Smart Online Coffee Roasting Process Control: Modelling Coffee Roast Degree and Brew Antioxidant Capacity for Real-Time Prediction by Resonance-Enhanced Multi-Photon Ionization Mass Spectrometric (REMPI-TOFMS) Monitoring of Roast Gases
Abstract
1. Introduction
2. Materials and Methods
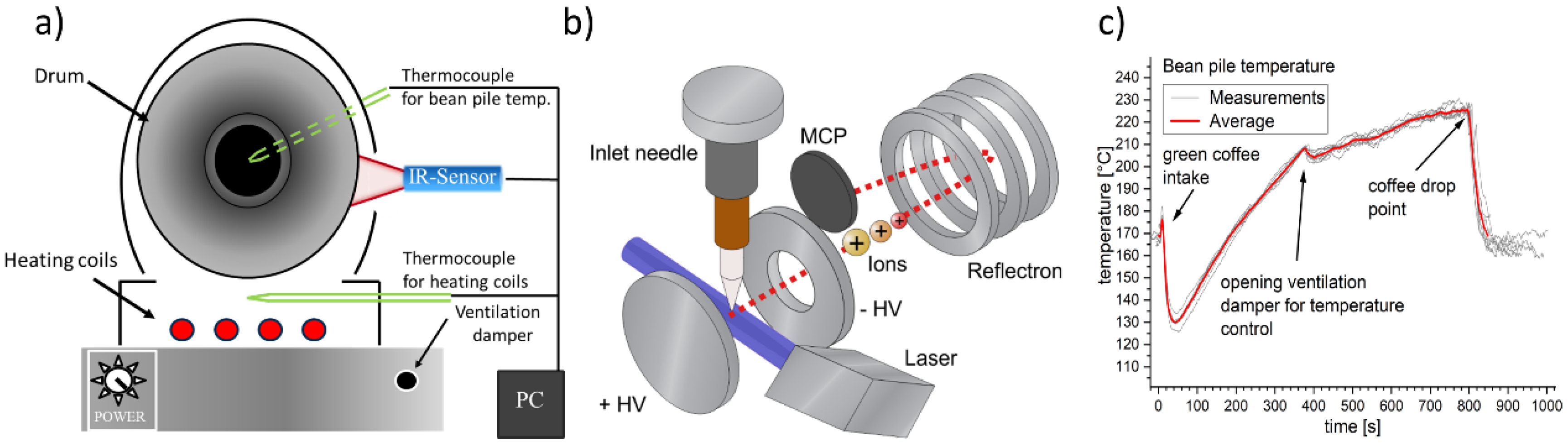
2.1. Drum Roaster and Coffee Beans
2.2. Resonance-Enhanced Multi-Photon Ionization Time-of-Flight Mass Spectrometry (REMPI-TOFMS)
2.3. Measurmeent of Bean Color by Colorette
2.4. Measurement of Coffee Brew Antioxidant Capacity by Folin–Ciocalteu (FC) Assay
2.5. Data Analysis
2.5.1. Data Pretreatment
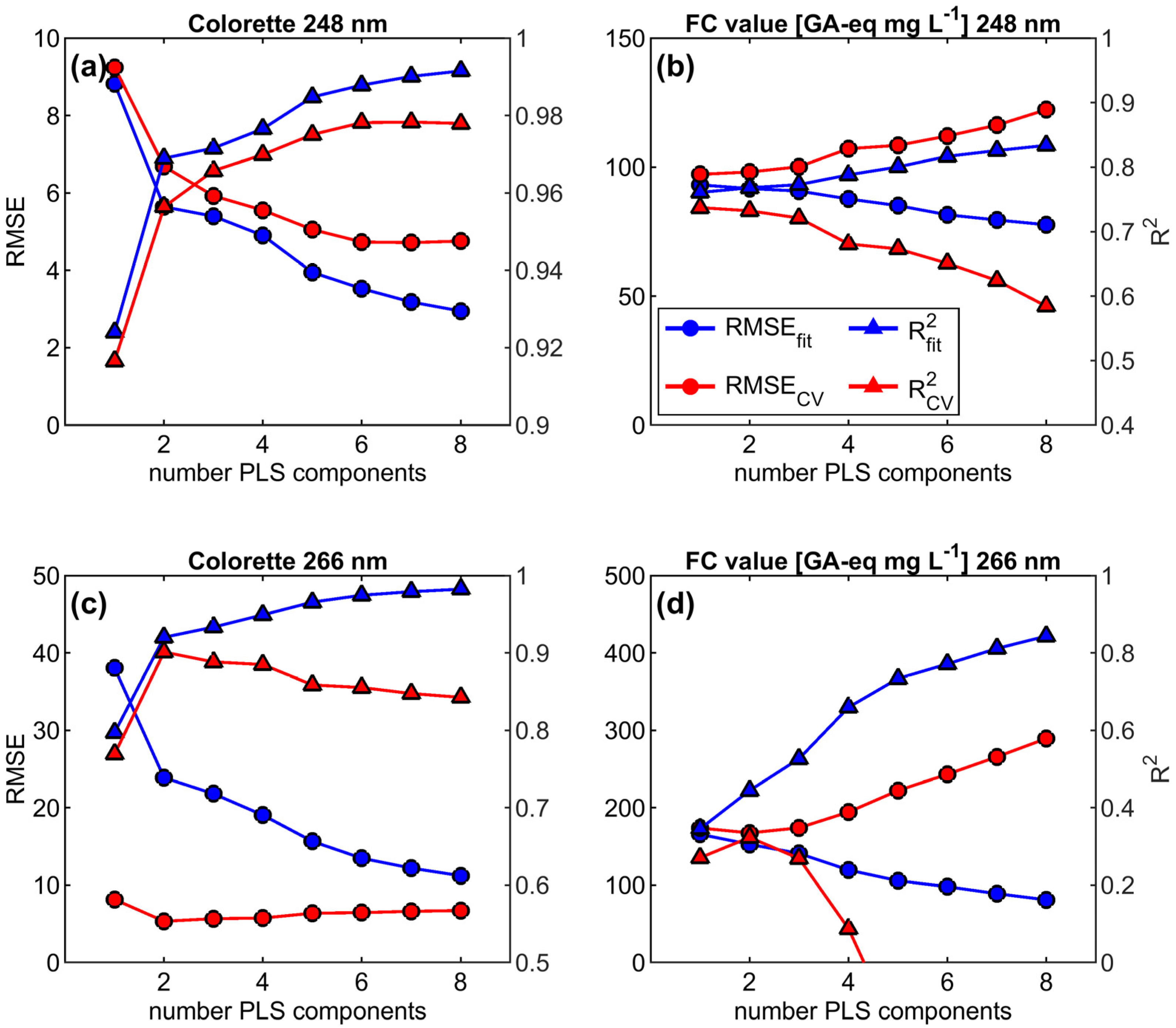
2.5.2. PLS Regression Modelling and Optimization
3. Results and Discussion
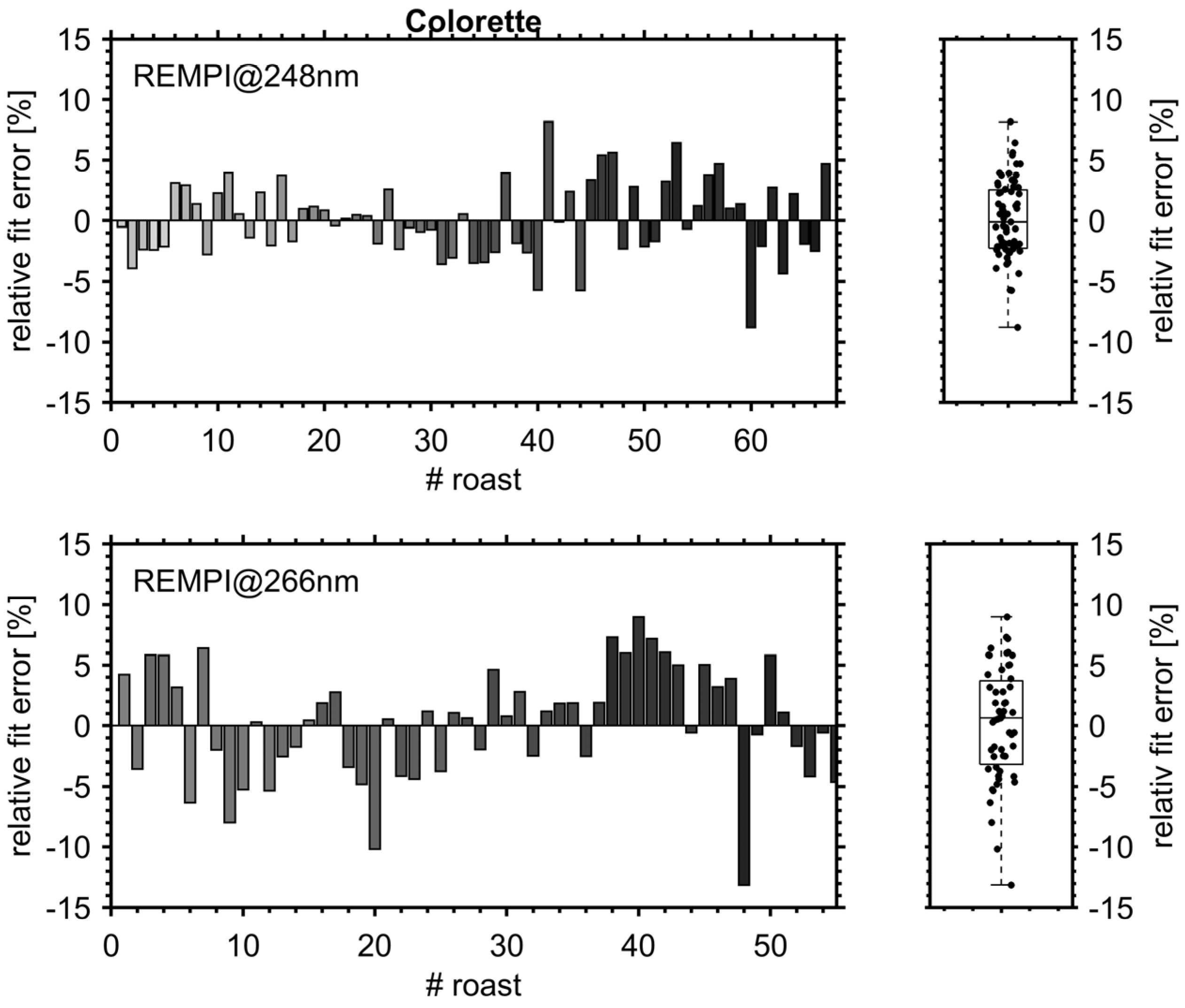
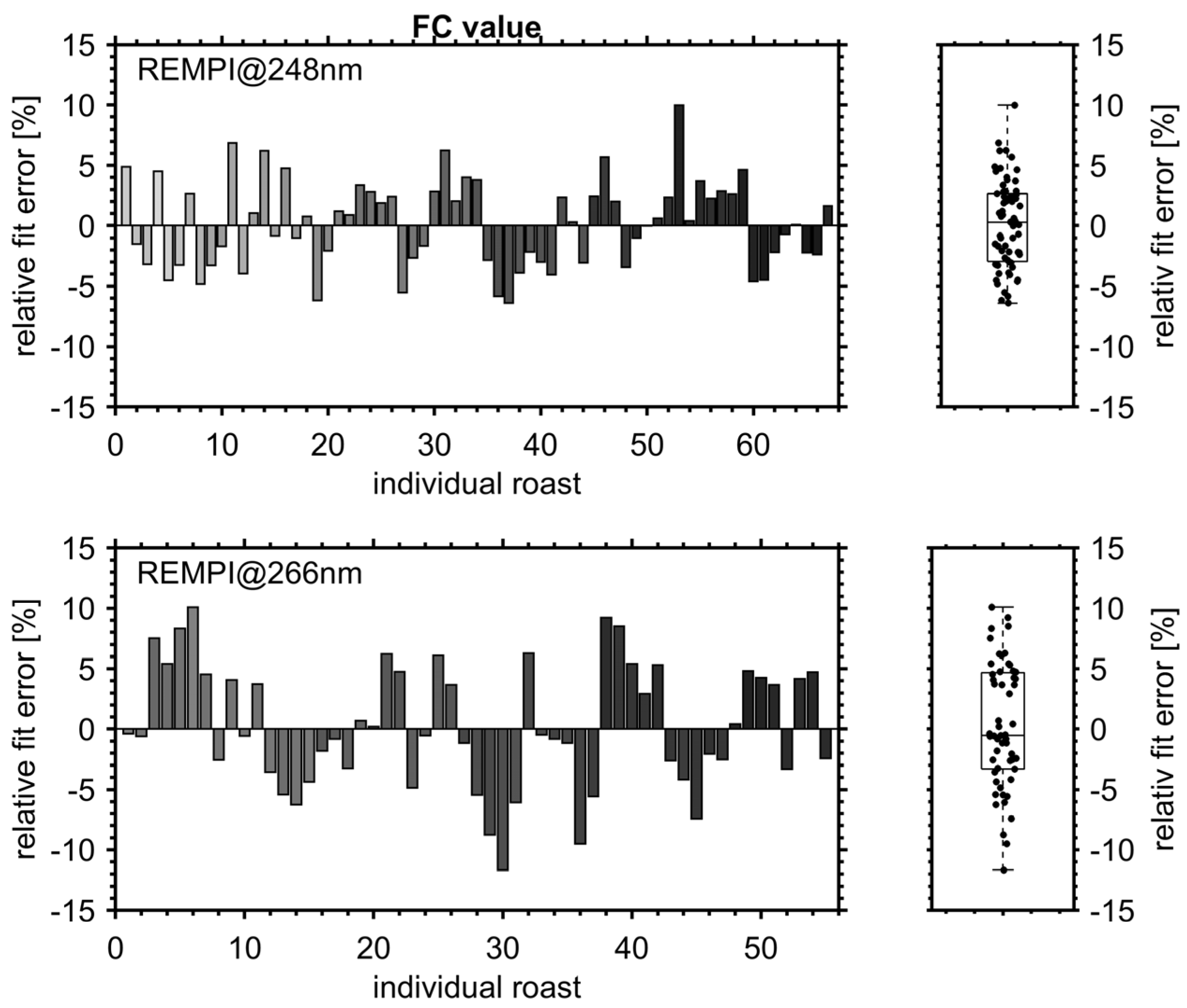
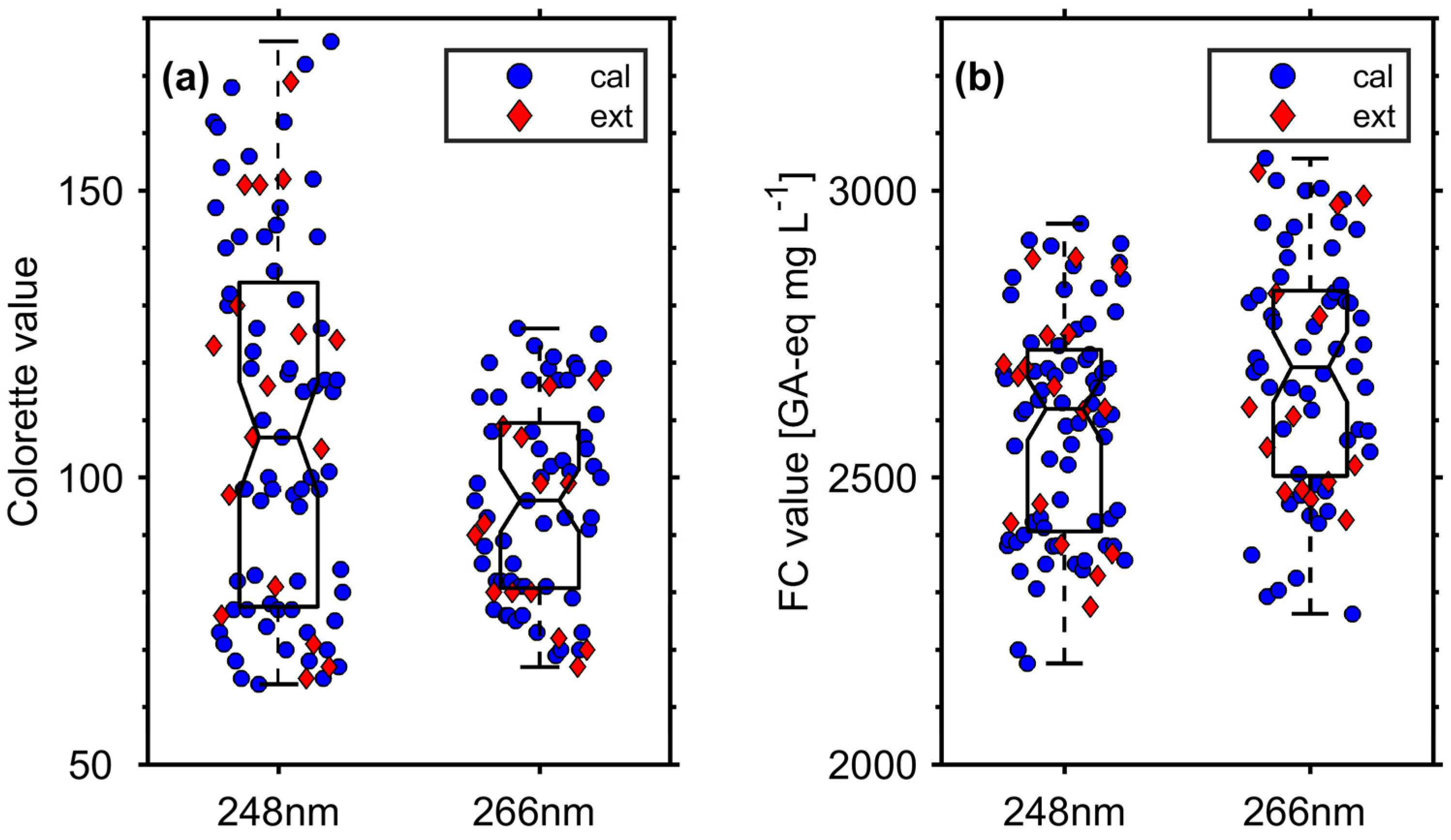
3.1. Roast Degree and FC Value of Roasted Coffee Beans
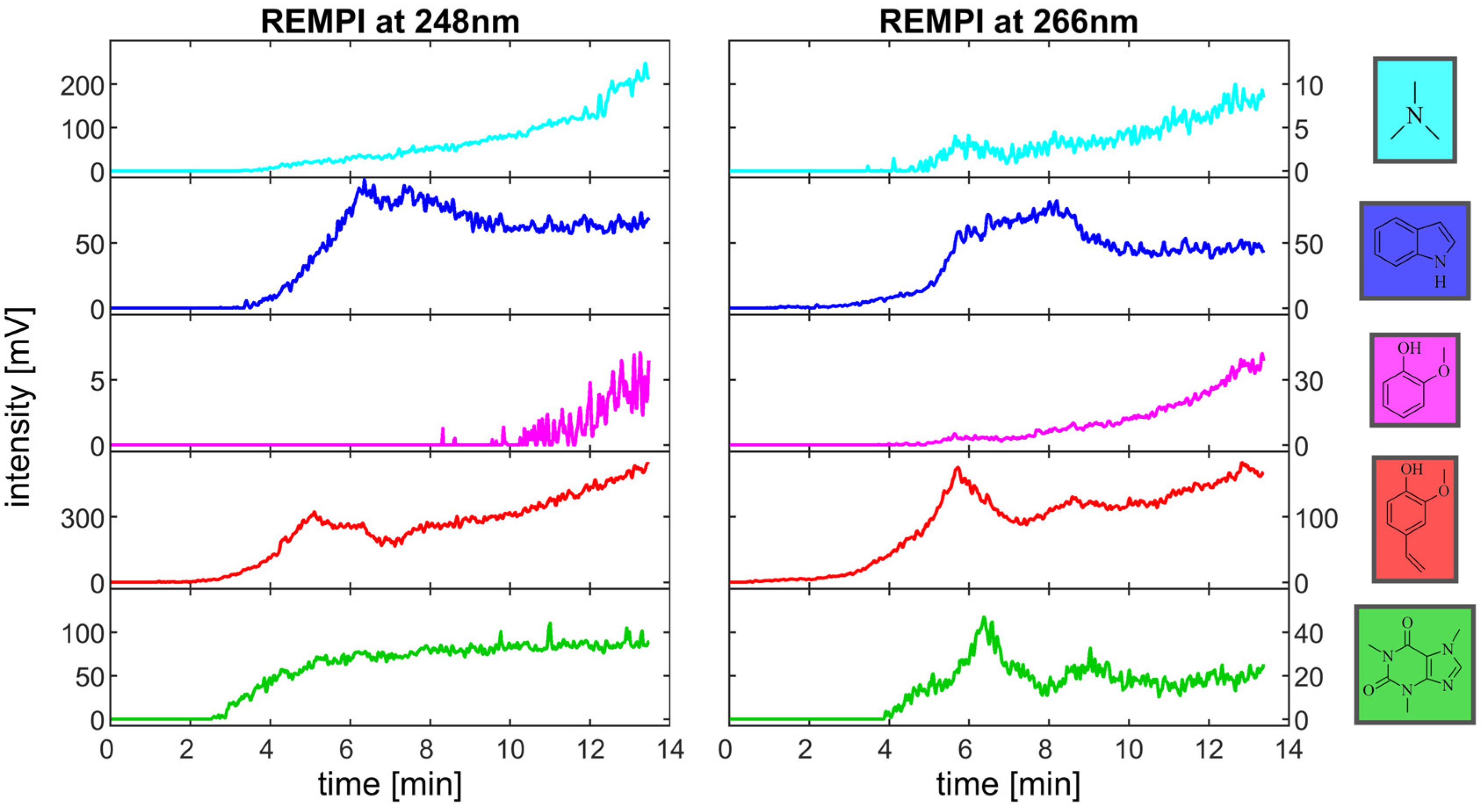
3.2. Components in the Roast Gas and Their Temporal Evolution
3.3. PLS Prediction Models for Roast Degree (Colorette) and Antioxidant Capacity (FC Value)
3.3.1. Figures of Merit
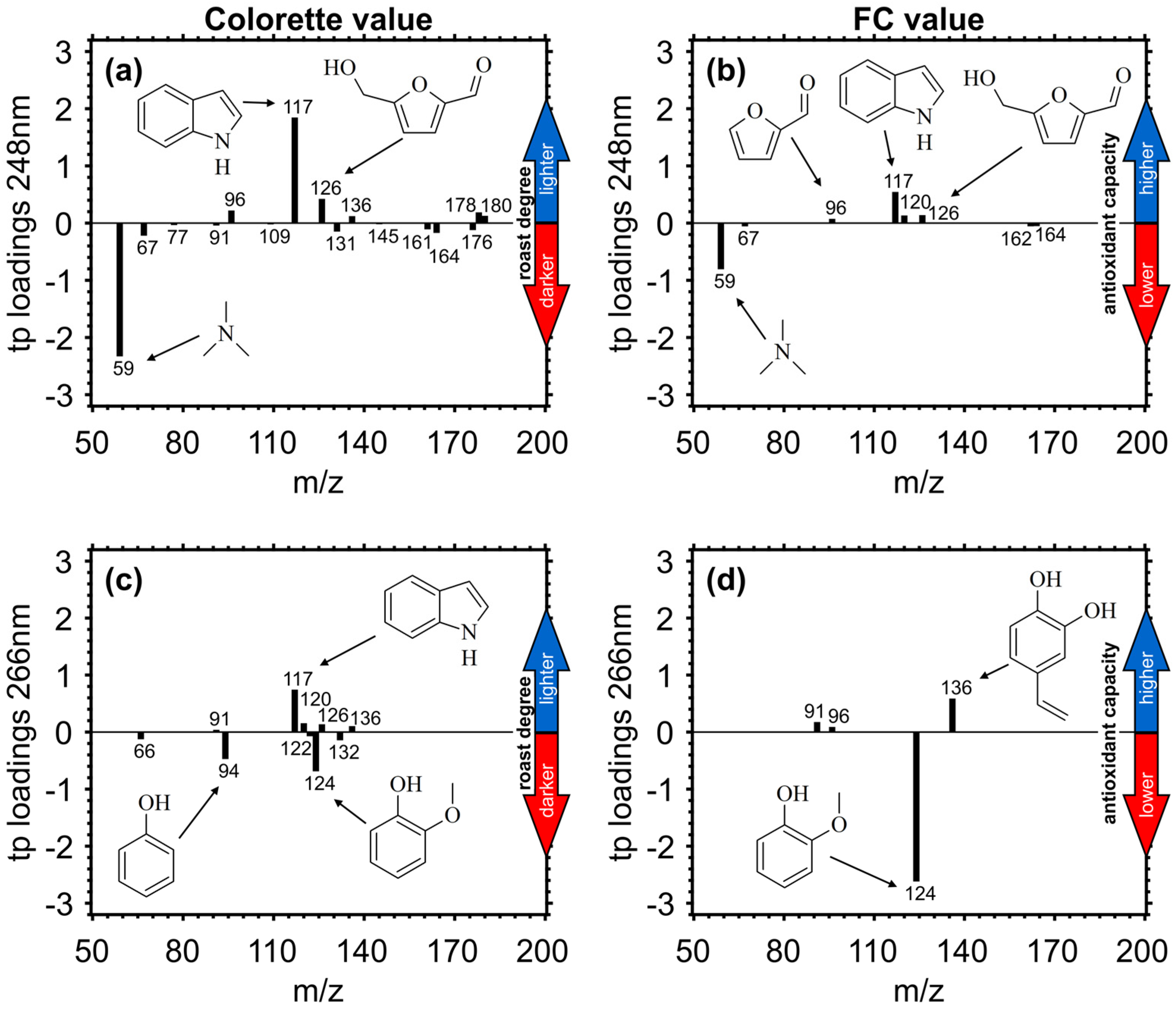
3.3.2. Target Projection (TP) Loadings
3.3.3. Toward Online Prediction of Colorette and FC Values in Real-Time
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
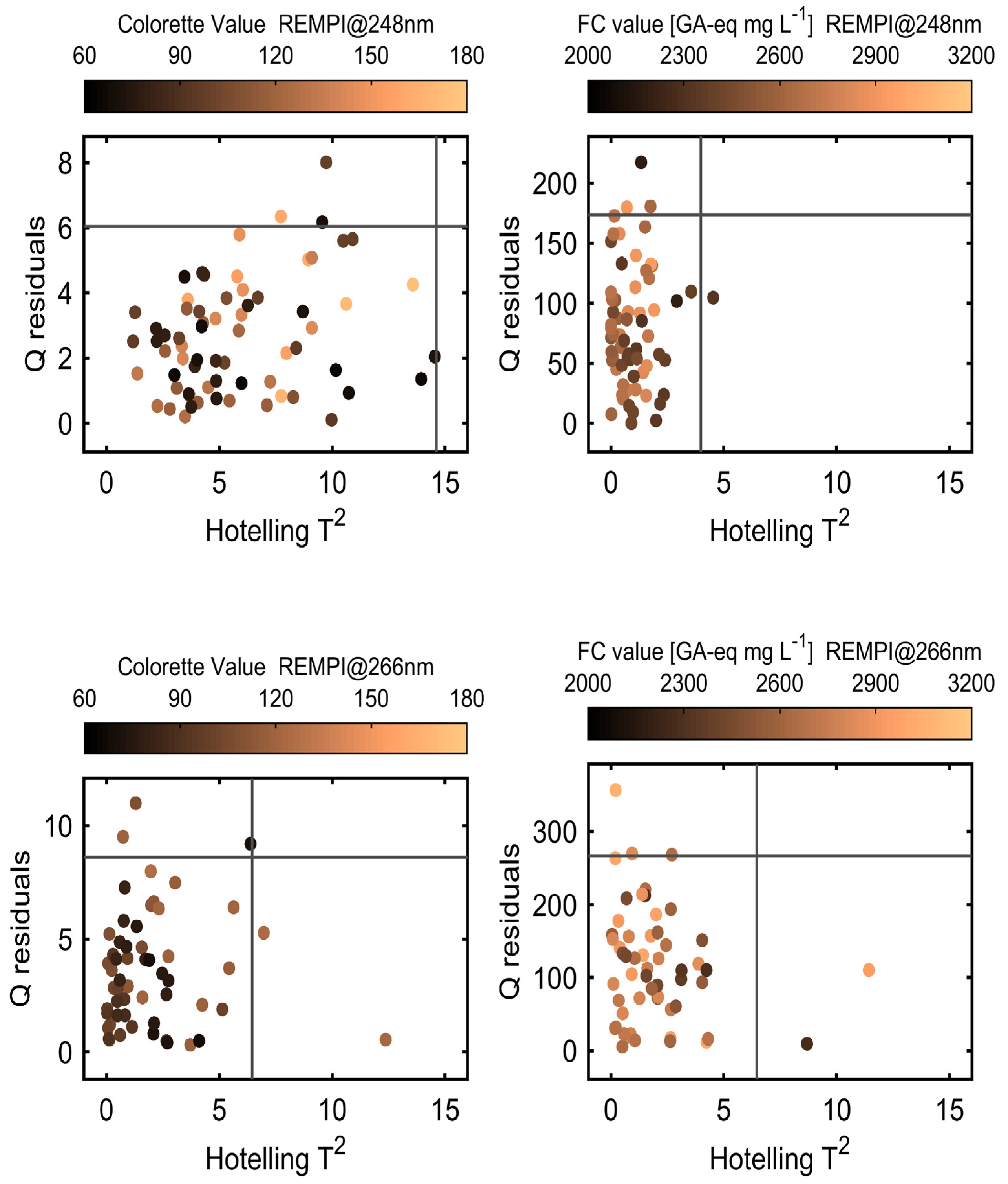
Appendix A
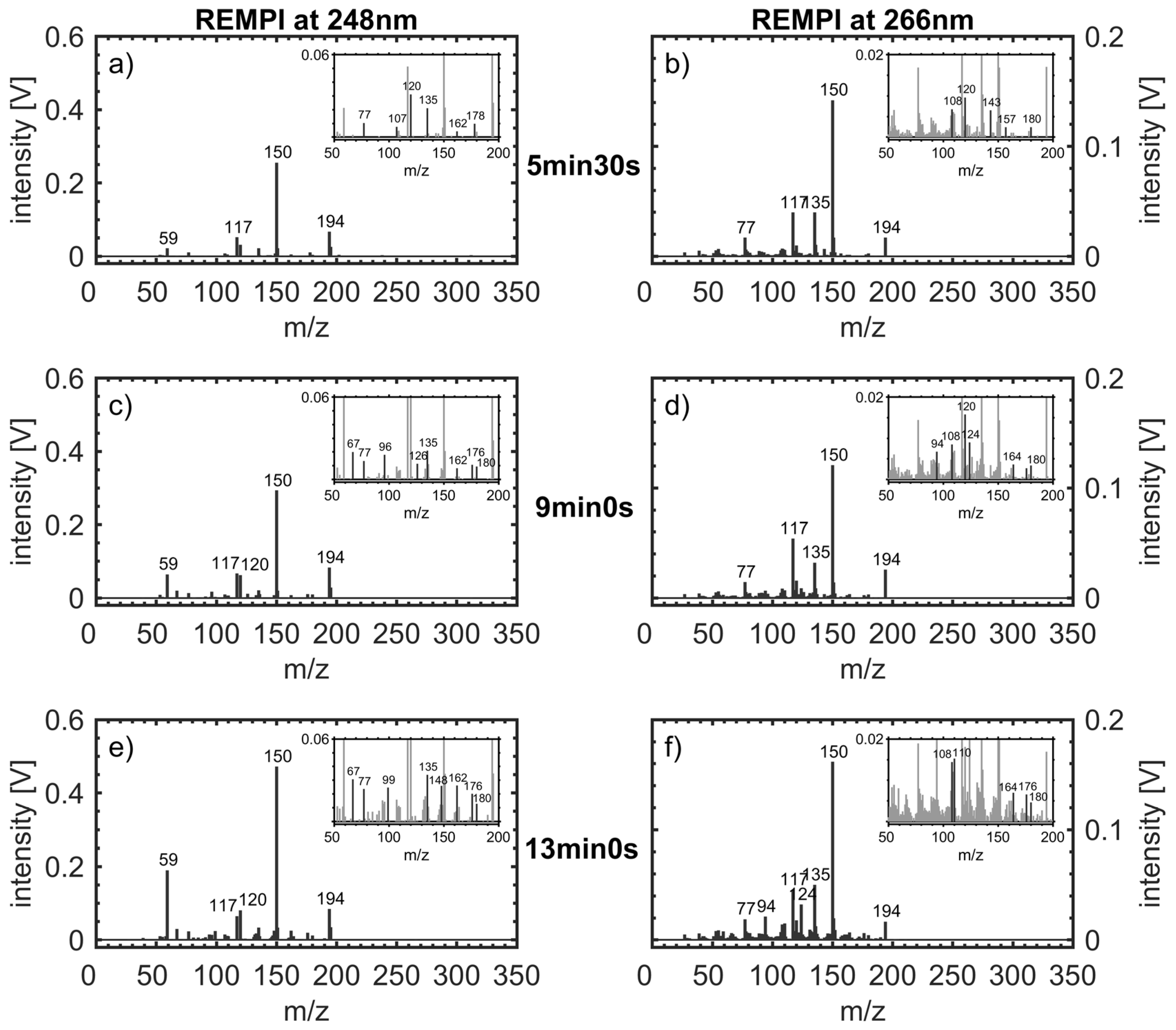
| m/z | Structure Assignment | ||
|---|---|---|---|
| 59 | C3-amines | 126 | hydroxymethylfurfural |
| 66 | fragment of phenolic species | 131 | unknown fragment |
| 67 | pyrrole | 132 | cinnamaldehyde, vinylbenzaldehyde |
| 77 | fragment of aromatic species | 135 | fragment from vinylguiacol |
| 91 | fragment of alkylated aromatics | 136 | vinyldihydroxybenzene |
| 94 | phenol | 143 | unknown fragment |
| 96 | furfural | 148 | 2,2’-methylen-bis-furan |
| 99 | succinimide, methylthiazole | 150 | vinylguaiacol |
| 107 | fragment of vinylguaiacol | 157 | unknown fragment |
| 108 | methylphenol | 162 | dihydroxy cinnamaldehyde |
| 109 | fragment of guaiacol | 164 | dimethoxystyrene |
| 110 | benzenediol, methylfurfural | 176 | 2,2’-methylen-bis(5-methylfuran) |
| 117 | indole | 178 | difurfurylether |
| 120 | phenylacetaldehyde | 180 | caffeic acid |
| 122 | dimethylphenol | 194 | caffeine, ferulic acid |
| 124 | guaiacol, methylctechol |
Appendix B
Appendix C
References
- Sunarharum, W.B.; Williams, D.J.; Smyth, H.E. Complexity of coffee flavor: A compositional and sensory perspective. Food Res. Int. 2014, 62, 315–325. [Google Scholar] [CrossRef]
- Ruosi, M.R.; Cordero, C.; Cagliero, C.; Rubiolo, P.; Bicchi, C.; Sgorbini, B.; Liberto, E. A further tool to monitor the coffee roasting process: Aroma composition and chemical indices. J. Agric. Food Chem. 2012, 60, 11283–11291. [Google Scholar] [CrossRef]
- Bressanello, D.; Liberto, E.; Cordero, C.; Rubiolo, P.; Pellegrino, G.; Ruosi, M.R.; Bicchi, C. Coffee aroma: Chemometric comparison of the chemical information provided by three different samplings combined with GC-MS to describe the sensory properties in cup. Food Chem. 2017, 214, 218–226. [Google Scholar] [CrossRef]
- Catelani, T.A.; Santos, J.R.; Páscoa, R.N.M.J.; Pezza, L.; Pezza, H.R.; Lopes, J.A. Real-time monitoring of a coffee roasting process with near infrared spectroscopy using multivariate statistical analysis: A feasibility study. Talanta 2018, 179, 292–299. [Google Scholar] [CrossRef]
- Gloess, A.N.; Vietri, A.; Wieland, F.; Smrke, S.; Schönbächler, B.; López, J.A.S.; Petrozzi, S.; Bongers, S.; Koziorowski, T.; Yeretzian, C. Evidence of different flavour formation dynamics by roasting coffee from different origins: On-line analysis with PTR-ToF-MS. Int. J. Mass Spectrom. 2014, 365–366, 324–337. [Google Scholar] [CrossRef]
- Wieland, F.; Gloess, A.N.; Keller, M.; Wetzel, A.; Schenker, S.; Yeretzian, C. Online monitoring of coffee roasting by proton transfer reaction time-of-flight mass spectrometry (PTR-ToF-MS): Towards a real-time process control for a consistent roast profile. Anal. Bioanal. Chem. 2012, 402, 2531–2543. [Google Scholar] [CrossRef]
- Hertz-Schünemann, R.; Streibel, T.; Ehlert, S.; Zimmermann, R. Looking into individual coffee beans during the roasting process: Direct micro-probe sampling on-line photo-ionisation mass spectrometric analysis of coffee roasting gases. Anal. Bioanal. Chem. 2013, 405, 7083–7096. [Google Scholar] [CrossRef]
- Dorfner, R.; Ferge, T.; Yeretzian, C.; Kettrup, A.; Zimmermann, R. Laser mass spectrometry as on-line sensor for industrial process analysis: Process control of coffee roasting. Anal. Chem. 2004, 76, 1386–1402. [Google Scholar] [CrossRef]
- Czech, H.; Schepler, C.; Klingbeil, S.; Ehlert, S.; Howell, J.; Zimmermann, R. Resolving Coffee Roasting-Degree Phases Based on the Analysis of Volatile Compounds in the Roasting Off-Gas by Photoionization Time-of-Flight Mass Spectrometry (PI-TOFMS) and Statistical Data Analysis: Toward a PI-TOFMS Roasting Model. J. Agric. Food Chem. 2016, 64, 5223–5231. [Google Scholar] [CrossRef]
- Hertz-Schünemann, R.; Dorfner, R.; Yeretzian, C.; Streibel, T.; Zimmermann, R. On-line process monitoring of coffee roasting by resonant laser ionisation time-of-flight mass spectrometry: Bridging the gap from industrial batch roasting to flavour formation inside an individual coffee bean. J. Mass Spectrom. 2013, 48, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Heger, H.-J.; Yeretzian, C.; Nagel, H.; Boesl, U. Application of Laser Ionization Mass Spectrometry for On-line Monitoring of Volatiles in the Headspace of Food Products: Roasting and Brewing of Coffee. Rapid Commun. Mass Spectrom. 1996, 10, 1975–1979. [Google Scholar] [CrossRef]
- Gloess, A.N.; Yeretzian, C.; Knochenmuss, R.; Groessl, M. On-line analysis of coffee roasting with ion mobility spectrometry–mass spectrometry (IMS–MS). Int. J. Mass Spectrom. 2018, 424, 49–57. [Google Scholar] [CrossRef]
- Fischer, M.; Wohlfahrt, S.; Varga, J.; Matuschek, G.; Saraji-Bozorgzad, M.R.; Walte, A.; Denner, T.; Zimmermann, R. Evolution of Volatile Flavor Compounds During Roasting of Nut Seeds by Thermogravimetry Coupled to Fast-Cycling Optical Heating Gas Chromatography-Mass Spectrometry with Electron and Photoionization. Food Anal. Methods 2017, 10, 49–62. [Google Scholar] [CrossRef]
- Yashin, A.; Yashin, Y.; Wang, J.Y.; Nemzer, B. Antioxidant and Antiradical Activity of Coffee. Antioxidants 2013, 2, 230–245. [Google Scholar] [CrossRef]
- Shen, T.; Han, X.-Z.; Wang, X.-N.; Fan, P.-H.; Ren, D.-M.; Lou, H.-X. Protective Effects of Dietary Polyphenols in Human Diseases and Mechanisms of Action. In Nutritional Antioxidant Therapies: Treatments and Perspectives; Al-Gubory, K.H., Laher, I., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 307–345. ISBN 978-3-319-67623-4. [Google Scholar]
- Del Rio, D.; Rodriguez-Mateos, A.; Spencer, J.P.E.; Tognolini, M.; Borges, G.; Crozier, A. Dietary (poly)phenolics in human health: Structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 2013, 18, 1818–1892. [Google Scholar] [CrossRef]
- Bøhn, S.K.; Blomhoff, R.; Paur, I. Coffee and cancer risk, epidemiological evidence, and molecular mechanisms. Mol. Nutr. Food Res. 2014, 58, 915–930. [Google Scholar] [CrossRef]
- Ludwig, I.A.; Clifford, M.N.; Lean, M.E.J.; Ashihara, H.; Crozier, A. Coffee: Biochemistry and potential impact on health. Food Funct. 2014, 5, 1695–1717. [Google Scholar] [CrossRef]
- Gollücke, A.P.B.; Peres, R.C.; Ribeiro, D.A.; Aguair, O. Polyphenols as Supplements in Foods and Beverages: Recent Discoveries and Health Benefits, an Update. In Polyphenols: Mechanisms of Action in Human Health and Disease, 2nd ed.; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Elsevier Academic Press: Amsterdam, The Netherland, 2018; pp. 12–18. ISBN 978-0-12-813006-3. [Google Scholar]
- Marfella, R.; D’Onofrio, N.; Sirangelo, I.; Rizzo, M.R.; Capoluongo, M.C.; Servillo, L.; Paolisso, G.; Luisa Balestrieri, M. Polyphenols, Oxidative Stress, and Vascular Damage in Diabetes. Diabetes: Oxidative Stress and Dietary Antioxidants; Elsevier: Amsterdam, The Netherland, 2014; pp. 145–156. ISBN 9780124058859. [Google Scholar]
- Hurtado-Barroso, S.; Quifer-Rada, P.; Rinaldi de Alvarenga, J.F.; Pérez-Fernández, S.; Tresserra-Rimbau, A.; Lamuela-Raventos, R.M. Changing to a Low-Polyphenol Diet Alters Vascular Biomarkers in Healthy Men after Only Two Weeks. Nutrients 2018, 10, 1766. [Google Scholar] [CrossRef]
- Ye, Y.; Ji, J.; Sun, Z.; Shen, P.; Sun, X. Recent advances in electrochemical biosensors for antioxidant analysis in foodstuff. TrAC 2020, 122, 115718. [Google Scholar] [CrossRef]
- Shivakumar, A.; Yogendra Kumar, M.S. Critical Review on the Analytical Mechanistic Steps in the Evaluation of Antioxidant Activity. Crit. Rev. Anal. Chem. 2018, 48, 214–236. [Google Scholar] [CrossRef]
- Santos, J.R.; Lopo, M.; Rangel, A.O.S.S.; Lopes, J.A. Exploiting near infrared spectroscopy as an analytical tool for on-line monitoring of acidity during coffee roasting. Food Control 2016, 60, 408–415. [Google Scholar] [CrossRef]
- Santos, J.R.; Viegas, O.; Páscoa, R.N.M.J.; Ferreira, I.M.P.L.V.O.; Rangel, A.O.S.S.; Lopes, J.A. In-line monitoring of the coffee roasting process with near infrared spectroscopy: Measurement of sucrose and colour. Food Chem. 2016, 208, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Catelani, T.A.; Páscoa, R.N.M.J.; Santos, J.R.; Pezza, L.; Pezza, H.R.; Lima, J.L.F.C.; Lopes, J.A. A Non-invasive Real-Time Methodology for the Quantification of Antioxidant Properties in Coffee During the Roasting Process Based on Near-Infrared Spectroscopy. Food Bioprocess. Technol. 2017, 10, 630–638. [Google Scholar] [CrossRef]
- Yergenson, N.; Aston, D.E. Monitoring coffee roasting cracks and predicting with in situ near-infrared spectroscopy. J. Food Process. Eng. 2020, 43. [Google Scholar] [CrossRef]
- Heide, J.; Czech, H.; Ehlert, S.; Koziorowski, T.; Zimmermann, R. Toward Smart Online Coffee Roasting Process Control: Feasibility of Real-Time Prediction of Coffee Roast Degree and Brew Antioxidant Capacity by Single-Photon Ionization Mass Spectrometric Monitoring of Roast Gases. J. Agric. Food Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Streibel, T.; Zimmermann, R. Resonance-enhanced multiphoton ionization mass spectrometry (REMPI-MS): Applications for process analysis. Annu. Rev. Anal. Chem. 2014, 7, 361–381. [Google Scholar] [CrossRef]
- Opitz, S.E.W.; Smrke, S.; Goodman, B.A.; Keller, M.; Schenker, S.; Yeretzian, C. Antioxidant Generation during Coffee Roasting: A Comparison and Interpretation from Three Complementary Assays. Foods 2014, 3, 586–604. [Google Scholar] [CrossRef]
- Li, H.-D.; Xu, Q.-S.; Liang, Y.-Z. libPLS: An integrated library for partial least squares regression and linear discriminant analysis. Chemometr. Intell. Lab. 2018, 176, 34–43. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemometr. Intell. Lab. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef]
- Todeschini, R.; Ballabio, D.; Grisoni, F. Beware of Unreliable Q(2)! A Comparative Study of Regression Metrics for Predictivity Assessment of QSAR Models. J. Chem. Inf. Model. 2016, 56, 1905–1913. [Google Scholar] [CrossRef] [PubMed]
- Consonni, V.; Ballabio, D.; Todeschini, R. Evaluation of model predictive ability by external validation techniques. J. Chemometr. 2010, 24, 194–201. [Google Scholar] [CrossRef]
- Yang, N.; Liu, C.; Liu, X.; Degn, T.K.; Munchow, M.; Fisk, I. Determination of volatile marker compounds of common coffee roast defects. Food Chem. 2016, 211, 206–214. [Google Scholar] [CrossRef]
- Flament, I. Coffee Flavour Chemistry; Wiley: Hoboken, NJ, USA, 2001; ISBN 978-0-471-72038-6. [Google Scholar]
- Dietz, W.; Neussner, H.J.; Boesl, U.; Schlag, E.W. A Model for Multiphoton Ionisation Mass Spectroscopy with Application to Benzene. Chem. Phys. 1982, 66, 105–127. [Google Scholar] [CrossRef]
- Filgueiras, P.R.; Alves, J.C.L.; Sad, C.M.S.; Castro, E.V.R.; Dias, J.C.M.; Poppi, R.J. Evaluation of trends in residuals of multivariate calibration models by permutation test. Chemometr. Intell. Lab. 2014, 133, 33–41. [Google Scholar] [CrossRef]
- Smrke, S.; Opitz, S.E.W.; Vovk, I.; Yeretzian, C. How does roasting affect the antioxidants of a coffee brew? Exploring the antioxidant capacity of coffee via on-line antioxidant assays coupled with size exclusion chromatography. Food Funct. 2013, 4, 1082–1092. [Google Scholar] [CrossRef]
- Bellon-Maurel, V.; Fernandez-Ahumada, E.; Palagos, B.; Roger, J.-M.; McBratney, A. Critical review of chemometric indicators commonly used for assessing the quality of the prediction of soil attributes by NIR spectroscopy. TrAC 2010, 29, 1073–1081. [Google Scholar] [CrossRef]
- Lorber, A.; Faber, N.M.; Kowalski, B.R. Net Analyte Signal Calculation in Multivariate Calibration. Anal. Chem. 1997, 69, 1620–1626. [Google Scholar] [CrossRef]
- Faber, N.M. Efficient computation of net analyte signal vector in inverse multivariate calibration models. Anal. Chem. 1998, 70, 5108–5110. [Google Scholar] [CrossRef]
- Ferré, J.; Brown, S.D.; Rius, F.X. Improved calculation of the net analyte signal in inverse multivariate calibration. J. Chemometr. 2001, 15, 537–553. [Google Scholar] [CrossRef]
- Olivieri, A.C. Analytical figures of merit: From univariate to multiway calibration. Chem. Rev. 2014, 114, 5358–5378. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.C.; Sarabia, L.A.; Herrero, A.; Sánchez, M.S.; Sanz, M.B.; Rueda, M.E.; Giménez, D.; Meléndez, M.E. Capability of detection of an analytical method evaluating false positive and false negative (ISO 11843) with partial least squares. Chemometr. Intell. Lab. 2003, 69, 21–33. [Google Scholar] [CrossRef]
- Allegrini, F.; Olivieri, A.C. IUPAC-consistent approach to the limit of detection in partial least-squares calibration. Anal. Chem. 2014, 86, 7858–7866. [Google Scholar] [CrossRef] [PubMed]
- Boqué, R.; Rius, F.X. Computing detection limits in multicomponent spectroscopic analysis. TrAC 1997, 16, 432–436. [Google Scholar] [CrossRef]
- Kvalheim, O.M.; Karstrang, T.V. Interpretation of Latent-Variable Regression Models. Chemometr. Intell. Lab. 1989, 7, 39–51. [Google Scholar] [CrossRef]
- Kvalheim, O.M. Interpretation of partial least squares regression models by means of target projection and selectivity ratio plots. J. Chemometr. 2010, 24, 496–504. [Google Scholar] [CrossRef]
- Thawabteh, A.; Juma, S.; Bader, M.; Karaman, D.; Scrano, L.; Bufo, S.A.; Karaman, R. The Biological Activity of Natural Alkaloids against Herbivores, Cancerous Cells and Pathogens. Toxins 2019, 11, 656. [Google Scholar] [CrossRef]
- Kanzler, C.; Haase, P.T.; Schestkowa, H.; Kroh, L.W. Antioxidant Properties of Heterocyclic Intermediates of the Maillard Reaction and Structurally Related Compounds. J. Agric. Food Chem. 2016, 64, 7829–7837. [Google Scholar] [CrossRef]
- Kamiyama, M.; Moon, J.-K.; Jang, H.W.; Shibamoto, T. Role of degradation products of chlorogenic acid in the antioxidant activity of roasted coffee. J. Agric. Food Chem. 2015, 63, 1996–2005. [Google Scholar] [CrossRef]
- Gehm, C.; Streibel, T.; Passig, J.; Zimmermann, R. Determination of Relative Ionization Cross Sections for Resonance Enhanced Multiphoton Ionization of Polycyclic Aromatic Hydrocarbons. Appl. Sci. 2018, 8, 1617. [Google Scholar] [CrossRef]
- Faber, N.M.; Song, X.-H.; Hopke, P.K. Sample-specific standard error of prediction for partial least squares regression. TrAC 2003, 22, 330–334. [Google Scholar] [CrossRef]
- Van der Voet, H. Pseudo-degrees of freedom for complex predictive models: The example of partial least squares. J. Chemometr. 1999, 13, 195–204. [Google Scholar] [CrossRef]

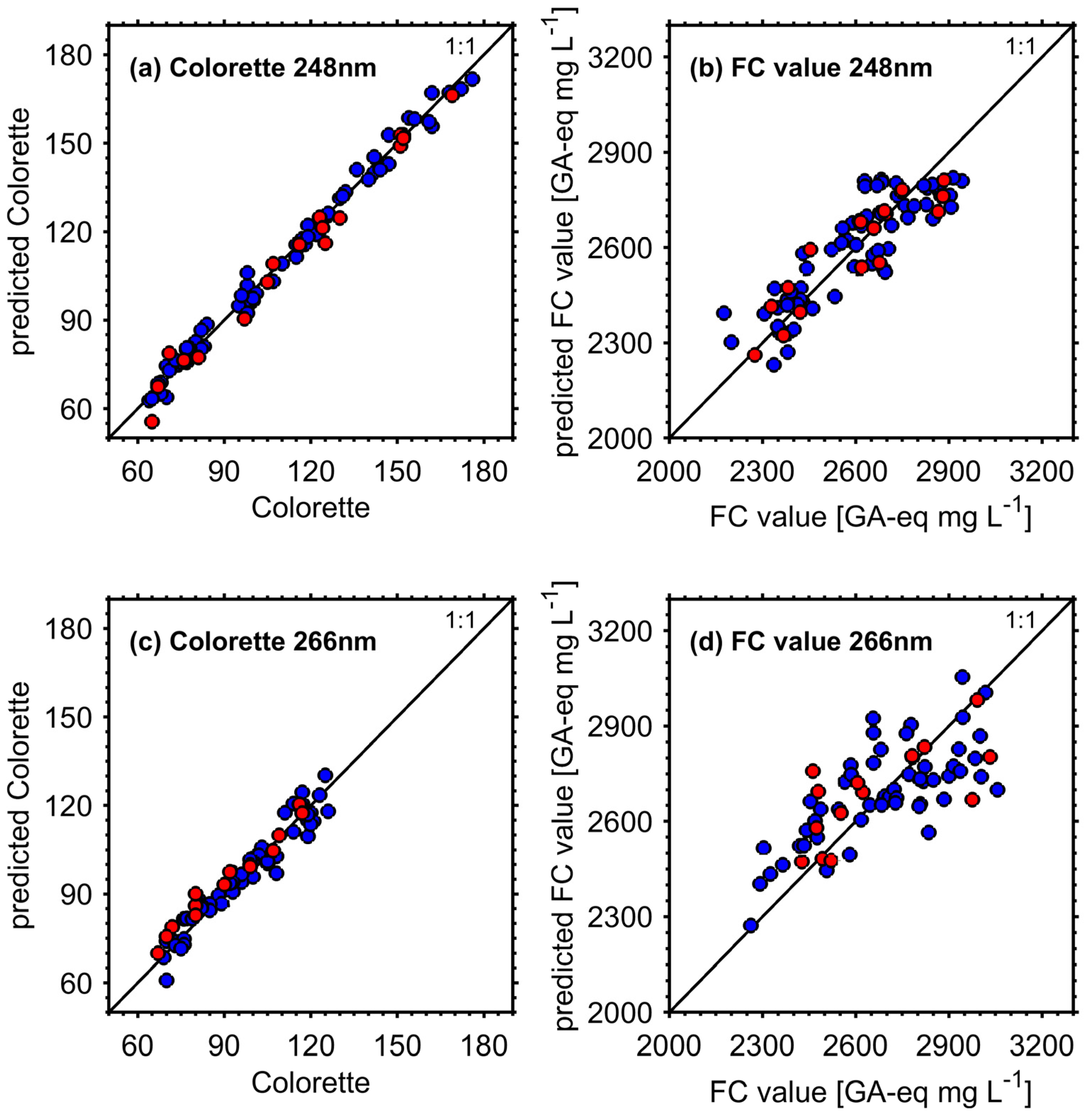

| REMPI Wavelength | Coffee Property | RMSECV | RMSEP | Rel. RMSEP | R2CV | R2P | #LV 2 | #Var 3 | RER |
|---|---|---|---|---|---|---|---|---|---|
| 248 nm | Colorette | 3.88 (0.09) | 4.54 | 0.040 | 0.985 (0.001) | 0.980 | 6 (1) | 16 (2) | 23 |
| 266 nm | Colorette | 4.69 (0.04) | 4.63 | 0.050 | 0.924 (0.001) | 0.926 | 2 (0) | 10 (2) | 11 |
| 248 nm | FC | 94.6 (0.26) 1 | 80.3 1 | 0.031 | 0.753 (0.001) | 0.822 | 1 (0) | 8 (4) | 7.6 |
| 266 nm | FC | 146 (4.2) 1 | 151 1 | 0.057 | 0.490 (0.029) | 0.454 | 2 (0) | 4 (2) | 4.0 |
| REMPI Wavelength | Coffee Property | SENb 1 | SENNAS 1 | ASEN 2 | (ASEN-1) 3 | SEL 4 |
|---|---|---|---|---|---|---|
| 248 nm | Colorette | 5.59·10−4 | 7.48·10−4 | 0.305 | 3.27 | 8.63 |
| 266 nm | Colorette | 1.41·10−3 | 8.70·10−4 | 0.359 | 2.79 | 16.1 |
| 248 nm | FC | 3.41·10−4 | 1.21·10−4 | 0.297 | 3.37 | 16.3 |
| 266 nm | FC | 3.7·10−5 | 1.31·10−5 | 0.013 | 77.7 | 19.6 |
| REMPI Wavelength | Coffee Property | LODpu | LOD3×RMSEP | LODNAS | LODmin | LODmax | LODss 2 |
|---|---|---|---|---|---|---|---|
| 248 nm | Colorette | 6.34 | 13.6 | 5.94 | 2.17 | 3.12 | 2.41 |
| 266 nm | Colorette | 11.6 | 13.9 | 10.6 | 3.91 | 4.46 | 3.89 |
| 248 nm | FC 1 | 216 | 241 | 35.7 | 127 | 297 | 146 |
| 266 nm | FC 1 | 487 | 454 | 608 | 138 | 492 | 214 |
| Colorette 248nm | 0.980 | 0.408 | 0.010 |
| 290 | FC Value 248nm | 0.383 | 0.002 |
| 18.2 | 16.0 | Colorette 266nm | 0.598 |
| 0.111 | 0.024 | 41.8 | FC Value 266nm |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czech, H.; Heide, J.; Ehlert, S.; Koziorowski, T.; Zimmermann, R. Smart Online Coffee Roasting Process Control: Modelling Coffee Roast Degree and Brew Antioxidant Capacity for Real-Time Prediction by Resonance-Enhanced Multi-Photon Ionization Mass Spectrometric (REMPI-TOFMS) Monitoring of Roast Gases. Foods 2020, 9, 627. https://doi.org/10.3390/foods9050627
Czech H, Heide J, Ehlert S, Koziorowski T, Zimmermann R. Smart Online Coffee Roasting Process Control: Modelling Coffee Roast Degree and Brew Antioxidant Capacity for Real-Time Prediction by Resonance-Enhanced Multi-Photon Ionization Mass Spectrometric (REMPI-TOFMS) Monitoring of Roast Gases. Foods. 2020; 9(5):627. https://doi.org/10.3390/foods9050627
Chicago/Turabian StyleCzech, Hendryk, Jan Heide, Sven Ehlert, Thomas Koziorowski, and Ralf Zimmermann. 2020. "Smart Online Coffee Roasting Process Control: Modelling Coffee Roast Degree and Brew Antioxidant Capacity for Real-Time Prediction by Resonance-Enhanced Multi-Photon Ionization Mass Spectrometric (REMPI-TOFMS) Monitoring of Roast Gases" Foods 9, no. 5: 627. https://doi.org/10.3390/foods9050627
APA StyleCzech, H., Heide, J., Ehlert, S., Koziorowski, T., & Zimmermann, R. (2020). Smart Online Coffee Roasting Process Control: Modelling Coffee Roast Degree and Brew Antioxidant Capacity for Real-Time Prediction by Resonance-Enhanced Multi-Photon Ionization Mass Spectrometric (REMPI-TOFMS) Monitoring of Roast Gases. Foods, 9(5), 627. https://doi.org/10.3390/foods9050627





