential applications of milkderived antioxidative peptides. Amino Acids 2013, 44, 797–820. [Google Scholar] [CrossRef]
- Coscueta, R.F.; Amorim, M.M.; Voss, G.B.; Nerli, B.B.; Picó, G.A.; Pintado, M.A. Bioactive properties of peptides obtained from Argentinian defatted soy flour protein by Corolase PP hydrolysis. Food Chem. 2016, 198, 36–44. [Google Scholar] [CrossRef]
- Ktari, N.; Nasri, R.; Mnafgui, K.; Hamden, K.; Belguith, O.; Boudaouara, T.; Nasri, M. Antioxidative and ACE inhibitory activities of protein hydrolysates from zebra blenny (Salaria basilisca) in alloxan-induced diabetic rats. Process Biochem. 2014, 5113, 73–77. [Google Scholar] [CrossRef]
- Ambigaipalan, P.; Al-Khalifa, A.S.; Shahidi, F. Antioxidant and angiotensin I converting enzyme (ACE) inhibitory activities of date seed protein hydrolysates prepared using Alcalase, Flavourzyme and Thermolysin. J. Funct. Foods 2015, 18, 1125–1137. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Bioactive peptides. J. AOAC Int. 2008, 91, 914–931. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Inhibition of Dipeptidyl Peptidase (DPP)-IV and α-Glucosidase Activities by Pepsin-Treated Whey Proteins. J. Agric. Food Chem. 2013, 61, 7500–7506. [Google Scholar] [CrossRef]
- Siow, H.L.; Lim, T.S.; Gan, C.Y. Development of a workflow for screening and identification of a-amylase inhibitory peptides from food source using an integrated Bioinformatics-phage display approach: Case study—Cumin seed. Food Chem. 2017, 214, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Meinert, L.; de Lichtenberg Broge, E.H.; Bejerholm, C.; Jensen, K. Application of hydrolyzed proteins of animal origin in processed meat. Food Sci. Nutr. 2015, 4, 290–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selamassakul, O.; Laohakunjit, N.; Kerdchoechuen, O.; Yang, L.; Maier, C. Isolation and characterisation of antioxidative peptides from bromelain-hydrolysed brown rice protein by proteomic technique. Process Biochem. 2018, 17, 359–391. [Google Scholar] [CrossRef] [PubMed]
- Soleymanzadeh, N.; Mirdamadi, S.; Mirzaei, M.; Kianirad, M. Novel β-casein derived antioxidant and ACE-inhibitory active peptide from camel milk fermented by Leuconostoc lactis PTCC1899: Identification and molecular docking. Int. Dairy J. 2019, 19, 12–46. [Google Scholar] [CrossRef]
- Jonker, J.T.; Wijngaarden, M.A.; Kloek, J.; Groeneveld, Y.; Gerhardt, C.; Brand, R.; Kies, A.K.; Romijn, J.A.; Smit, J.W.A. Effects of low doses of casein hydrolysate on postchallenge glucose and insulin levels. Eur. J. Intern. Med. 2011, 22, 245–248. [Google Scholar] [CrossRef]
- Yang, H.J.; Kwon, D.Y.; Kim, M.J.; Kang, S.; Park, S. Meju, unsalted soybeans fermented with Bacillus subtilis and Aspergilus oryzae, potentiates insulinotropic actions and improves hepatic insulin sensitivity in diabetic rats. Nutr. J. 2012, 9, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.M.; Fonseca, R.A.D.S.; Prentice, C. Comparing the hydrolysis degree of industrialization byproducts of Withemouth croaker (Micropogonias furnieri) using microbial enzymes. Int. Food Res. J. 2014, 21, 1757–1761. [Google Scholar]
- Oveisipour, M.; Abedian, A.M.; Motamedzadegan, A.; Rasco, B.; Safari, R.; Shahiri, H. The effect of enzymatic hydrolysis time and temperature on the properties of protein hydrolysates from the Persian sturgeon (Acipenser persicus) viscera. Food Chem. 2009, 115, 238–242. [Google Scholar] [CrossRef]
- Maqsoudlou, A.; Sadeghi Mahoonak, A.; Mora, L.; Mohebodini, H.; Toldrá, F.; Ghorbani, M. Peptide identification in alcalase hydrolysated pollen and comparison of its bioactivity with royal jelly. Food Res. Int. 2018, 88, 963–974. [Google Scholar] [CrossRef]
- Kim, S.Y.; Je, J.Y.; Kim, S.K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J. Nutr. Biochem. 2007, 18, 31–38. [Google Scholar] [CrossRef]
- Yang, B.; Yang, H.; Li, J.; Li, Z.; Jiang, Y. Amino acid composition, molecular weight distribution and antioxidant activity of protein hydrolysates of soy sauce lees. Food Chem. 2010, 124, 551–555. [Google Scholar] [CrossRef]
- Ruiz, J.; Dávila-Ortíz, G.; Chel-Guerrero, L.; Betancur-Ancona, D. Angiotensin iconverting enzyme inhibitory and antioxidant peptide fractions from hard-to-cook bean enzymatic hydrolysates. J. Food Biochem. 2013, 37, 26–35. [Google Scholar] [CrossRef]
- Lassoued, I.; Mora, L.; Barkia, A.; Aristoy, M.C.; Nasri, M.; Toldrá, F. Angiotensin I-converting enzyme inhibitory peptides FQPSF and LKYPI identified in Bacillus subtilisA26 hydrolysate of thornback ray muscle. Int. J. Food Sci. Technol. 2016, 51, 1604–1615. [Google Scholar] [CrossRef]
- Meshginfar, N.; Sadeghi Mahoonak, A.; Hosseinian, F.; Ghorbani, M.; Tsopmo, A. Production of antioxidant peptide fractions from a by-product of tomato processing: Mass spectrometry identification of peptides and stability to gastrointestinal digestion. Int. J. Food Sci. Technol. 2018, 18, 274–287. [Google Scholar] [CrossRef]
- Torkova, A.; Kononikhin, A.; Bugrova, A.; Khotchenkov, V.; Tsentalovich, M.; Medvedeva, U. Effect of in vitro gastrointestinal digestion on bioactivity of poultry protein hydrolysate. CRNFSJ 2016, 4, 77–86. [Google Scholar]
- Salema, R.B.; Ktaria, N.; Bkhairiaa, I.; Nasria, R.; Morab, L.; Kallelc, R.; Hamdid, S.; Jamoussid, K.; Boudaouarac, T.; El-Fekie, A.; et al. In vitro and in vivo anti-diabetic and anti-hyperlipidemic effects of protein hydrolysates from Octopus vulgaris in alloxanic rats. Food Res. 2018, 106, 952–963. [Google Scholar]
- Liu, R.; Zheng, W.; Li, J.; Wang, L.; Wu, H.; Wang, X.; Shi, L. Rapid identification of bioactive peptides with antioxidant activity from the enzymatic hydrolysate of Mactra veneriformis by UHPLC–Q-TOF mass spectrometry. Food Chem. 2014, 167, 484–489. [Google Scholar] [CrossRef]

| Amino Acid Name | Concentration (mg/g of OSPH) ± SD |
|---|---|
| Asp | 0.1276 |
| Glu | 0.1434 |
| Ser | 0.1087 |
| Asn | 0.13 ± 0.0001 |
| Gly | 0.0742 |
| Gln | 0.1278 ± 0.0001 |
| β Ala | 0.0885 ± 0.0001 |
| His | 0.1616 ± 0.0001 |
| Thr | 0.1172 |
| Ala | 0.0871 ± 0.0003 |
| Arg | 0.1682 |
| Pro | 0.1103 |
| Tyr | 0.1665 |
| Val | 0.1158 |
| Met | 0.1439 |
| Ile | 0.1245 |
| Leu | 0.1305 |
| Phe | 0.153 |
| Trp | 0.1637 |
| Orn | 0.0812 |
| Lys | 0.1417 |
| Total | 2.6657 ± 0.001 |
| Sequence | Protein | Bioactivity (Sequence Previously Described as Responsible for the Bioactivity) | MW |
|---|---|---|---|
| ATREQRQQQQRFQTQ | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Inhibition of prolyl endopeptidase (IHPFAQTQ) | 1933.3 |
| EQEQEFQGSGD | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | ACE-inhibitory (RKSGDPLGR) | 1253.36 |
| HNINDPSGA KHNINDPSGA | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Antimicrobial activities (GYVSGAVIEIPDEILDSAR) | 924.06 1052.51 |
| HNINDPSGAD KHNINDPSGAD | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Celiac toxic (GWFGGADWHA) | 1039.16 1166.53 |
| HNINDPSGADA | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Kinases inhibitor, (KKALRRQEAADAL) Antioxidative (LLPHHADADY) Antithrombotic, Antimicrobial (DNIADAVACAKRVVRDPQGIR) | 1110.25 |
| PSGADAYNPR | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Antioxidative (PRHVFYRWFLSNPRI) | 1052.25 |
| QESQQRSSESQSRSQDQHQKVR | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Antimicrobial (TKCFQWQRNMRKVRGPPVSCIKR) | 1167.35 |
| QQQQRFQTQ TREQRQQQQRFQTQ | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | ACE inhibitor (QTQSLVYP) | 1047.21 1903.93 |
| SSESQSRSQDQHQKVR FQSSKSQDQHQKVR | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Antimicrobial (AGRGKQGGKVRAKAKTRSSRA) ACE inhibitor (KVREGTTY) | 2644.05 1701.84 |
| LRHNIDKPSHAD | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Antimicrobial (LLPHHADADY) | 1191.41 |
| QDSQQQQSFQSS | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Neuropeptide (FSEFMRQYLVLSMQSSQ) | 1887.18 |
| SFQSSKSQDQHQKVR | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | ACE inhibitor (KVREGTTY) | 1862.21 |
| SQGGRSQGSQGSDDRRAGN SQGSQGSDDRRAGN | Citrin OS = Citrus sinensis Q39627_CITSI Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | VLKMAGNSFQEN (Celiac Toxic coeliac toxic peptide) YLAGNQ, EVMAGNYLPG, EVMAGNLYPG (ACE inhibitor) | 1918.83 1433.59 |
| SQSQGGRSQGSQGSDDRRAGNL GSQGSDDGRGGNL SQGGRSQGSQGSDDGRGGNL SQSQGGRSQGSQGSDDGRGGNL | Citrin OS = Citrus sinensis Q39627_CITSI | Antimicrobial (GLFDAIGNLLGGLGLG) Antiamnestic (PEP inhibitor) (RYDWWPYGNLFGGHTFISP) | 2247.01 1218.51 1918.83 2133.91 |
| VFPGCAETFQDSQQQQSFQSSKSQDQHQKVR | Uncharacterized protein OS = Penicillium digitatum tr|K9FZ18|K9FZ18_PEND1 | Antimicrobial (AGRGKQGGKVRAKAKTRSSRAGLQFPV) ACE inhibitor (KVREGTTY) | 3549.70 |
| VTDITREGKQQ | Citrin OS = Citrus sinensis tr|Q39627|Q39627_CITSI | Regulating (QQQKQQQQPSSQVS) | 1790.14 |
| GTQDHPHDDYAE | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Anti-inflammatory peptide (KGHYAERVG) ACE inhibitor (YAEERYPIL) | 1920.17 |
| GTQDHPHDDYAEAK | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Kinases inhibitor (VTCDILSVEAKGVKLG) | 1434.59 |
| AGDTHLGGED | Uncharacterized protein OS = Penicillium digitatum tr|K9FZ18|K9FZ18_PEND1 | Antithrombotic (EAGEDCDCGSPANPCCDAATCKLIPGAQCGEGLCCDQCSFIEEGTVCRIARGDDLDDYCNGRSAGCPRNPFH) | 2135.41 |
| DQGNRITPS | Uncharacterized protein OS = Penicillium digitatum tr|K9FZ18|K9FZ18_PEND1 | Antimicrobial (GIFSSRKCKTPSKTFKGICTRDSNCDTSCRYEGYPAGDCKGIRRRCMCSKPC) Neuropeptide (NKLASVYALTPSLRVG) TPSPR (Alpha-glucosidase inhibitor) | 2248.59 |
| SEGTEPIQSK GQKDAYVGDEAQSK | Protein disulfide-isomerase OS = Citrus limon tr|V9HXG3|V9HXG3_CITLI | Antimicrobial (FVPYNPPRPGQSKPFPSFPGHGPFNPKIQWPYPLPNPGH) | 3585.3 |
| DYDKPVQQ | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Coeliac toxic peptide (QQPFVQQQQPFVQQ) | 1219.38 |
| GADDSADNKSSNAPTRTY | Uncharacterized protein OS = Citrus clementina tr|V4UJF9|V4UJF9_9ROSI | Erythropoietin receptor agonist peptide (YQRRPAIAINNPYVPRTYYANPAVVRPHAQIPQRQYLPNSHPPTVVRRPNLHPSF) | 1920.2 |
| GETGGPHPGYETR | Uncharacterized protein OS = Citrus clementina tr|V4SQX0|V4SQX0_9ROSI | Alpha amylase inhibitory (DETRL) | 2135.44 |
| IVPRKAASSEE | Uncharacterized protein OS = Citrus clementina tr|V4S6W0|V4S6W0_9ROSI | Antioxidative (RELEELNVPGEIVESLSSSEESITR) | 1274.56 |
| EVHNPATGE | Succinate-semialdehyde dehydrogenase OS = Citrus clementina tr|V4RMN0|V4RMN0_9ROSI | ACE inhibitor (VLSPPFTGE) | 1384.5 |
| QDQGPMVK | Uncharacterized protein OS = Citrus unshiu tr|A0A2H5NKL3|A0A2H5NKL3_CITUN | Neuropeptide (SPTISITAPIDVLRKTWEQERARKQMVKNREFLNSLN) Membrane-active peptides (MVKSKIGSWILVLFVAMWSDVGLCKKRPKP) | 1583.78 |
| GQMNEPPGAR | ATP synthase subunit beta (Fragment) OS =Poncirus trifoliata tr|Q9THW1|Q9THW1_PONTR | Antimicrobial, (GICACRRRFCPNSERFSGYCRVNGARYVRCCSRR)Opioid, (GICACRRRFCPNSERFSGYCRVNGARYVRCCSRR, YGGFTGARKSARKLANQ, FGGFTGARKSA) Neuropeptide (YGGFLGARKSARKLANQ) Antioxidative (QGAR) | 971.09 |
| FYLGGNPQPQLQ | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Neuropeptide (MPRVRSLFQEQEEPEPGMEEAGEMEQKQLQ) coeliac toxic peptide (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) | 987.16 |
| QQQRFQTQ | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | ACE inhibitor (QTQSLVYP) | 1075.28 |
| GSAKESGDKAEQGS | ABC transporter solute-binding protein OS = Streptomyces sp. tr|A0A3L8QSZ7|A0A3L8QSZ7_9ACTN | coeliac toxic peptide, (QQQQPSSQVSFQQPLQQYPLGQGSFRPSQQNPQA, QGSFRPSQQNPQAQ, QYPLGQGSFRPS, LGQGSFRPSQQN) ACE inhibitor, (YQGS) Antioxidative (AWEEREQGSR) | 992.16 |
| SQSRSQDQHQKVRQIRE | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Neuropeptide (SENFTPWAYIILNGEAPIIREVHYSPRL) | 1870.1 |
| HNIDKPSHAD | Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | Antioxidative (LLPHHADADY) | 1495.76 |
| QSFQSSKSQDQHQKVR | Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | ACE inhibitor (KVREGTTY, KVREGT) | 1357.59 |
| QDSQQQQSFQS | Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | Antimicrobial (GLLSRLRDFLSDRGRRLGEKIERIGQKIKDLSEFFQS) | 1186.47 |
| QPAGKRGEGPPKRAGQVR | REVERSED Uncharacterized protein OS = Curtobacterium sp. RRRRRtr|A0A1E5MMN3|A0A1E5MMN3_9MICO | Celiac toxic (VQGQGHQPPQQPAQL, VQGQGIIQPQQPAQL) | 953.1 |
| QKVESEAGVT | Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | Antioxidative (DSGVT) | 902.15 |
| VIGTVLAFALIASAESLF | Sulfate transporter OS = Streptomyces sp. tr|A0A3L8QBZ9|A0A3L8QBZ9_9ACTN | Antimicrobial (SLFSLIKAGAKFLGKNLLKQGAQYAACKVSKECG, SLFSLIKAGAKFLGKNLLKQGACYAACKASKQC) Neuropeptide (MPRVRSLFQEQEEPEPGMEEAGEMEQKQLQ) | 1056.3 |
| TAAEEVLQKAEAAP | CHAD domain-containing protein OS = Methylobacterium mesophilicum tr|M7YUM6|M7YUM6_9RHIZ | Antimicrobial (WNPFKELERAGQRVRDAVISAAPAVATVGQAAAIARG) | 1361.71 |
| SAEKGNLYQN | Uncharacterized protein OS = Citrus clementina tr|V4S993|V4S993_9ROSI | Regulating (EPFYQNVPD) | 1063.26 |
| NALEPR | Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | Peptide stimulating insulin release Antioxidative (GFGSFLGKALKA ALKIGANALGGSPQQ) | 1350.55 |
| QNPANPVTLAQAIEGEPK | DNA-directed DNA polymerase OS = Methylobacterium mesophilicum tr|M7Y0I8|M7Y0I8_9RHIZ | ACE Inhibitor (EPKAIP, HSGIQSEPKAIP)Antioxidative (APIRMWYMYRKLTDMEPKPVA) | 2110.53 |
| SSDTIDNVKAK | Polyubiquitin OS = Avena fatua UBIQP_AVEFA | Antimicrobial (GCASRCKAKCAGRRCJGWASASFRGRCYCKCFRC) | 1133.32 |
| QSDVTTTS | Transcription factor MYB21 OS = Arabidopsis thaliana MYB21_ARATH | Antimicrobial (QLKTADLPAGRDETTSFVLV) Antioxidative (PIAAEVYEHTEGSTTSY) | 1918.29 |
| DESTGTIGKR | Fructose-bisphosphate aldolase, cytoplasmic isozyme 1 OS = Pisum sativum ALF1_PEA | Neuropeptide (KRQHPGKR) | 1310.46 |
| SAPKKEKSQGFLQ | Type II inositol polyphosphate 5-phosphatase 14 OS = Arabidopsis thaliana IP5PE_ARATH | ACE Inhibitor, DPP IV inhibitor (TPVVVPPFLQP, FLQP) | 1219.35 |
| FASTRRRQCPQ | Lignin-forming anionic peroxidase OS = Nicotiana sylvestris PERX_NICSY | Antimicrobial (CPQLQPQNPSQQQPQEQG) | 1889.41 |
| GSGAKPGTKPTKCT | Chaperone protein dnaJ A6, chloroplastic OS = Arabidopsis thaliana DNJA6_ARATH | Antimicrobial (RSVCRQIKICRRRGGCYYKCTNRPY) | 1047.27 |
| TNADKATTVS | Soluble starch synthase 3, chloroplastic/amyloplastic OS = Solanum tuberosum SSY3_SOLTU | Antimicrobial (GILDTLKQFAKGVGKDLVKGAAQGVLSTVSCKLAKTC, GIFSSRKCKTVSKTFRGICTRNANC) | 1822.44 |
| WTAEHSVNAALGQFE | Pyruvate, phosphate dikinase regulatory protein, chloroplastic OS = Zea mays PDRP1_MAIZE | Celiac Toxic (QQLPQFEEIRNL, GSVQPQQQLPQFEIR) | 1427.76 |
| GSEEPNVEEDS | FHA domain-containing protein DDL OS = Arabidopsis thaliana DDL_ARATH | Antimicrobial (YPGPQAKEDSEGPSQGPASREK) | 1123.33 |
| Sequence | Protein | Bioactivity | MW |
|---|---|---|---|
| SQGSQGSDDGRGGNL | Citrin OS = Citrus sinensis Q39627_CITSI | PEP inhibitor (Antiamnestic), (RYDWWPYGNLFGGHTFISP) Antibacterial, (LLPIVGNLLKSLL, GLLDMVTGLLGNLG, GLFDAIGNLLGGLGLG)Antioxidative, (QLGNLGV)ACE inhibitor (EVMAGNLYPG) | 1219.38 |
| GSQGSDDGRGGNL | Citrin OS = Citrus sinensis Q39627_CITSI | PEP inhibitor (Antiamnestic), (RYDWWPYGNLFGGHTFISP)Antibacterial, (LLPIVGNLLKSLL, GLLDMVTGLLGNLG, GLFDAIGNLLGGLGLG) Antioxidative, (QLGNLGV)ACE inhibitor (EVMAGNLYPG) | 1434.62 |
| GSQGSDDGRGGNL | Citrin OS = Citrus sinensis Q39627_CITSI | PEP inhibitor (Antiamnestic), (RYDWWPYGNLFGGHTFISP)Antibacterial, (LLPIVGNLLKSLL, GLLDMVTGLLGNLG, GLFDAIGNLLGGLGLG)Antioxidative, (QLGNLGV)ACE inhibitor (EVMAGNLYPG) | 1219.38 |
| SQGSQGSDDRRAGN | Uncharacterized protein OS = Citrus clementina tr|V4U2L8|V4U2L8_9ROSI | Celiac Toxic, (VLKMAGNSFQEN) ACE inhibitor (YLAGNQ, EVMAGNYLPG, EVMAGNLYPG) | 1434.59 |
| IDLPQPQ | Serine/threonine-protein kinase TOR OS = Arabidopsis TOR_ARATH | ACE inhibitor, (QPQPLIYP) Potential coeliac toxic peptide, (PQNPSQQQPQEQVP) Celiac Toxic, (SQPQAFP) Antibacterial (PYPQPQPF) | 1219.38 |
| RLAIEEAISITTTLVAQY | Protein RETICULATA-RELATED 5, chloroplastic OS = Arabidopsis thaliana RER5_ARATH | Antibacterial, (NLLKQGAQYAACKVSKECG) ACE inhibitor (VLAQYK) | 1434.59 |
| SSCPVINVD | Uncharacterized GPI-anchored protein At1g61900 OS = Arabidopsis thaliana UGPI6_ARATH | Antibacterial, (CSCRTSSCRFGERL) Natriuretic (RSSCFGGRIDRIGAC) | 810.02 |
| RHSWMMN | Phosphoenolpyruvate carboxylase kinase 2 OS = Arabidopsis thaliana PPCK2_ARATH | Antibacterial, (VPMPKGRSSRGRRHS) Antioxidative (RHS) | 1992.57 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazloomi, S.N.; Mora, L.; Aristoy, M.-C.; Mahoonak, A.S.; Ghorbani, M.; Houshmand, G.; Toldrá, F. Impact of Simulated Gastrointestinal Digestion on the Biological Activity of an Alcalase Hydrolysate of Orange Seed (Siavaraze, Citrus sinensis) by-Products. Foods 2020, 9, 1217. https://doi.org/10.3390/foods9091217
Mazloomi SN, Mora L, Aristoy M-C, Mahoonak AS, Ghorbani M, Houshmand G, Toldrá F. Impact of Simulated Gastrointestinal Digestion on the Biological Activity of an Alcalase Hydrolysate of Orange Seed (Siavaraze, Citrus sinensis) by-Products. Foods. 2020; 9(9):1217. https://doi.org/10.3390/foods9091217
Chicago/Turabian StyleMazloomi, Seyadeh Narges, Leticia Mora, M-Concepción Aristoy, Alireza Sadeghi Mahoonak, Mohammad Ghorbani, Gholamreza Houshmand, and Fidel Toldrá. 2020. "Impact of Simulated Gastrointestinal Digestion on the Biological Activity of an Alcalase Hydrolysate of Orange Seed (Siavaraze, Citrus sinensis) by-Products" Foods 9, no. 9: 1217. https://doi.org/10.3390/foods9091217