Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters
Abstract
1. Introduction
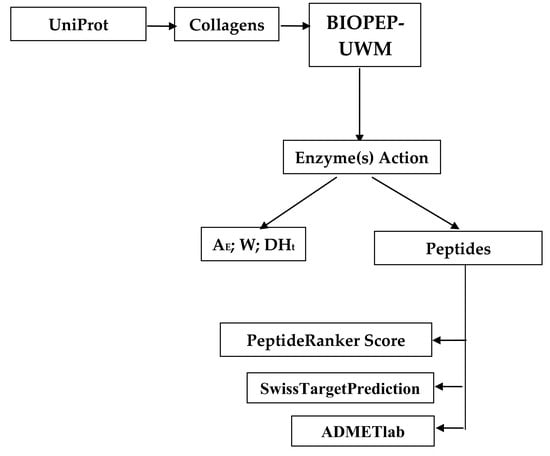
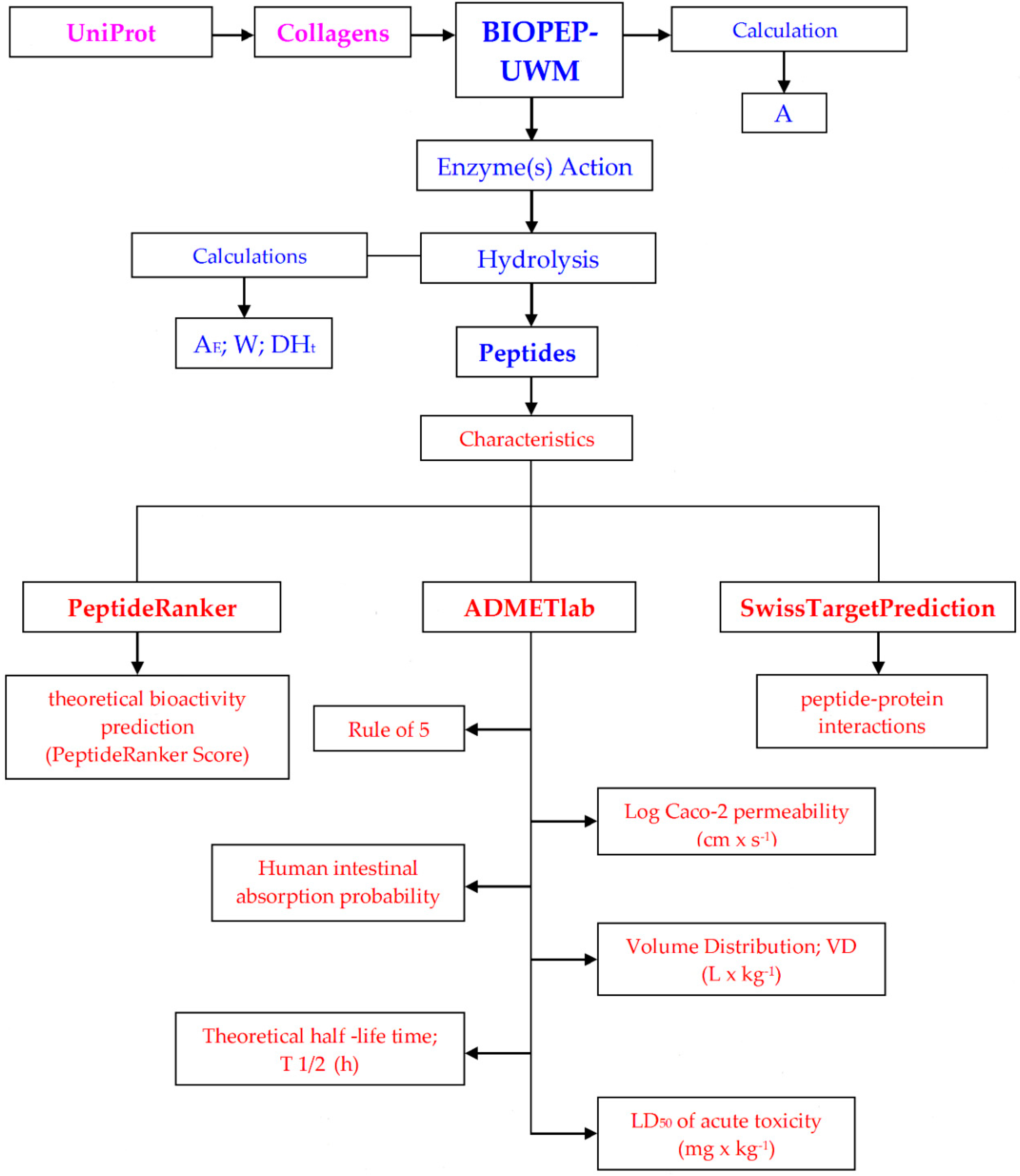
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACE | angiotensin converting enzyme (EC 3.4.15.1) |
| ACEi | angiotensin converting enzyme inhibitor |
| ADMET | absorption, distribution, metabolism, excretion, toxicity |
| BIOPEP-UWM | database of protein and bioactive peptide sequences (http://www.uwm.edu.pl/biochemia) [22] |
| A | depending on the context: alanine or the frequency of the occurrence of bioactive fragments in a protein sequence [22] described by the following equation: A = a/N where a—the number of fragments with a given activity, N—the number of amino acid residues in a protein |
| AE | The frequency of release of fragments with a given activity by selected enzymes [22,23] described by the following equation: AE = d/N where d-the number of peptides with a given activity (e.g., ACE inhibitors) released by a given enzyme (e.g., trypsin) N-the number of amino acid residues in a protein |
| B | stem bromelain (bromelain) (EC 3.4.22.32) |
| CaMPDE | calmodulin-dependent cyclic nucleotide phosphodiesterase (EC 3.1.4.17) |
| CH | collagen hydrolysate |
| Ch | chymotrypsin (EC 3.4.21.1) |
| ChEMBL | ChEMBL database of molecules with drug-like properties (https://www.ebi.ac.uk/chembl) [66] |
| CoA | coenzyme A |
| Complement factor B | alternative-complement-pathway C3/C5 convertase (EC 3.4.21.47) |
| COX-2 | cyclooxygenase-2 (prostaglandin-endoperoxide synthase; EC 1.14.99.1) |
| DHt | theoretical degree of hydrolysis (%) [22] described by the following equation: DHt = (d/D) × 100% where d—the number of hydrolyzed peptide bonds in a protein/peptide chain D—the total number of peptide bonds in a protein/peptide chain |
| DPP-III | dipeptidyl peptidase III (EC 3.4.14.4) |
| DPP-IV | dipeptidyl peptidase IV (EC 3.4.14.5) |
| DPP-IVi | dipeptidyl peptidase IV inhibitor |
| F | depending on the context: phenylalanine or ficin (EC 3.4.22.3) |
| G | glycine |
| HMG-CoA | 3-hydroxy-3-methyl-glutaryl-CoA reductase (EC 1.1.1.34) |
| HT | high throughput technology |
| Hyp | hydroxyl-proline/hydroxyl-lysine |
| IC50 | concentration of a peptide corresponding to its half-inhibitory effect (μM) |
| IUBMB | International Union of Biochemistry and Molecular Biology |
| LD50 | dose of a compound which kills 50% tested animals (mg × kg −1) |
| P | proline |
| Pap | papain (EC 3.4.22.2) |
| Pep | pepsin (EC 3.4.23.1) |
| QSAR | Quantitative Structure-Activity Relationship [19] |
| SMILES | Simplified Molecular Input Line Entry Specification [25] |
| T | depending on the context: treonine or trypsin (EC 3.4.21.4) |
| T1/2 | theoretical half-life time (h) |
| VD | volume distribution (L × kg −1) |
| W | depending on the context: tryptophan or the relative frequency of release of fragments with a given activity by selected enzymes [22] described by the following equation: W = AE/A where AE—the frequency of release of fragments with a given activity by selected enzymes (see above) A—the frequency of bioactive fragments occurrence in a protein sequence (see above). |
References
- Song, H.; Li, B. Beneficial Effects of Collagen Hydrolysate: A Review on Recent Developments. Biomed. J. Sci. Tech. Res. 2017, 1, 458–461. [Google Scholar] [CrossRef]
- Zdzieblik, D.; Oesser, S.; Baumstark, M.W.; Gollhoffer, M.; König, D. Collagen peptide supplementation in combination with resistance training improves body composition and increases muscle strength in elderly sarcopenic men: A randomised controlled trial. Br. J. Nutr. 2015, 114, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Offengenden, M.; Chakrabarti, S.; Wu, J. Chicken collagen hydrolysates differentially mediate anti-inflammatory activity and type I collagen synthesis on human dermal fibroblasts. Food Sci. Hum. Wellness 2018, 7, 138–147. [Google Scholar] [CrossRef]
- Raman, M.; Gopakumar, K. Fish Collagen and its Applications in Food and Pharmaceutical Industry: A Review. EC Nutr. 2018, 13, 752–767. [Google Scholar]
- Gómez-Guillén, M.C.; Giménez, B.; López-Caballero, M.E.; Montero, M.P. Functional and bioactive properties of collagen and gelatin from alternative sources: A review. Food Hydrocoll. 2011, 8, 1813–1827. [Google Scholar] [CrossRef]
- Sylvipriya, K.S.; Kumar, K.K.; Bhat, A.R.; Kumar, B.D.; John, A.; Iakshmanan, P. Collagen: Animal sources and biomedical application. J. Appl. Pharm. Sci. 2015, 5, 123–127. [Google Scholar] [CrossRef]
- León-López, A.; Vargas-Torres, A.; Zeugolis, D.I.; Aguirre-Álvarez, G. Hydrolyzed Collagen-Sources and Applications. Molecules 2019, 24, 4031. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive peptides from muscle sources: Meat and fish. Nutrients 2011, 3, 765. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein-originating peptides as tastants–Physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Darewicz, M.; Minkiewicz, P. Peptides Derived from Foods as Supportive Diet Components in the Prevention of the Metabolic Syndrome. Compr. Rev. Food Sci. Food Saf. 2018, 17, 63–81. [Google Scholar] [CrossRef]
- Awosika, T.O.; Aluko, R.E. Inhibition of the in vitro activities of α-amylase, α-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int. J. Food Sci. Technol. 2019, 54, 2021–2034. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-Originating ACE Inhibitors, Including Antihypertensive Peptides, as Preventive Food Components in Blood Pressure Reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- Girija, A.R. Peptide nutraceuticals. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Cambridge, UK, 2018; pp. 157–181. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Miciński, J.; Darewicz, M.; Bucholska, J. Biological and chemical databases for research into the composition of animal source foods. Food Rev. Int. 2013, 29, 321–351. [Google Scholar] [CrossRef]
- Agyei, D.; Bambarandage, E.; Udenigwe, C.C. The role of bioinformatics in the discovery of bioactive peptides. In Encyclopedia of Food Chemistry; Melton, L., Shahidi, F., Valeris, P., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 337–344. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocol Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Protasiewicz, M.; Mogut, D. Chemometrics and cheminformatics in the analysis of biologically active peptides from food sources. J. Funct. Foods 2015, 16, 334–351. [Google Scholar] [CrossRef]
- He, R.; Ma, H.; Zhao, W.; Qu, W.; Zhao, J.; Luo, L.; Zhu, W. Modeling the QSAR of ACE-Inhibitory Peptides with ANN and Its Applied Illustration. Int. J. Pept. 2012, 620609. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trend Anal. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- The UniProt Consortium, UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Dziuba, J.; Michalska, J. Bovine meat proteins as potential precursors of biologically active peptides—A computational study based on the BIOPEP database. Food Sci. Technol. Int. 2011, 7, 39–45. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. Annotation of peptide structures using SMILES and other chemical codes–practical solutions. Molecules 2017, 22, 2075. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Wang, N.-N.; Dong, J.; Deng, Y.-H.; Zhu, M.-F.; Wen, M.; Yao, Z.-J.; Lu, A.-P.; Wang, J.-B.; Cao, D.-S. ADME properties evaluation in drug discovery: Prediction of Caco-2 cell permeability using a combination of NSGA-II and boosting. J. Chem. Inf. Model. 2016, 56, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.-N.; Huang, C.; Dong, J.; Yao, Z.-J.; Zhu, M.-F.; Deng, Z.-K.; Lv, B.; Lu, A.-P.; Chen, A.F.; Cao, D.-S. Predicting human intestinal absorption with modified random forest approach: A comprehensive evaluation of molecular representation, unbalanced data, and applicability domain issues. RSC Adv. 2017, 7, 19007–19018. [Google Scholar] [CrossRef]
- Kerns, E.H.; Di, L. Drug-like properties: Concepts, Structure Design and Methods: From ADME to Toxicity Optimization; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Lei, T.; Li, Y.; Song, Y.; Li, D.; Sun, H.; Hou, T. ADMET evaluation in drug discovery: 15. Accurate prediction of rat oral acute toxicity using relevance vector machine and consensus modeling. J. Cheminform. 2016, 8, 6. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Panjaitan, F.C.A.; Gomez, H.L.R.; Chang, Y.-W. In Silico Analysis of Bioactive Peptides Released from Giant Grouper (Epinephelus lanceolatus) Roe Proteins Identified by Proteomics Approach. Molecules 2018, 23, 2910. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Murray, B.A.; Walsh, D.J. Hypotensive peptides from milk proteins. J. Nutr. 2004, 134, 980S–988S. [Google Scholar] [CrossRef] [PubMed]
- Vermeirssen, V.; van der Bent, A.; Van Camp, J.; van Amerongen, A.; Verstraete, W. A quantitative in silico analysis calculates angiotensin I converting enzyme (ACE) inhibitory activity in pea and whey protein digests. Biochimie 2004, 86, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef]
- Nassa, M.; Anand, P.; Jain, A.; Chhabra, A.; Jaiswal, A.; Malhotra, U.; Rani, V. Analysis of human collagen sequences. Bioinformation 2012, 8, 26–33. [Google Scholar] [CrossRef]
- Fu, U.; Therkildsen, M.E.; Aluko, R.E.; Lametsch, R. Exploration of collagen recovered from animal by-products as a precursor of bioactive peptides: Successes and challenges. Crit. Rev. Food Sci. Nutr. 2019, 59, 2011–2027. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Darewicz, M.; Mogut, D.; Minkiewicz, P. Elucidation of the role of in silico methodologies in approaches to studying bioactive peptides derived from foods. J. Funct. Foods 2019, 61, 1–14. [Google Scholar] [CrossRef]
- Yu, D.; Wang, C.; Song, Y.; Zhu, J.; Zhang, X. Discovery of Novel Angiotensin-Converting Enzyme Inhibitory Peptides from Todarodes pacificus and Their Inhibitory Mechanism: In Silico and In Vitro Studies. Int. J. Mol. Sci. 2019, 20, 4159. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Pliszka, M. Carp proteins as a source of bioactive peptides—An in silico approach. Czech. J. Food Sci. 2016, 34, 111–117. [Google Scholar] [CrossRef]
- Borawska, J.; Darewicz, M.; Vegarud, G.E.; Iwaniak, A.; Minkiewicz, P. Ex vivo digestion of carp muscle tissue – ACE inhibitory and antioxidant activities of obtained hydrolysates. Food Funct. 2015, 6, 211–218. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef]
- Iwaniak, A.; Mogut, D. Metabolic Syndrome-Preventive Peptides Derived from Milk Proteins and Their Presence in Cheeses: A Review. Appl. Sci. 2020, 10, 2772. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Hrynkiewicz, M.; Bucholska, J.; Darewicz, M. Hybrid Approach in the Analysis of Bovine Milk Protein Hydrolysates as a Source of Peptides Containing Di- and Tripeptide Bitterness Indicators. Pol. J. Food Nutr. Sci. 2020, 70, 139–150. [Google Scholar] [CrossRef]
- Iwaniak, A.; Hrynkiewicz, M.; Minkiewicz, P.; Bucholska, J.; Darewicz, M. Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches. Appl. Sci. 2020, 10, 2514. [Google Scholar] [CrossRef]
- Song, J.J.; Wang, Q.; Du, M.; Ji, X.M.; Mao, X.Y. Identification of dipeptidyl peptidase-IV inhibitory peptides from mare whey protein hydrolysates. J. Dairy Sci. 2017, 100, 6885–6894. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhang, F.; Ma, Z.; Chen, S.; Ding, G.; Tian, X.; Feng, R. Isolation and identification of the angiotensin-I converting enzyme (ACE) inhibitory peptides derived from cottonseed protein: Optimization of hydrolysis conditions. Int. J. Food Prop. 2019, 22, 1296–1309. [Google Scholar] [CrossRef]
- Lin, H.-C.; Alashi, A.M.; Aluko, R.E.; Pan, B.S.; Chang, Y.-W. Antihypertensive properties of tilapia (Oreochromis spp.) frame and skin enzymatic protein hydrolysates. Food Nutr. Res. 2017, 61, 1391666. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, W.; Zhu, M.; Xiao, Z. In silico assessment of the potential of the patatin as a precursor of bioactive peptides. J. Food Biochem. 2016, 40, 366–370. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Toldrá, F. The relevance of dipeptides and tripeptides in the bioactivity and taste of dry-cured ham. Food Prod. Process. Nutr. 2019, 1, 2. [Google Scholar] [CrossRef]
- Byun, H.-G.; Kim, S.-K. Structure and activity of angiotensin I converting enzyme inhibitory peptides derived from Alaskan Pollack skin. J. Biochem. Mol. Biol. 2001, 35, 239–243. [Google Scholar] [CrossRef]
- Suetsuna, K. Isolation and characterization of angiotensin I-converting enzyme inhibitor dipeptides derived from Allium sativum L (garlic). J. Nutr. Biochem. 1998, 9, 415–419. [Google Scholar] [CrossRef]
- Lan, V.T.T.; Ito, K.; Ohno, M.; Motoyama, T.; Ito, S.; Kawarasaki, Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015, 175, 66–73. [Google Scholar] [CrossRef]
- Nogata, Y.; Nagamine, T.; Yanaka, M.; Ohta, H. Angiotensin I Converting Enzyme Inhibitory Peptides Produced by Autolysis Reactions from Wheat Bran. J. Agric. Food Chem. 2009, 57, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Hu, J.; Aita, D.Q.; Maruyama, S. Angiotensin I-converting enzyme inhibitory activity and insulin secretion stimulative activity of fermented fish sauce. J. Biosci. Bioeng. 2003, 95, 496–499. [Google Scholar] [CrossRef]
- Sentandreu, M.A.; Toldrá, F. Evaluation of ACE inhibitory activity of dipeptides generated by the action of porcine muscle dipeptidyl peptidases. Food Chem. 2007, 102, 511–515. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Meisel, H. Lactokinins: Whey protein-derived ACE inhibitory peptides. Nahrung 1999, 43, 165–167. [Google Scholar] [CrossRef]
- Välimaa, A.-L.; Mäkinen, S.; Mattila, P.; Marnila, P.; Pihlanto, A.; Mäki, M.; Hiidenhovi, J. Fish and fish side streams are valuable sources of high-value components. Food Qual. Saf. 2019, 3, 209–226. [Google Scholar] [CrossRef]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T. Quantitative structure-activity relationship modeling of ACE-inhibitory peptides derived from milk proteins. Eur. Food Res. Technol. 2004, 219, 579–583. [Google Scholar] [CrossRef]
- Yi, Y.; Lv, Y.; Zhang, L.; Yang, Y.; Shi, Q. High throughput identification of antihypertensive peptides from fish proteome datasets. Mar. Drugs 2020, 16, 365. [Google Scholar] [CrossRef]
- Udenigwe, C.C. Bioinformatic approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 137–143. [Google Scholar] [CrossRef]
- Gogktug, A.N.; Chai, S.C.; Chen, T. Data analysis approaches in high throughput screening. In Drug Discovery; El-Shemy, H., Ed.; IntechOpen: Rijeka, Croatia, 2013; pp. 201–226. [Google Scholar] [CrossRef]
- Fu, Y.; Young, J.F.; Løkke, M.M.; Lametsch, R.; Aluko, R.E.; Therkildsen, M. Revalorisation of bovine collagen as a potential precursor of angiotensin I-converting enzyme (ACE) inhibitory peptides based on in silico and in vitro protein digestions. J. Funct. Foods 2016, 24, 196–206. [Google Scholar] [CrossRef]
- Rajendran, S.R.C.K.; Mason, B.; Udenigwe, C.C. Peptidomics of peptic digest of selected potato tuber proteins: Post-translational modifications and limited cleavage specificity. J. Agric. Food Chem. 2016, 64, 2432–2437. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Martinez, M.; Gonzalez de Mejia, E.; García-Lara, S.; Aguilar, O.; Lopez-Castillo, L.M.; Otero-Pappatheodorou, J.T. Antiproliferative effect of peptide fractions isolated from a quality protein maize, a white hybrid maize, and their derived peptides on hepatocarcinoma human HepG2 cells. J. Funct. Foods 2017, 34, 36–48. [Google Scholar] [CrossRef]
- Mojica, L.; Luna-Vital, D.A.; Gonzalez de Mejia, E. Black bean peptides inhibit glucose uptake in Caco-2 adenocarcinoma cells by blocking the expression and translocation pathway of glucose transporters. Toxicol. Rep. 2018, 5, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Dókus, L.E.; Yousef, M.; Bánóczi, Z. Modulators of calpain activity: Inhibitors and activators as potential drugs. Expert Opin. Drug Discov. 2020, 15, 471–486. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene 2010, 29, 781–788. [Google Scholar] [CrossRef]
- Sheng, J.; Sun, H.; Yu, F.-B.; Li, B.; Zhang, Y.; Zhu, Y.-T. The role of cyclooxygenase-2 in colorectal cancer. Int. J. Med. Sci. 2020, 17, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liu, Y.; Zhang, Z.; Yang, G.-Y. Significance of complement system in ischemic stroke: A comprehensive review. Aging Dis. 2019, 10, 429–462. [Google Scholar] [CrossRef]
- Noris, M.; Donadelli, R.; Remuzzi, G. Autoimmune abnormalities of the alternative complement pathway in membranoproliferative glomerulonephritis and C3 glomerulopathy. Pediatr. Nephrol. 2019, 4, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.; Rojas-Quintero, J.; Cano, C.; Pérez, J.L.; Ramírez, P.; Carrasquero, R.; Torres, W.; Espinoza, C.; Chacín-González, M.; Bermúdez, V. Neprilysin: A potential therapeutic target of arterial hypertension? Curr. Cardiol. Rev. 2020, 16, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Książczyk, M.; Lelonek, M. Angiotensin receptor/neprilysin inhibitor—A breakthrough in chronic heart failure therapy: Summary of subanalysis on PARADIGM-HF trial findings. Heart Fail. Rev. 2020, 25, 393–402. [Google Scholar] [CrossRef]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-L.; Schimmel, P.; Ewalt, K.L. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem. Sci. 2004, 29, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Saiyasit, N.; Sripetchwandee, J.; Chattipakorn, N.; Chattipakorn, S.C. Potential roles of neurotensin on cognition in conditions of obese-insulin resistance. Neuropeptides 2018, 72, 12–22. [Google Scholar] [CrossRef]
- Herrera-Ruiz, D.; Knipp, G.T. Current perspectives on established and putative mammalian oligopeptide transporters. J. Pharmaceut. Sci. 2003, 92, 691–714. [Google Scholar] [CrossRef]
- Hessler, G.; Baringhaus, K.-H. Artificial intelligence in drug design. Molecules 2020, 23, 2520. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Mayorga, K.; Madariaga-Mazon, A.; Medina-Franco, J.L.; Maggiora, G. The impact of chemoinformatics on drug discovery in the pharmaceutical industry. Expert Opin. Drug Discov. 2020, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Naveja, J.J.; Rico-Hidalgo, M.P.; Medina-Franco, J.L. Analysis of a large food chemical database: Chemical space, diversity, and complexity. F1000 Res. 2018, 7, 993. [Google Scholar] [CrossRef]
- Santibáñez-Morán, M.G.; Rico-Hidalgo, M.P.; Manallack, D.T.; Medina-Franco, J.L. The acid/base profile of a large food chemical database. Mol. Inf. 2019, 38, 1800171. [Google Scholar] [CrossRef]
- Santibáñez-Morán, M.G.; Medina-Franco, J.L. Analysis of the acid/base profile of natural products from different sources. Mol. Inf. 2020, 39, 1900099. [Google Scholar] [CrossRef]
- Yu, Z.; Fan, Y.; Zhao, W.; Ding, L.; Li, J.; Liu, L. Novel angiotensin-converting enzyme inhibitory peptides derived from Oncorhynchus mykiss nebulin: Virtual screening and in silico molecular docking study. J. Food Sci. 2018, 83, 2375–2383. [Google Scholar] [CrossRef]
- Zhao, W.; Xue, S.; Yu, Z.; Ding, L.; Li, J.; Liu, J. Novel ACE inhibitors derived from soybean proteins using in silico and in vitro studies. J. Food Biochem. 2019, 43, e12975. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, D.; Yu, Z.; Ding, L.; Liu, J. Novel membrane peptidase inhibitory peptides with activity against angiotensin converting enzyme and dipeptidyl peptidase IV identified from hen eggs. J. Funct. Foods 2020, 64, 103649. [Google Scholar] [CrossRef]
- Fan, Y.; Yu, Z.; Zhao, W.; Ding, L.; Zheng, F.; Li, J.; Liu, J. Identification and molecular mechanism of angiotensin-converting enzyme inhibitory peptides from Larimichthys crocea titin. Food Sci. Hum. Wellness 2020. [Google Scholar] [CrossRef]
- Capecchi, A.; Awale, M.; Probst, D.; Reymond, J.-L. PubChem and ChEMBL beyond Lipinski. Mol. Inf. 2019, 38, 1900016. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Matsui, T. Intestinal absorption of small peptides: A review. Int. J. Food Sci. Technol. 2019, 54, 1942–1948. [Google Scholar] [CrossRef]

| Source of Collagen | Major A (A ≥ 0.500) | Moderate A (A = 0.1002−0.499) | Minor A (A = 0.001−0.099) | ||
|---|---|---|---|---|---|
| cow (Bos taurus) | 0.834 ah 1 0.854 dpp | 0.214 am;re 0.238 at | 0.002 apr;35pd | 0.003 im | 0.006 ren |
| 0.009 ne | 0.014 emb | 0.037 che | |||
| 0.041 glui | 0.057 inh | 0.059 ao | |||
| 0.069 st | 0.073 dpp3 | ||||
| pig (Sus scrofa) | 0.846 dpp 0.847 ah | 0.216 am;re 0.238 at | 0.001 emb | 0.002 apr;35pd | 0.003 im |
| 0.006 ren | 0.007 st | 0.008 ne | |||
| 0.038 che | 0.042 glui | 0.058 inh | |||
| 0.060 ao | 0.076 dpp3 | ||||
| sheep (Ovis aries) | 0.833 ah 0.845 dpp | 0.198 am;re 0.215 at | 0.001 emb;im | 0.002 35pd | 0.003 apr |
| 0.006 ren | 0.007 st | 0.008 ne | |||
| 0.033 che | 0.045 glui | 0.052 inh | |||
| 0.059 ao | 0.073 dpp3 | ||||
| chicken (Gallus gallus) | 0.843 ah 0.852 dpp | 0.210 am;re 0.223 at | 0.001 lig | 0.002 apr;35pd;emb | 0.003 im |
| 0.006 ren;is;st | 0.008 ne | 0.040 glui;che | |||
| 0.057 inh | 0.061 ao | 0.075 dpp3 | |||
| duck (Anas platyrhynchos platyrhynchos) | 0.847 ah 0.870 dpp | 0.240 at 0.210 am;re | 0.002 hypl;35pd | 0.003 lig | 0.004 emb |
| 0.007 apr | 0.011 ren | 0.033 glui;che | |||
| 0.046 inh | 0.057 ao | 0.083 dpp3 | |||
| horse (Equus caballus) | 0.843 ah;dpp | 0.215 am;re 0.238 at | 0.001 emb | 0.00235pd | 0.003 apr;im |
| 0.006 ren | 0.007 st | 0.009 ne | |||
| 0.037 che | 0.041 glui | 0.058 inh;ao | |||
| 0.073 dpp3 | |||||
| salmon (Salmo salar) | 0.798 dpp 0.799 ah | 0.170 at;am;re | 0.002 emb;is | 0.00335pd;lig;apr | 0.005 st |
| 0.008 ne;ren | 0.023 che | 0.029 glui | |||
| 0.053 ao | 0.074 dpp3 | ||||
| rainbow trout (Oncorhynchus mykiss) | 0.810 dpp 0.846 ah | 0.161 am 0.162 re 0.181 at | 0.001 hypl | 0.002 lig | 0.003 emb;35pd |
| 0.006 apr | 0.008 st | 0.009 ne | |||
| 0.013 ren | 0.015 glui | 0.018 che | |||
| 0.020 inh | 0.034 ao | 0.071 dpp3 | |||
| goat (Capra hircus) | 0.842 ah 0.849 dpp | 0.213 am;re 0.273 at | 0.001 emb | 0.002 apr;35pd | 0.003 im |
| 0.006 ren | 0.007 st | 0.009 ne | |||
| 0.037 che | 0.041 glui | 0.057 inh | |||
| 0.058 ao | 0.074 dpp3 | ||||
| rabbit (Oryctolagus cuniculus) | 0.834 ah 0.849 dpp | 0.199 am;re 0.215 at | 0.001 emb;im | 0.002 35pd | 0.003 apr |
| 0.006 ren | 0.007 st | 0.008 ne | |||
| 0.034 che | 0.045 glui | 0.052 inh | |||
| 0.059 ao | 0.073 dpp3 | ||||
| turkey (Meleagris gallopavo) | 0.822 dpp 0.841 ah | 0.192 am;re 0.220 at | 0.002 hypl | 0.003 lig;35pd | 0.004 emb |
| 0.007 apr | 0.009 ne | 0.012 st | |||
| 0.014 ren | 0.027 che | 0.030 glui | |||
| 0.041 inh | 0.059 ao | 0.082 dpp3 | |||
| Source of Collagen | Enzyme | AE | W | DHt (%) |
|---|---|---|---|---|
| cow (Bos taurus) | B 1 | 0.158 dpp2 0.097 ah 0.042 am;at;re 0.014 dpp3 0.005 glui 0.001 ao | 0.25 im 0.195 am;re 0.191 dpp3 0.186 dpp 0.175 at 0.120 glui 0.115 ah 0.113 ren 0.024 ao | 55.55 |
| F | 0.122 dpp 0.071 ah 0.021 am;at;re 0.008 dpp3 0.001 ao;ren | 0.226 ren 0.143 dpp 0.118 am;re 0.114 dpp3 0.105 at 0.084 ah 0.024 ao | 45.69 | |
| Pap | 0.151 dpp 0.105 ah 0.018 am;at;re 0.008 dpp3 0.001 ren | 1.000 lig 0.226 ren 0.176 dpp 0.124 ah 0.104 dpp3 0.084 am;re 0.076 at 0.012 ao | 46.11 | |
| Pep | 0.004 ah;dpp 0.002 ren 0.001 dpp3 | 0.339 ren 0.019 dpp3 0.004 ah;dpp | 4.38 | |
| T | 0.002 ah 0.001 dpp;ao;at | 0.012 ao 0.006 at 0.003 ah 0.002 dpp | 8.75 | |
| Pep + T | 0.013 ah;dpp 0.005 dpp3 0.002 ren 0.001 at | 0.338 ren 0.067 dpp3 0.016 dpp 0.012 ao 0.006 at | 13.13 | |
| Pep + T + Ch | 0.141 ah 0.107 dpp 0.055 re;am;at 0.029 dpp3 0.005 ne 0.003 ren 0.002 ao 0.001 35pd | 0.667 35pd 0.544 ne 0.452 ren 0.409 dpp3 0.256 re;at 0.230 am 0.161 ah 0.125 dpp 0.036 ao | 45.63 | |
| pig (Sus scrofa) | B | 0.158 dpp 0.098 ah 0.041 am;at;re 0.013 dpp3 0.006 glui 0.001 ao | 0.250 im 0.192 am;re 0.187 dpp 0.176 dpp3 0.174 at 0.133 glui 0.116 ah 0.111 ren 0.024 ao | 55.43 |
| F | 0.124 dpp 0.721 ah 0.025 am;at;re 0.008 dpp3 0.001 ao;ren | 0.117 re 0.106 at 0.085 ah 0.117 am 0.024 ao 0.147 dpp 0.222 ren 0.102 dpp3 | 45.91 | |
| Pap | 0.152 dpp 0.109 ah 0.018 am;at;re 0.007 dpp3 0.001 ren | 1.000 lig 0.222 ren 0.180 dpp 0.128 ah 0.093 dpp3 0.084 am;re 0.076 at 0.012 ao | 45.83 | |
| Pep | 0.003 ah;dpp 0.002 ren | 0.333 ren 0.009 dpp3 0.004 ah 0.003 dpp | 4.41 | |
| T | 0.001 at;ah | 0.012 ao 0.006 at 0.002 ah | 8.76 | |
| Pep + T | 0.013 ah 0.012 dpp 0.004 dpp3 0.002 ren 0.001 at | 0.333 ren 0.056 dpp3 0.015 ah 0.014 dpp 0.012 ao 0006 at | 13.17 | |
| Pep + T + Ch | 0.147 ah 0.109 dpp 0.055 am;at;re 0.031 dpp3 0.005 ne 0.003 ao;ren 0.001 35pd | 0.667 35pd 0.583 ne 0.444 ren 0.407 dpp3 0.256 am;re 0.232 at 0.174 ah 0.128 dpp 0.100 st 0.005 ao | 45.83 | |
| sheep (Ovis aries) | B | 0.158 dpp 0.095 ah 0.041 am;at;re 0013 dpp3 0.005 glui 0.001 ao | 0.500 im 0.205 am;re 0.189 at 0.186 dpp 0.180 dpp3 0.114 ah 0.107 glui 0.023 ao | 55.69 |
| F | 0.118 dpp 0.070 ah 0.023 am;at;re 0.007 dpp3 0.003 ao | 0.139 dpp 0.127 ren 0.115 am;re 0.106 at 0.095 dpp3 0.084 ah 0.047 ao | 45.69 | |
| Pap | 0.145 dpp 0.103 ah 0.019 am;at;re 0.006 dpp3 | 1.000 lig 0.175 dpp 0.127 ren 0.124 ah 0.094 am;re 0.087 at 0.075 dpp3 0.016 glui 0.012 ao | 45.62 | |
| Pep | 0.003 ah;dpp 0.001 dpp3;ren | 0.255 ren 0.019 dpp3 0.004 ah 0.003 dpp | 4.55 | |
| T | 0.001 ah | 0.012 ao 0.003 at 0.002 ah | 9.10 | |
| Pep + T | 0.012 ah;dpp 0.005 dpp3 0.001 ren | 0.255 ren 0.066 dpp3 0.015 ah 0.014 dpp 0.012 ao 0.003 at | 13.65 | |
| Pep + T + Ch | 0.137 ah 0.107 dpp 0.050 am;at;re 0.030 dpp3 0.004 ne 0.003 ren 0.002 ao 0.001 35pd | 0.667 35pd 0.509 ren 0.494 ne 0.410 dpp3 0.250 am;re 0.231 at 0.165 ah 0.127 dpp 0.101 st 0.036 ao | 46.04 | |
| chicken (Gallus gallus) | B | 0.154 dpp 0.096 ah 0.043 am;at,re 0.012 dpp3 0.006 glui 0.001 ao;im | 0.500 im 0.203 am;re 0.185 at 0.180 dpp 0.159 dpp3 0.141 glui 0.114 ah 0.111 ren 0.023 ao | 55.59 |
| F | 0.120 dpp 0.068 ah 0.025 am;at;re 0.008 dpp3 0.003 ao 0.001 ren | 0.222 ren 0.141 dpp 0.116 am;re 0.107 at 0.103 dpp3 0.080 ah 0.047 ao | 44.97 | |
| Pap | 0.158 dpp 0.108 ah 0.019 am;at;re 0.007 dpp3 0.001 ren | 0.500 lig 0.250 im 0.222 ren 0.185 dpp 0.128 ah 0.094 dpp3 0.090 am;re 0.082 at 0.012 ao | 46.43 | |
| Pep | 0.003 ah;dpp 0.002 ren | 0.333 ren 0.009 dpp3 0.003 ah;dpp | 4.55 | |
| T | 0.002 ah 0.001 dpp | 0.012 ao 0.003 ah;at 0.001 dpp | 8.60 | |
| Pep + T | 0.013 ah;dpp 0.004 dpp3 0.002 ren | 0.333 ren 0.056 dpp3 0.016 ah 0.012 ao 0.003 at | 13.15 | |
| Pep + T + Ch | 0.143 ah 0.112 dpp 0.055 am;at;re 0.033 dpp3 0.005 ao 0.004 ne 0.003 ren 0.001 35pd | 0.667 35pd 0.500 ne 0.444 ren 0.439 dpp3 0.259 am;re 0.237 at 0.170 ah 0.131 dpp 0.125 st 0.082 ao | 45.38 | |
| duck (Anas platyrhynchos platyrhynchos) | B | 0.146 dpp 0.091 ah 0.034 am;at;re 0.015 dpp3 0.005 ren 0.003 glui 0.002 ao; st 0.001 35pd | 0.652 35pd 0.533 hyp 0.397 ren 0.247 st 0.178 dpp3 0.172 dpp 0.163 am;re 0.144 at 0.104 ah 0.096 glui 0.041 ao | 57.79 |
| F | 0.123 dpp 0.083 ah 0.025 am;at;re 0.012 dpp3 0.005 ren 0.002 st 0.001 ao; hyp,35pd | 0.533 hyp 0.397 ren 0.348 35pd 0.248 st 0.146 dpp 0.130 dpp3 0.126 re 0.119 am 0.105 at 0.095 ah 0.014 ao | 48.57 | |
| Pap | 0.154 dpp 0.113 ah 0.019 am;at;re 0.010 dpp3 0.004 ren 0.002 ao; st 0.001 hyp;lig;35pd | 0.533 hyp 0.348 35pd 0.336 ren 0.258 lig 0.182 dpp 0.161 st 0.130 ah 0.122 dpp3 0.092 am;re 0.082 at 0.027 ao | 47.87 | |
| Pep | 0.005 ah 0.004 dpp 0.001 dpp3;ren | 0.070 ren 0.010 dpp3 0.006 ah 0.005 dpp | 5.58 | |
| T | 0.001 ah;at;dpp | 0.034 at 0.001 ah;dpp | 8.83 | |
| Pep + T | 0.013 ah;dpp 0.002 at;dpp3 0.001 ren | 0.070 ren 0.018 dpp3 0.016 dpp 0.015 ah 0.006 at | 14.41 | |
| Pep + T + Ch | 0.024 dpp 0.023 ah 0.003 at 0.001 ao;dpp3;glui;reg;ren | 0.070 ren 0.028 dpp 0.027 ah;ao 0.025 glui 0.013 at 0.010 dpp3 0.004 re | 20.99 | |
| horse (Equus caballus) | B | 0.155 dpp 0.096 ah 0.042 am;at;re 0.013 dpp3 0.006 glui 0.001 ao;im;ren | 0.25 im 0.193 am;re 0.184 dpp 0.181 dpp3 0.175 at 0.115 glui 0.114 ah 0.113 ren 0.024 ao | 55.55 |
| F | 0.124 dpp 0.074 ah 0.026 am;at;re 0.008 dpp3 0.001 ao;ren | 0.226 ren 0.147 dpp 0.123 am;re 0.112 at 0.104 dpp3 0.088 ah 0.012 ao | 45.83 | |
| Pap | 0.151 dpp 0.108 ah 0.018 am;at;re 0.007 dpp3 0.001 ao;lig;ren | 1.000 lig 0.226 ren 0.179 dpp 0.128 ah 0.095 dpp3 0.084 am;re 0.077 at 0.012 ao | 45.90 | |
| Pep | 0.004 ah 0.003 dpp 0.021 ren 0.001 dpp3 | 0.339 ren 0.010 dpp3 0.004 ah 0.003 dpp | 4.31 | |
| T | 0.001 dpp;ah;ao,at | 0.012 ao 0.006 at 0.002 ah 0.001 dpp | 8.82 | |
| Pep + T | 0.013 ah 0.012 dpp 0.004 dpp3 0.002 ren 0.001 ao;at | 0.339 ren 0.058 dpp3 0.015 ah 0.014 dpp 0.012 ao 0.006 at | 13.13 | |
| Pep + T + Ch | 0.022 dpp 0.019 ah 0.005 dpp3 0.003 at;ren 0.002 ao 0.001 glui; st;35pd | 0.452 ren 0.333 35pd 0.101 st 0.067 dpp3 0.036 ao 0.026 dpp 0.023 ah 0.014 glui 0.012 at | 17.85 | |
| salmon (Salmo salar) | B | 0.143 dpp 0.093 ah 0.036 am;at;re 0.008 glui 0.006 dpp3 0.003 ren 0.002 ao 0.001 st;35pd | 0.333 ren 0.250 35pd 0.223 glui 0.219 at 0.218 re 0.198 at 0.179 dpp 0.143 st 0.116 ah 0.086 dpp3 0.040 ao | 57.85 |
| F | 0.115 dpp 0.071 ah 0.023 am;at;re 0.006 dpp3 0.004 ao;ren 0.001 HMGi;35pd | 1.000 HMGi 0.417 ren 0.250 35pd 0.144 dpp 0.139 am 0.139 re 0.126 at 0.089 ah 0.086 dpp3 0.067 ao | 47.48 | |
| Pap | 0.144 dpp 0.110 ah 0.0203 am;at;re 0.006 dpp3 0.003 ren 0.001 ao;HMGi;35pd | 1.000 HMGi 0.333 ren 0.250 35pd 0.181 dpp 0.138 ah 0.122 am;re 0.112 at 0.076 dpp3 0.027 ao | 46.91 | |
| Pep | 0.003 ah 0.002 dpp 0.001 ao;dpp3;ren | 0.167 ren 0.013 ao 0.010 dpp3 0.003 ah;dpp | 4.07 | |
| T | 0.002 ah 0.001 dpp | 0.003 ah 0.002 dpp | 8.77 | |
| Pep + T | 0.010 dpp 0.009 ah 0.006 dpp3 0.001 ren | 0.167 ren 0.076 dpp3 0.012 dpp 0.001 ah | 12.83 | |
| Pep + T + Ch | 0.019 ah 0.018 dpp 0.008 dpp3 0.003 ao 0.002 at;ren 0.001 glui; is;35pd | 0.333 is 0.250 ren;35pd 0.105 dpp3 0.053 ao 0.024 ah 0.023 dpp 0.021 glui 0.011 at | 18.93 | |
| rainbow trout (Oncorhynchus mykiss) | B | 0.142 dpp 0.096 ah 0.034 am;at;re 0.013 dpp3 0.003 ao;glui;ren 0.002 35pd 0.001 hyp; st | 0.500 35pd 0.467 hyp 0.236 ren 0.209 am 0.208 re 0.187 at 0.178 dpp3 0.143 glui 0.113 ah 0.093 st 0.082 ao | 61.89 |
| F | 0.118 dpp 0.098 ah 0.027 am;at;re 0.008 dpp3 0.003 ao 0.002 ren 0.001 HMGi; st;35pd | 1.000 HMGi 0.333 35pd 0.173 ren 0.171 re 0.168 am 0.149 at 0.147 dpp 0.115 ah;dpp3 0.093 st 0.082 ao | 52.44 | |
| Pap | 0.153 dpp 0.137 ah 0.025 am;at;re 0.008 dpp3 0.004 ren 0.002 ao;glui;35pd 0.001 HMGi; hyp; st;lig | 1.000 HMGi 0.500 35pd 0.467 hyp 0.318 lig 0.291 ren 0.190 dpp 0.162 ah 0.153 am;re 0.138 at 0.115 dpp3 0.105 glui 0.093 st 0.060 ao | 51.46 | |
| Pep | 0.003 ah;dpp 0.001 dpp3;ren | 0.055 ren 0.010 dpp3 0.004 ah;dpp | 5.48 | |
| T | 0.002 at 0.001 ah | 0.008 at 0.001 ah | 8.93 | |
| Pep + T | 0.011 ah;dpp 0.002 at;dpp3 0.001 ren | 0.055 ren 0.031 dpp3 0.014 dpp 0.013 ah 0.008 at | 14.40 | |
| Pep + T + Ch | 0.022 ah 0.020 dpp 0.002 ao;at;dpp3 0.001 glui; st;re;ren | 0.093 st 0.060 ao;ren 0.033 glui 0.026 ah 0.021 dpp3 0.008 at 0.004 re | 21.61 | |
| go at (Capra hircus) | B | 0.156 dpp 0.096 ah 0.042 am;at;re 0.014 dpp3 0.005 glui 0.001 ao;im;ren | 0.250 im 0.195 am;re 0.189 dpp3 0.184 dpp 0.175 at 0.114 ah 0.113 ren 0.104 glui 0.024 ao | 55.55 |
| F | 0.120 dpp 0.069 ah 0.024 am;at;re 0.008 dpp3 0.001 ao;ren | 0.226 ren 0.114 dpp 0.113 dpp3 0.111 am;re 0.010 at 0.082 ah 0.024 ao | 45.55 | |
| Pap | 0.152 dpp 0.105 ah 0.019 am;at;re 0.008 dpp3 0.001 ao;lig;ren | 1.000 lig 0.226 ren 0.179 dpp 0.125 ah 0.103 dpp3 0.088 am;re 0.079 at 0.012 ao | 46.18 | |
| Pep | 0.040 ah;dpp 0.002 ren 0.001 dpp3 | 0.339 ren 0.019 dpp3 0.004 ah;dpp | 4.38 | |
| T | 0.01 ah;ao;at;dpp | 0.117 ao 0.006 at 0.02 ah 0.001 dpp | 8.75 | |
| Pep + T | 0.013 ah;dpp 0.005 dpp3 0.002 ren 0.001 ao;at | 0.339 ren 0.067 dpp3 0.015 ah;dpp 0.012 ao 0.006 at | 13.13 | |
| Pep + T + Ch | 0.023 dpp 0.021 ah 0.006 dpp3 0.003 am;at;re 0.001 glui; st;35pd | 0.452 ren 0.333 35pd 0.101 st 0.008 dpp3 0.045 ao 0.027 dpp 0.025 ah 0.015 glui 0.012 at | 18.06 | |
| rabbit (Oryctolagus cuniculus) | B | 0.158 dpp 0.095 ah 0.041 am;at;re 0.013 dpp3 0.005 glui 0.001 ao;im | 0.500 im 0.205 am;re 0.189 at 0.186 dpp 0.179 dpp3 0.114 ah 0.097 glui 0.024 ao | 55.72 |
| F | 0.118 dpp 0.070 ah 0.023 am;at;re 0.007 dpp3 0.003 ao 0.001 ren | 0.134 dpp 0.127 ren 0.114 am;re 0.106 at 0.094 dpp3 0.084 ah 0.047 ao | 45.72 | |
| Pap | 0.149 dpp 0.103 ah 0.019 am;at;re 0.006 dpp3 0.001 ao;glui;lig;ren | 1.000 lig 0.175 dpp 0.127 ren 0.124 ah 0.094 am;re 0.087 at 0.075 dpp3 0.014 glui 0.012 ao | 45.66 | |
| Pep | 0.003 ah;dpp 0.001 ren;dpp3 | 0.255 ren 0.019 dpp3 0.004 ah 0.003 dpp | 4.55 | |
| T | 0.001 ah;ao;at;dpp | 0.012 ao 0.003 at 0.002 at 0.001 dpp | 9.10 | |
| Pep + T | 0.012 ah;dpp 0.005 dpp3 0.001 ao;at;ren | 0.255 ren 0.066 dpp3 0.015 ah 0.014 dpp 0.012 ao 0.003 at | 13.66 | |
| Pep + T + Ch | 0.021 dpp 0.020 ah 0.006 dpp3 0.003 ao 0.002 at;ren 0.001 glui; st;35pd | 0.382 ren 0.333 35pd 0.101 st 0.075 dpp3 0.057 ao 0.025 dpp 0.024 ah 0.014 glui 0.001 at | 18.83 | |
| turkey (Meleagris gallopavo) | B | 0.141 dpp 0.092 ah 0.032 am;at;re 0.016 dpp3 0.005 ren 0.004 ao 0.003 glui 0.002 st;35pd 0.001 hyp | 0.767 35pd 0.533 hyp 0.387 ren 0.196 dpp3 0.166 am;re 0.146 at 0.110 ah 0.080 glui 0.065 ao | 57.74 |
| F | 0.118 dpp 0.082 ah 0.022 re 0.021 am;at 0.011 dpp3 0.005 ren 0.003 st 0.002 ao 0.001 hyp;35pd | 0.533 hyp 0.500 35pd 0.336 ren 0.246 st 0.144 dpp 0.131 dpp3 0.115 reg 0.120 am 0.098 ah 0.097 at 0.039 ao | 48.82 | |
| Pap | 0.147 dpp 0.110 ah 0.019 am;at;re 0.011 dpp3 0.005 ren 0.003 ao 0.002 glui; st;35pd 0.001 hyp;lig | 0.533 hyp 0.500 35pd 0.336 ren 0.267 lig 0.179 dpp 0.131 ah;dpp3 0.123 st 0.100 re 0.087 at 0.087 glui 0051 ao | 47.52 | |
| Pep | 0.006 ah 0.005 dpp 0.001 ao;ren;dpp3 | 0.058 ren 0.018 dpp3 0.014 ao 0.007 ah 0.006 dpp | 6.18 | |
| T | 0.002 ah 0.001 dpp | 0.003 ah 0.002 dpp | 9.15 | |
| Pep + T | 0.017 dpp 0.016 ah 0.003 dpp3 0.001 ao;at;ren | 0.058 ren 0.037 dpp3 0.204 dpp 0.019 ah 0.014 ao 0.004 at | 15.33 | |
| Pep + T + Ch | 0.136 ah 0.106 dpp 0.047 re 0.046 am;at 0.030 dpp3 0.003 ne 0.002 ao 0.001 ren; st | 0.364 dpp3 0.330 ne 0.241 re 0.238 am 0.208 at 0.161 ah 0.129 dpp 0.066 st 0.058 ren 0039 ao | 45.69 |
| Peptide Sequence | Peptideranker Score | Collagen Source | Enzyme Applied |
|---|---|---|---|
| GF ACEi;DPP-IVi 1 | 0.994 | cow (Bos Taurus)/sheep (Ovis aries) | Pep 2 |
| SFACEi;DPP-IVi | 0.948 | cow (Bos taurus)/pig (Sus scrofa)/sheep (Ovis aries)/chicken (Gallus gallus)/horse (Equus caballus) | Pep |
| QF DPP-IVi | 0.946 | cow (Bos taurus)/chicken (Gallus gallus)/ | Pep |
| DF ACEi | 0.942 | horse (Equus caballus) | Pep |
| PGL ACEi | 0.855 | cow (Bos taurus)/pig (Sus scrofa)/chicken (Gallus gallus)/horse (Equus caballus)/salmon (Salmo salar) 3 | Pep |
| TF ACEi;DPP-IVi | 0.826 | cow (Bos taurus)/pig (Sus scrofa)/sheep (Ovis aries)/chicken (Gallus gallus)/horse (Equus caballus)/salmon (Salmo salar) | Pep |
| GR ACEi | 0.766 | rainbow trout (Oncorhynchus mykiss) | T 4 |
| RL ACEi;DPP-IVi 4 | 0.626 | cow (Bos taurus)/pig (Sus scrofa)/sheep (Ovis aries)/chicken (Gallus gallus)/horse (Equus caballus)/salmon (Salmo salar) | Pep |
| DR DPP-IVi | 0.289 | horse (Equus caballus) | T |
| Peptide Sequence | Protein 1 | Protein 2 | Protein 3 |
|---|---|---|---|
| PGL | Dipeptidyl peptidase IV (UniProt—P27487 1; ChEMBL—CHEMBL284 2) Probability: 0.526 3 | Angiotensin converting enzyme (UniProt—P12821; ChEMBL—CHEMBL1808) Probability: 0.445 | Cyclooxygenase-2 (UniProt—P35354; ChEMBL—CHEMBL230) Probability: 0.420 |
| RL | Neurotensin receptor 2 (UNiProt—O95665; ChEMBL—CHEMBL2514) Probability: 0.166 | Complement factor B (UniProt—P00751; ChEMBL—CHEMBL573) Probability: 0.166 | Subtilisin/kexin type 6 (UniProt—P29122; ChEMBL—CHEMBL2951) Probability: 0.133 |
| GF | Oligopeptide transporter small intestine isoform (UniProt—P46059; ChEMBL—CHEMBL4605) Probability: 0.130 | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL389) Probability: 0.112 | Neprilysin (UniProt—P08473; ChEMBL—CHEMBL1944) Probability: 0.104 |
| SF | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.081 | Oligopeptide transporter small intestine isoform (UniProt—P46059; ChEMBL—CHEMBL4605) Probability: 0.072 | Cyclooxygenase-2 (UniProt—P35354; ChEMBL—CHEMBL230) Probability: 0.063 |
| TF | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.238 | Tyrosyl-tRNA synthetase (UniProt—P54577; ChEMBL—CHEMBL3179) Probability: 0.143 | Cyclooxygenase-2 (UniProt—P35354; ChEMBL—CHEMBL230) Probability: 0.143 |
| QF | Angiotensin converting enzyme (UniProt—P12821; ChEMBL—CHEMBL1808) Probablity: 0.238 | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.230 | Tyrosyl-tRNA synthetase (UniProt—P54577; ChEMBL—CHEMBL3179) Probability: 0.140 |
| DF | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.150 | Angiotensin converting enzyme (UniProt—P12821; ChEMBL—CHEMBL1808) Probablity: 0.117 | Neprilysin (UniProt—P08473; ChEMBL—CHEMBL1944) Probability: 0.109 |
| DR | Complement factor B (UniProt—P00751; ChEMBL—CHEMBL5731) Probability: 0.109 | Furin (UniProt—P09958; ChEMBL—CHEMBL2611) Probability: 0.109 | Integrin alpha-IIb/beta-3 (UniProt—P08514; P05106; ChEMBL—CHEMBL2093869) Probability: 0.109 |
| GR | Complement factor B (UniProt—P00751; ChEMBL—CHEMBL5731) Probability: 0.112 | Furin (UniProt—P09958; ChEMBL—CHEMBL2611) Probability: 0.104 | Neurotensin receptor 2 (UNiProt—O95665; ChEMBL—CHEMBL2514) Probability: 0.086 |
| Sequence | Rule of 5 | Log Caco-2 Permeability (Permeability Expressed in cm × s −1) | Human Intestinal Absorption Probability | VD 1 (L × kg −1) | T 1/2 2 (h) | LD50 3 of Acute Toxicity (mg × kg −1) |
|---|---|---|---|---|---|---|
| PGL | + | −5.643 | 0.309 | 0.149 | 0.701 | 1589 |
| RL | + | −6.203 | 0.398 | 0.160 | 1.184 | 45,963 |
| GF | + | −5.354 | 0.482 | 0.209 | 0.691 | 1344 |
| SF | + | −5.818 | 0.281 | 0.130 | 0.663 | 1513 |
| TF | + | −5.781 | 0.310 | 0.103 | 0.660 | 1385 |
| QF | + | −5.929 | 0.368 | 0.090 | 0.578 | 1592 |
| DF | + | −5.625 | 0.385 | 0.072 | 0.580 | 1672 |
| DR | + | −6.407 | 0.275 | 0.054 | 0.811 | 1494 |
| GR | + | −6.292 | 0.335 | 0.150 | 0.962 | 1140 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaniak, A.; Minkiewicz, P.; Pliszka, M.; Mogut, D.; Darewicz, M. Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods 2020, 9, 965. https://doi.org/10.3390/foods9070965
Iwaniak A, Minkiewicz P, Pliszka M, Mogut D, Darewicz M. Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods. 2020; 9(7):965. https://doi.org/10.3390/foods9070965
Chicago/Turabian StyleIwaniak, Anna, Piotr Minkiewicz, Monika Pliszka, Damir Mogut, and Małgorzata Darewicz. 2020. "Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters" Foods 9, no. 7: 965. https://doi.org/10.3390/foods9070965
APA StyleIwaniak, A., Minkiewicz, P., Pliszka, M., Mogut, D., & Darewicz, M. (2020). Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods, 9(7), 965. https://doi.org/10.3390/foods9070965