Application of 1H and 13C NMR Fingerprinting as a Tool for the Authentication of Maltese Extra Virgin Olive Oil
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. 1H and 13C NMR Spectra Acquisition
2.3. Data Analysis
3. Results and Discussion
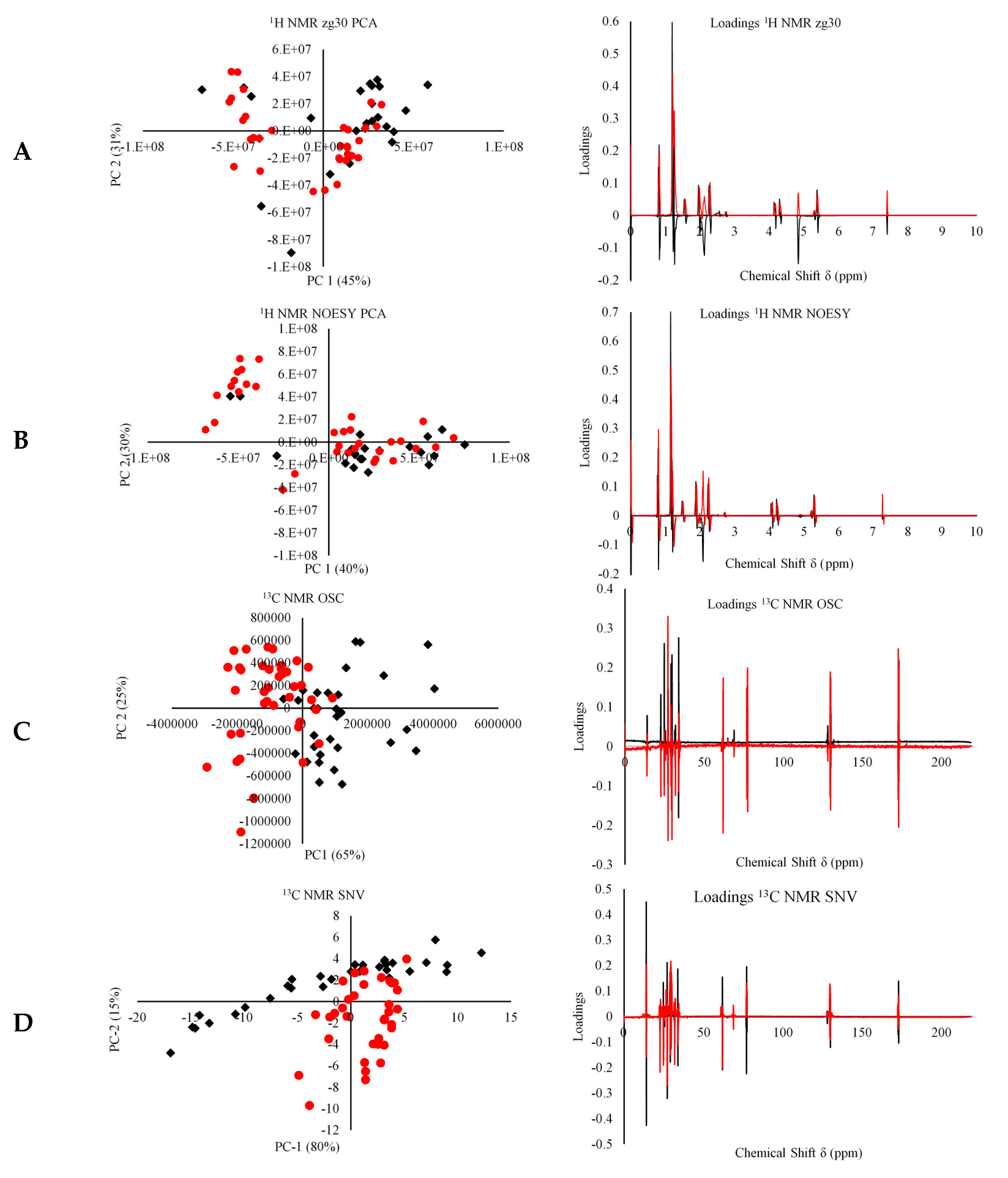
3.1. Geographical Classification of EVOO Using NMR Spectroscopy
3.2. Application of PLS-DA for the Discrimination of Maltese EVOOs
3.3. Whole 1H and 13C-NMR Modeling Using Feed-Forward Predictive Artificial Neural Networks
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Community Commission Regulation (EEC) no. 2568/1991 on the characteristics of olive and olive pomace oils and their analytical methods. Off. J. Eur. Communities 1991, L248, 1–83.
- International Olive Council (IOC). Trade Standard Applying to Olive Oils and Olive-Pomace Oils, COI/T.15/NC No. 3/Rev 6. 2011. Available online: http://www.internationaloliveoil.org (accessed on 11 February 2020).
- Ceci, L.N.; Carelli, A.A. Relation between oxidative stability and composition in Argentinian olive oils. J. AOCS 2010, 87, 1189–1197. [Google Scholar] [CrossRef]
- Frankel, E.N. Chemistry of extra virgin olive oil: Adulteration, oxidative stability, and antioxidants. J. Agric. Food Chem. 2010, 58, 5991–6006. [Google Scholar] [CrossRef] [PubMed]
- Canabate-Diaz, B.; Segura Carretero, A.; Fernandez-Gutierrez, A.; Belmonte Vega, A.; Garrido Frenich, A.; Martinez Vidal, J.L. Separation and determination of sterols in olive oil by HPLC-MS. Food Chem. 2007, 102, 593–598. [Google Scholar] [CrossRef]
- Murkovic, M.; Lechmer, S.; Pietzka, A.; Bratacos, M.; Katzoiamnos, E. Analysis of minor components in olive oil. J. Biochem. Biophys. Methods 2004, 61, 155–160. [Google Scholar] [CrossRef]
- Suárez, M.; Macià, A.; Romero, M.P.; Motilva, M.J. Improved liquid chromatography tandem mass spectrometry method for the determination of phenolic compounds in virgin olive oil. J. Chromatogr. A 2008, 1214, 90–99. [Google Scholar] [CrossRef]
- Morales, M.T.; Luna, G.; Aparicio, R. Comparative study of virgin olive oil sensory defects. Food Chem. 2005, 91, 293–301. [Google Scholar] [CrossRef]
- Vlahov, G. Application of NMR to the study of olive oils. Prog. Nucl. Magn. Reson. Spectrosc. 1999, 35, 341–357. [Google Scholar] [CrossRef]
- Sacchi, R.; Patumi, M.; Fontanazza, G.; Barone, P.; Fiordiponti, P.; Mannina, L.; Segre, A.L. A high-field 1H nuclear magnetic resonance study of the minor components in virgin olive oils. J. AOCS 1996, 23, 747–758. [Google Scholar]
- Guillen, M.D.; Ruiz, A. High resolution 1H nuclear magnetic resonance in the study of edible oils and fats. Trends Food Sci. Technol. 2001, 12, 328–338. [Google Scholar] [CrossRef]
- Hidalgo, F.J.; Gómez, G.; Navarro, J.L.; Zamora, R. Oil stability prediction by high-resolution 13C nuclear magnetic resonance spectroscopy. J. Agric. Food Chem. 2002, 50, 5825–5831. [Google Scholar] [CrossRef] [PubMed]
- Mannina, L.; Marini, F.; Gobbino, M.; Sobolev, A.P.; Capitani, D. NMR and chemometrics in tracing European olive oils: The case study of Ligurian samples. Talanta 2010, 80, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Mannina, L.; Patumi, M.; Proietti, N.; Bassi, D.; Segre, A.L. Geographical characterization of Italian extra virgin olive oils using high field 1H-NMR spectroscopy. J. Agric. Food Chem. 2001, 49, 2687–2696. [Google Scholar] [CrossRef] [PubMed]
- Spyros, A.; Dais, P. Application of 31P NMR spectroscopy in food Analysis, Quantitative determination of the mono- and di-glyceride composition of olive oils. J. Agric. Food Chem. 2000, 48, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Rezzi, S.; Axelson, D.E.; Heberger, K.; Reniero, F.; Mariani, C.; Guillou, C. Classification of olive oils using high throughput flow 1H-NMR fingerprinting with principal component analysis, linear discriminant analysis and probabilistic neural networks. Anal. Chim. Acta 2005, 55, 13–24. [Google Scholar] [CrossRef]
- Alonso-Salces, R.M.; Héberger, K.; Holland, M.V.; Moreno-Rojas, J.M.; Mariani, C.; Bellan, G.; Guillou, C. Multivariate analysis of NMR fingerprint of the unsaponifiable fraction of virgin olive oils for authentication purposes. Food Chem. 2010, 118, 956–965. [Google Scholar] [CrossRef]
- Longobardi, F.; Ventrella, A.; Napoli, C.; Humpfer, E.; Schütz, B.; Schäfer, H.; Sacco, A. Classification of olive oils according to geographical origin by using 1H NMR fingerprinting combined with multivariate analysis. Food Chem. 2012, 130, 177–183. [Google Scholar] [CrossRef]
- Preedy, V.R.; Watson, R.R. Olives and Olive Oil in Health and Disease Prevention; Elsevier Academic Press: Amsterdam, The Netherlans, 2010. [Google Scholar]
- Lia, F.; Farrugia, C.; Zammit-Mangion, M. Application of Elemental Analysis via Energy Dispersive X-Ray Fluorescence (ED-XRF) for the Authentication of Maltese Extra Virgin Olive Oil. Agriculture 2020, 10, 71. [Google Scholar] [CrossRef]
- Lia, F.; Farrugia, C.; Zammit-Mangion, M. A First Description of the Phenolic Profile of EVOOs from the Maltese Islands Using SPE and HPLC: Pedo-Climatic Conditions Modulate Genetic Factors. Agriculture 2019, 9, 107. [Google Scholar] [CrossRef]
- Lia, F.; Formosa, J.P.; Zammit-Mangion, M.; Farrugia, C. The First Identification of the Uniqueness and Authentication of Maltese Extra Virgin Olive Oil Using 3D-Fluorescence Spectroscopy Coupled with Multi-Way Data Analysis. Foods 2020, 9, 498. [Google Scholar] [CrossRef]
- Merchak, N.; Bacha, E.L.; Bou Khouzam, R.; Rizk, T.; Akoka, S.; Bejjani, J. Geoclimatic, morphological, and temporal effects on Lebanese olive oils composition and classification: A 1H NMR metabolomic study. Food Chem. 2017, 217, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.E. Standardization of chemical shifts of TMS and solvent signals in NMR solvents. Magn. Reson. Chem. 2006, 44, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, R.; Addeo, F.; Paolillo, L. 1H and13C NMR of virgin olive oil. An overview. Magn. Reson. Chem. 1997, 35, 133–145. [Google Scholar] [CrossRef]
- Nam, A.; Bighelli, A.; Tomi, F. Quantification of Squalene in Olive Oil Using 13C Nuclear Magnetic Resonance Spectroscopy. Magnetochemistry 2017, 3, 34. [Google Scholar] [CrossRef]
- Sacco, A.; Brescia, M.A.; Liuzzi, V.; Reniero, F.; Guillou, C.; Ghelli, S.; van der Meer, P. Characterization of Italian olive oils based on analytical and nuclear magnetic resonance determinations. JAOCS 2000, 77, 619–625. [Google Scholar] [CrossRef]
- Vlahov, G.; Del Re, P.; Simone, N. Determination of geographical origin of olive oils using 13C nuclear magnetic resonance spectroscopy. I—Classification of olive oils of the Puglia region with denomination of protected origin. J. Agric. Food Chem. 2003, 51, 5612–5615. [Google Scholar] [CrossRef]
- Shaw, A.D.; di Camillo, A.; Vlahov, G.; Jones, A. Discrimination of the variety and region of origin of extra virgin olive oil using 13C NMR and multivariate calibration with variable reduction. Anal. Chim. Acta 1997, 348, 357–374. [Google Scholar] [CrossRef]
- Alonso-Salces, R.M.; Moreno-Rojas, J.M.; Holland, M.V.; Reniero, F.; Guillou, C.; Heberger, K. Virgin Olive Oil Authentication by Multivariate Analyses of 1H NMR Fingerprints and 13C and 2H Data. J. Agric. Food Chem. 2010, 58, 5586–5596. [Google Scholar] [CrossRef]
- McKenzie, J.M.; Koch, K.R. Rapid analysis of major components and potential authentication of South African olive oils by quantitative 13C nuclear magnetic resonance. S. Afr. J. Sci. 2004, 100, 349–354. [Google Scholar]
- Harwood, J.L.; Aparicio, R. Handbook of Olive Oil: Analysis and Properties; Aspen. Henna: Gaithersburg, MD, USA, 2000. [Google Scholar]
- D’Imperio, M.; Dugo, G.; Alfa, M.; Mannina, L.; Segre, A.L. Statistical analysis on Sicilian olive oils. Food Chem. 2007, 102, 956–965. [Google Scholar] [CrossRef]
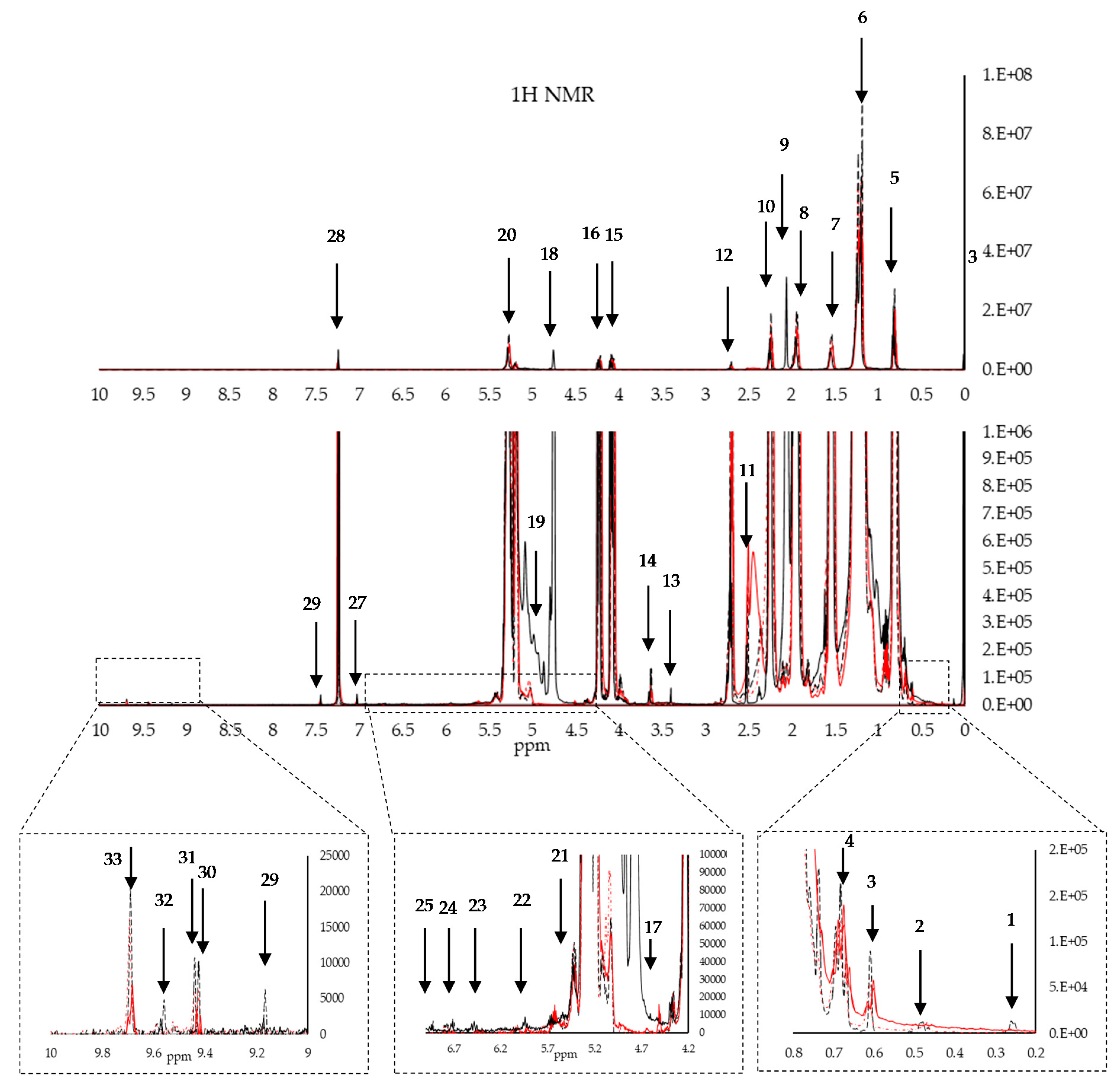
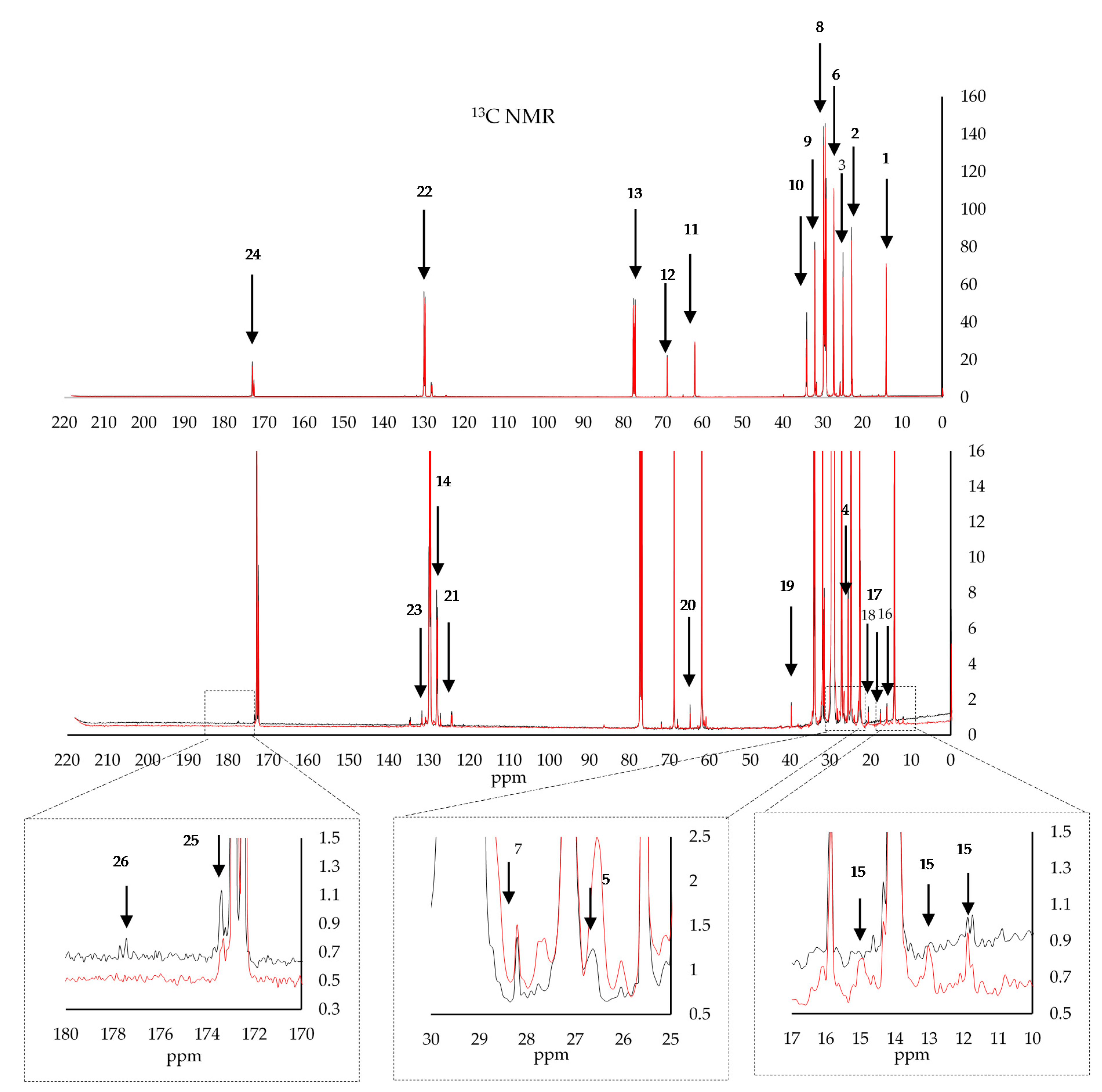
| 1H NMR | |||||
|---|---|---|---|---|---|
| Chemical Shift | Compound Functional Group | Chemical Shift | Compound Functional Group | ||
| 1 | 0.29 | -CH2-(cyclopropanic ring) cycloartenol | 17 | 4.53 | Terpene |
| 2 | 0.54 | -CH2-(cyclopropanic ring) cycloartenol | 18 | 4.65 | Terpene |
| 3 | 0.62 | -CH3(C18-steroid group) β-sitosterol | 19 | 4.95 | Terpene |
| 4 | 0.69 | -CH3(C18-steroid group) β-sitosterol | 20 | 5.28 | >CHOCOR (glyceryl group) |
| 5 | 0.81 | -CH3(acyl group) | 21 | 5.55 | Unk 2-Tocopherols |
| 6 | 1.19 | -(CH2)n-(acyl group) | 22 | 5.91 | -CH=CH-CH=CH-(cis, trans conjugated dienediene system) |
| 7 | 1.54 | -OCO-CH2-CH2-(acyl group) | 23 | 6.56 | -CH=CH-CH=CH-(cis, trans conjugated dienediene system) |
| 8 | 1.95 | -CH2-CH=CH-(acyl group) | 24 | 6.72 | -Ph-H (phenolic ring) |
| 9 | 2.08 | -CH2-CH=CH-(acyl group) | 25 | 6.95 | -Ph-H (phenolic ring) |
| 10 | 2.26 | CH-CH2-CH=(acyl group) | 26 | 7.02 | Chloroform 13C satellite |
| 11 | 2.54 | CH-CH2-CH=(acyl group) satellite | 27 | 7.24 | Chloroform |
| 12 | 2.71 | CH-CH2-CH=(acyl group) | 28 | 7.44 | Chloroform 13C satellite |
| 13 | 3.39 | Unk 1-alcohol | 29 | 9.17 | Unk 4-hydrocarbon |
| 14 | 3.71 | -CH2OCOR (glyceryl group) | 30 | 9.46 | Unk 5-hydrocarbon |
| 15 | 4.10 | -CH2OCOR (glyceryl group) | 31 | 9.47 | Unk 5-hydrocarbon |
| 16 | 4.22 | -CH2OCOR (glyceryl group) | 32 | 9.58 | Unk 6-hydrocarbon |
| 33 | 9.70 | Hexanal | |||
| 13C NMR | |||||
|---|---|---|---|---|---|
| Chemical Shift | Compound Functional Group | Chemical Shift | Compound Functional Group | ||
| 1 | 14 | C18(ω1) terminal carbon of fatty acyl chain | 14 | 128.01 129.82 | C9, C10 oleoyl unsaturated carbons between 2- and 1(3) of glycerol |
| 2 | 22.64 | C17(ω2) penultimate carbons from the fatty acyl chains | 15 | 11.97,13.12,14.95 | Unk 1 possibly being attributed to waxes |
| 3 | 24.74 | C3 methylenic group in β position with respect to the carbonylic group | 16 | 15.95 | Unk 2 possibly C8′a and C4′a of tocopherols |
| 4 | 25.53 | C11 Linoleyl Linolenyl | 17 | 17.51 | Unk 3 possibly C12′a of tocopherols |
| 5 | 26.61 | C8 allylic methylenes of sqaulene | 18 | 20.47 | C17(ω2) all acyl chains |
| 6 | 27.12 | C8 allylic carbons of oleoyl chains | 19 | 39.68 | Unk 4-C1 of tocopherol |
| 7 | 28.6 | C12 allylic methylenes of sqaulene | 20 | 64.89 | Unk 5 possibly C2 of elenolic acid derivative of tyrosol or hydroxytyrosol |
| 8 | 29.28 | C4–C7, C12–C15, C8–C15, C8–C13 methylenic groups in fatty acid central chain | 21 | 124.40 | C3′, C7′, C11′ of tocopherols |
| 9 | 31.88 | C16 methylenic acylic chains ω | 22 | 131.70 | C9 Linoleyl and linolenyl, C13 Linoleyl |
| 10 | 33.91 | C2, sn-2 acyl chains | 23 | 134.80 | C4′, C8′ of tocopherols |
| 11 | 61.93 | CH2O-1(3) glycerol carbons of triglycerides | 24 | 172.8 | C1, sn-2 2-glycerol chain |
| 12 | 68.86 | CH2O-2-glycerol carbon of triglycerides resonates | 25 | 173.2 | C1, sn-1,3 1(3) glycerol chain positions |
| 13 | 77.39 | CDCl3 Solvent | 26 | 177.92 | Unk 6-COOCH3 of elenolic acid |
| (a) | ||||
|---|---|---|---|---|
| Pretreatment | zg30 1H NMR | |||
| Whole Spectrum | VIP > 0.8 | |||
| % Accuracy | % Predictability | % Accuracy | % Predictability | |
| Raw | 77.59 | 27.27 | 82.76 | 45.45 |
| Normalised | 91.38 | 63.64 | 94.83 | 90.91 |
| Q Norm | 93.10 | 72.73 | 94.83 | 72.73 |
| Detrend | 70.69 | 36.36 | 68.97 | 36.36 |
| Deresolve | 87.93 | 63.64 | 82.76 | 45.45 |
| SNV | 93.10 | 81.82 | 60.34 | 36.36 |
| MSC | 91.38 | 81.82 | 67.24 | 63.64 |
| OSC | 72.41 | 45.45 | 94.83 | 72.73 |
| Savitzky-Golay | 74.14 | 54.55 | 98.28 | 90.91 |
| 1st Derivative | 68.97 | 45.45 | 94.83 | 72.73 |
| 2nd Derivative | 77.59 | 63.64 | 93.10 | 63.64 |
| Pretreatment | NOESY 1H NMR | |||
| Whole Spectrum | VIP > 0.8 | |||
| % Accuracy | % Predictability | % Accuracy | % Predictability | |
| Raw | 82.76 | 45.45 | 93.10 | 75.00 |
| Normalised | 94.83 | 90.91 | 94.83 | 83.33 |
| Q Norm | 94.83 | 72.73 | 96.55 | 91.67 |
| Detrend | 68.97 | 36.36 | 74.14 | 66.67 |
| Deresolve | 82.76 | 45.45 | 94.83 | 83.33 |
| SNV | 60.34 | 36.36 | 70.69 | 75.00 |
| MSC | 67.24 | 63.64 | 68.97 | 75.00 |
| OSC | 94.83 | 72.73 | 96.55 | 83.33 |
| Savitzky-Golay | 98.28 | 90.91 | 89.66 | 75.00 |
| 1st Derivative | 94.83 | 72.73 | 93.10 | 91.67 |
| 2nd Derivative | 93.10 | 63.64 | 93.10 | 91.67 |
| (b) | ||||
| Pretreatment | 13C NMR | |||
| Whole Spectrum | VIP > 0.8 | |||
| % Accuracy | % Predictability | % Accuracy | % Predictability | |
| Raw | 100.00 | 73.33 | 100.00 | 80.00 |
| Normalised | 100.00 | 86.68 | 94.64 | 100.00 |
| Q Norm | 100.00 | 80.00 | 100.00 | 100.00 |
| Detrend | 100.00 | 66.67 | 100.00 | 100.00 |
| Deresolve | 78.57 | 73.33 | 91.07 | 100.00 |
| SNV | 100.00 | 86.67 | 100.00 | 100.00 |
| MSC | 100.00 | 86.67 | 100.00 | 100.00 |
| OSC | 100.00 | 100.00 | 100.00 | 100.00 |
| Savitzky-Golay | 100.00 | 73.33 | 100.00 | 100.00 |
| 1st Derivative | 100.00 | 80.00 | 100.00 | 100.00 |
| 2nd Derivative | 100.00 | 86.67 | 100.00 | 100.00 |
| ANN | ||||||
|---|---|---|---|---|---|---|
| Pretreatment | Holdback | CV-10 | Excluded Row | |||
| Training | Validation | Training | Validation | Training | Validation | |
| (a) | ||||||
| zg30 1H NMR | ||||||
| Raw | 81.03 | 81.82 | 96.55 | 81.82 | 86.21 | 90.91 |
| Normalised | 94.83 | 81.82 | 94.83 | 100 | 81.03 | 63.64 |
| Q Norm | 98.28 | 90.91 | 98.28 | 90.91 | 82.76 | 81.82 |
| Detrend | 77.59 | 54.55 | 91.38 | 90.91 | 75.86 | 45.45 |
| Deresolve | 91.38 | 90.91 | 93.1 | 90.91 | 79.31 | 81.82 |
| SNV | 96.55 | 90.91 | 98.28 | 100.00 | 74.14 | 72.73 |
| MSC | 98.28 | 100.00 | 96.55 | 90.91 | 93.10 | 90.91 |
| OSC | 77.59 | 36.36 | 89.66 | 63.64 | 84.48 | 81.82 |
| 2nd Derivative | 96.55 | 90.91 | 98.28 | 100.00 | 86.21 | 45.45 |
| 1st Derivative | 84.48 | 90.91 | 98.28 | 90.91 | 81.03 | 63.64 |
| Savitzky-Golay | 91.38 | 81.82 | 98.28 | 90.91 | 85.00 | 90.91 |
| NOESY 1H NMR | ||||||
| Raw | 93.33 | 91.67 | 93.33 | 91.67 | 93.33 | 91.67 |
| Normalised | 95.00 | 91.67 | 95.00 | 100.00 | 93.33 | 100.00 |
| Q Norm | 98.33 | 100.00 | 98.33 | 100.00 | 93.33 | 100.00 |
| Detrend | 70.00 | 83.33 | 96.67 | 91.67 | 95.00 | 83.33 |
| Deresolve | 96.67 | 100.00 | 96.67 | 100.00 | 91.67 | 83.33 |
| SNV | 93.33 | 91.67 | 98.33 | 100.00 | 98.33 | 100.00 |
| MSC | 93.33 | 100.00 | 93.33 | 83.33 | 95.00 | 91.67 |
| OSC | 81.67 | 75.00 | 91.67 | 75.00 | 90.00 | 91.67 |
| 2nd Derivative | 96.67 | 100 | 98.33 | 100.00 | 96.67 | 91.67 |
| 1st Derivative | 93.33 | 91.67 | 98.33 | 100.00 | 93.33 | 100.00 |
| Savitzky-Golay | 88.33 | 100 | 98.33 | 100.00 | 90.00 | 91.67 |
| (b) | ||||||
| 13C NMR | ||||||
| Raw | 83.93 | 60.00 | 100.00 | 80.00 | 92.62 | 80.00 |
| Normalised | 91.07 | 46.67 | 100.00 | 100.00 | 100.00 | 100.00 |
| Q Norm | 78.57 | 80.00 | 98.21 | 86.67 | 98.21 | 100.00 |
| Detrend | 91.07 | 80.00 | 100.00 | 80.00 | 98.21 | 86.67 |
| Deresolve | 78.57 | 73.33 | 100.00 | 40.00 | 87.50 | 86.67 |
| SNV | 94.64 | 86.67 | 98.21 | 93.33 | 92.86 | 100.00 |
| MSC | 80.36 | 53.33 | 100.00 | 73.33 | 100.00 | 86.67 |
| OSC | 75.00 | 66.67 | 75.00 | 66.67 | 100.00 | 73.33 |
| 2nd Derivative | 85.71 | 40.00 | 100.00 | 66.67 | 96.43 | 86.67 |
| 1st Derivative | 94.64 | 73.33 | 100.00 | 46.67 | 100.00 | 73.33 |
| Savitzky-Golay | 83.93 | 40.00 | 100.00 | 73.33 | 83.93 | 93.33 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lia, F.; Vella, B.; Zammit Mangion, M.; Farrugia, C. Application of 1H and 13C NMR Fingerprinting as a Tool for the Authentication of Maltese Extra Virgin Olive Oil. Foods 2020, 9, 689. https://doi.org/10.3390/foods9060689
Lia F, Vella B, Zammit Mangion M, Farrugia C. Application of 1H and 13C NMR Fingerprinting as a Tool for the Authentication of Maltese Extra Virgin Olive Oil. Foods. 2020; 9(6):689. https://doi.org/10.3390/foods9060689
Chicago/Turabian StyleLia, Frederick, Benjamin Vella, Marion Zammit Mangion, and Claude Farrugia. 2020. "Application of 1H and 13C NMR Fingerprinting as a Tool for the Authentication of Maltese Extra Virgin Olive Oil" Foods 9, no. 6: 689. https://doi.org/10.3390/foods9060689
APA StyleLia, F., Vella, B., Zammit Mangion, M., & Farrugia, C. (2020). Application of 1H and 13C NMR Fingerprinting as a Tool for the Authentication of Maltese Extra Virgin Olive Oil. Foods, 9(6), 689. https://doi.org/10.3390/foods9060689





