The First Identification of the Uniqueness and Authentication of Maltese Extra Virgin Olive Oil Using 3D-Fluorescence Spectroscopy Coupled with Multi-Way Data Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. EEM Spectra Acquisition
2.3. The PARAFAC Model
2.4. Discriminant Multi-Way Partial Least Squares Regression (DN-PLSR)
3. Results
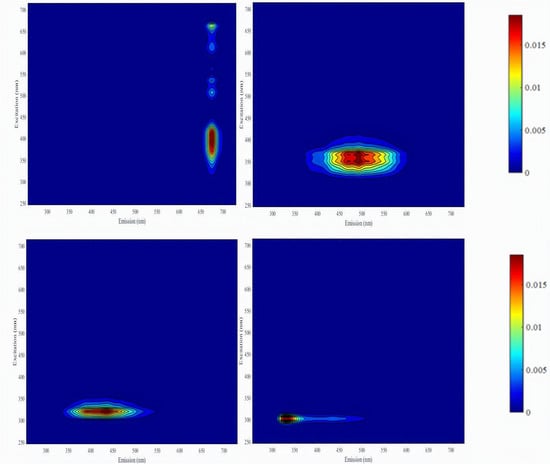
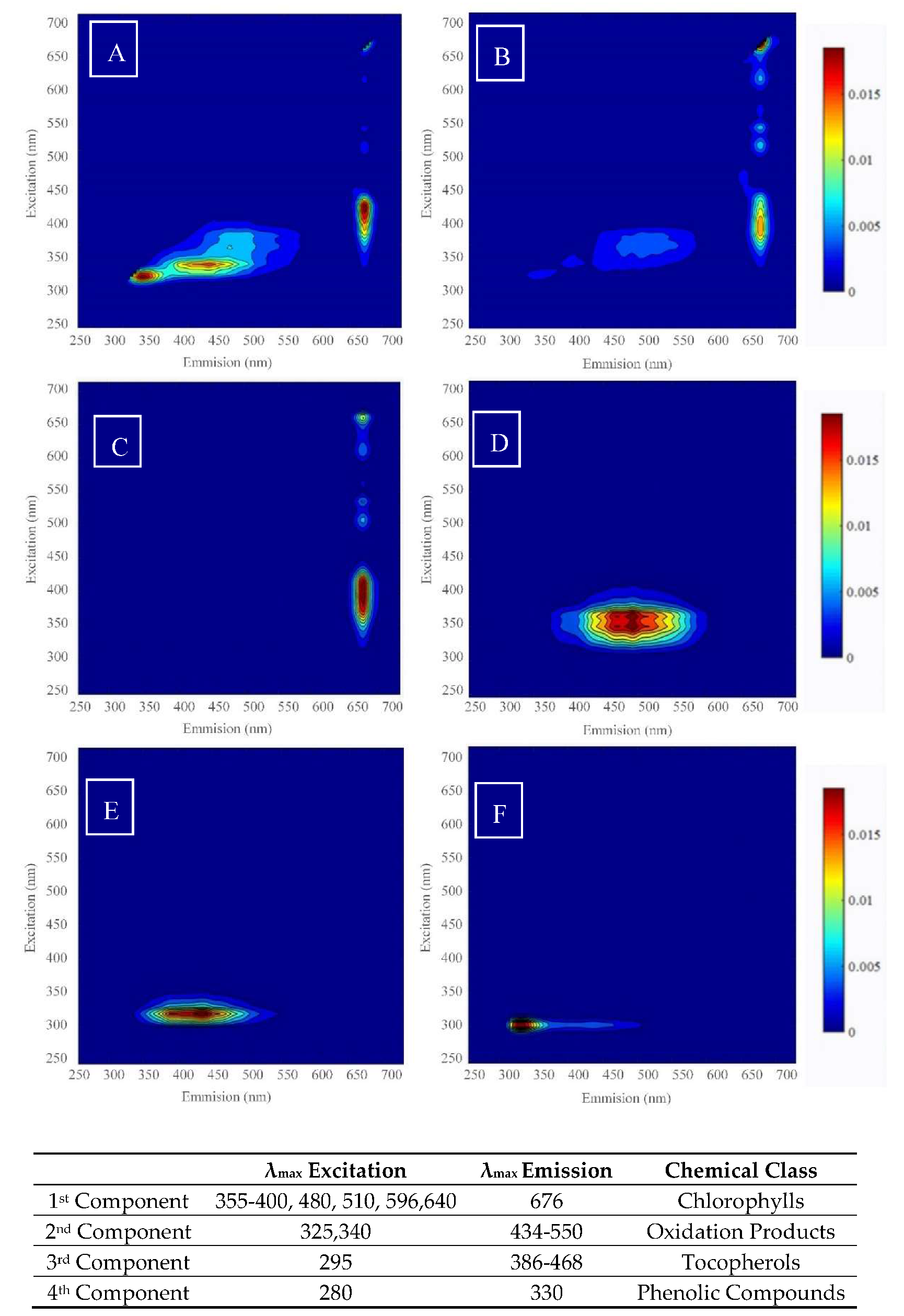
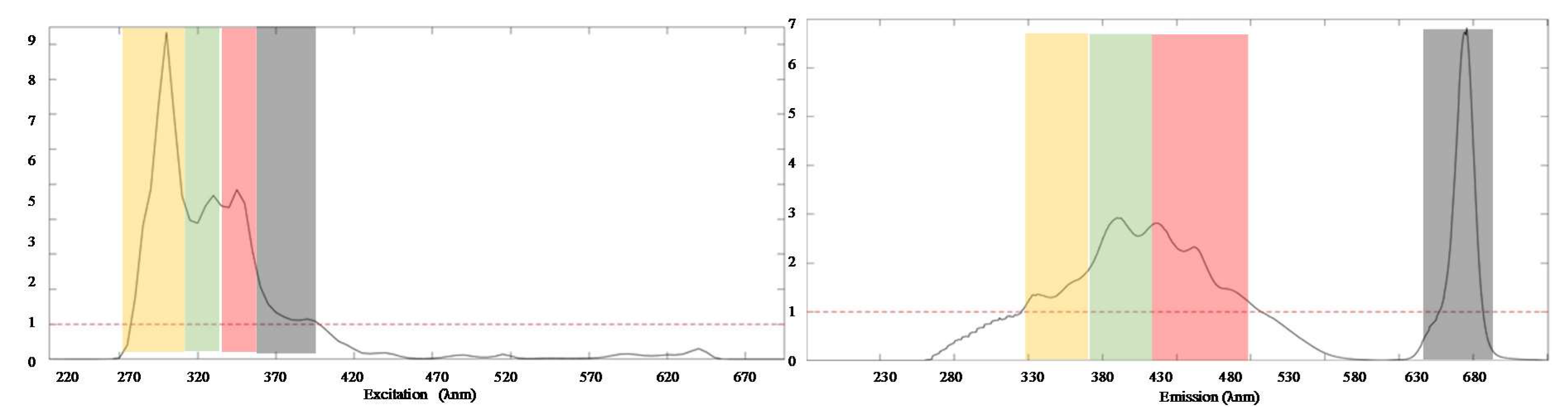
3.1. Extraction of PARAFAC Components
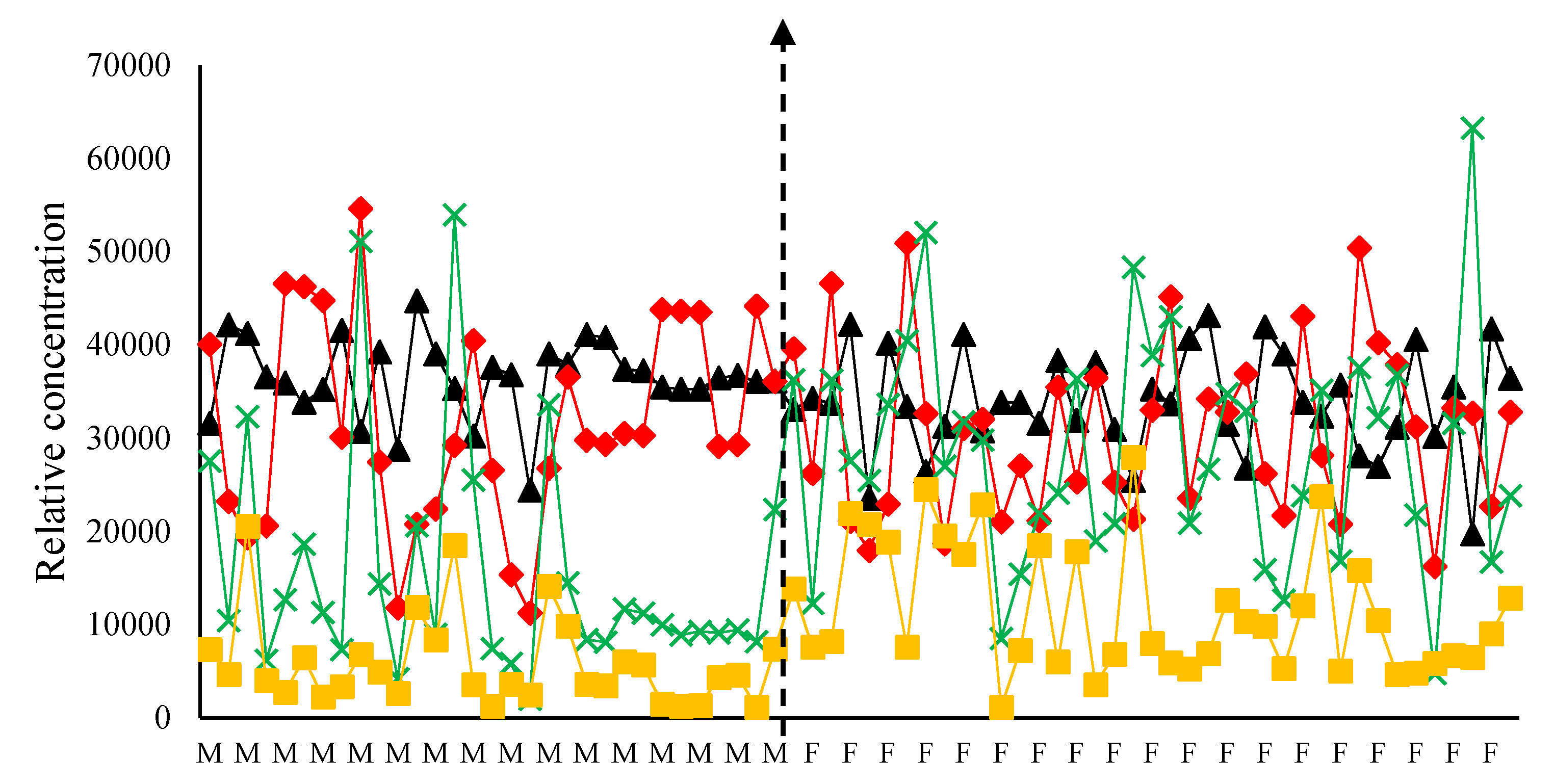
3.2. Linear Discrminate Analysis on Mode 1 of the PARAFAC Compontents PARAFAC-LDA
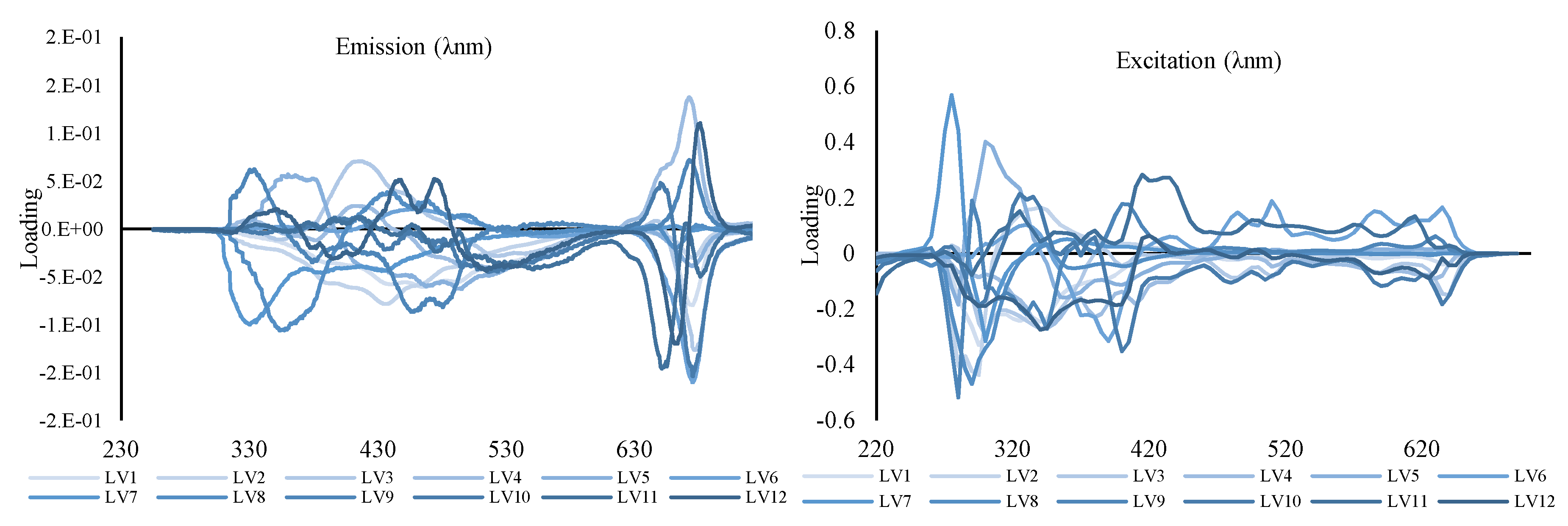
3.3. Discriminant Multi-Way Partial Least Squares Regression Performance
4. Discussion
4.1. Extraction of the Optimal Number of PARAFAC Components
4.2. Application of Univariate and Multivariate Analysis on the PARAFAC Components in Mode 1
4.3. Discrimination of Maltese EVOOs through DN-PLSR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lloyd, J.B.F. Synchronized Excitation of Fluorescence Emission Spectra. Nat. Phys. Sci. 1971, 231, 64–65. [Google Scholar] [CrossRef]
- Kyriakidis, N.B.; Skarkalis, P. Fluorescence Spectra Measurement of Olive Oil and Other Vegetable Oils. J. AOAC Int. 2000, 83, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, E.; Romaniuk, A.; Khmelinskii, I.V.; Herance, R.; Bourdelande, J.L.; Sikorski, M.; Kozioł, J. Characterization of Edible Oils Using Total Luminescence Spectroscopy. J. Fluoresc. 2004, 14, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, E.; Górecki, T.; Silorski, M.; Khmelinskii, I.V.M.; Koziol, J. Classification of edible oils using synchronous scanning fluorescence spectroscopy. Food Chem. 2005, 89, 217–225. [Google Scholar] [CrossRef]
- Poulli, K.I.; Mousdis, G.A.; Georgiou, C.A. Classification of Edible and Lampante Virgin Olive Oil Based on Synchronous Fluorescence and Total Luminescence Spectroscopy. Anal. Chim. Acta 2005, 542, 151–156. [Google Scholar] [CrossRef]
- Sayago, A.; Morales, M.T.; Aparicio, R. Detection of Hazelnut Oil in Virgin Olive Oil by a Spectrofluorimetric Method. Eur. Food Res. Technol. 2004, 218, 480–483. [Google Scholar] [CrossRef]
- Merás, I.D.; Manzano, J.D.; Rodríguez, D.A.; De La Peña, A.M. Detection and quantification of extra virgin olive oil adulteration by means of autofluorescence excitation-emission profiles combined with multi-way classification. Talanta 2018, 178, 751–762. [Google Scholar] [CrossRef]
- Tena, N.; Garcia-Gonzalez, D.L.; Aparicio, R. Evaluation of Virgin Olive Oil Thermal Deterioration by Fluorescence Spectroscopy. J. Agric. Food Chem. 2009, 57, 10505–10511. [Google Scholar] [CrossRef]
- Cabrera-Bañegil, M.; Martín-Vertedor, D.; Boselli, E.; Durán-Merás, I. Control of olive cultivar irrigation by front-face fluorescence excitation-emission matrices in combination with PARAFAC. J. Food Compos. Anal. 2018, 69, 189–196. [Google Scholar] [CrossRef]
- Sikorska, E.; Khmelinskii, I.V.; Sikorski, M.; Caponio, F.; Bilancia, M.T.; Pasqualone, A.; Gomes, T. Fluorescence Spectroscopy in Monitoring of Extra Virgin Olive Oil During Storage. Int. J. Food Sci. Technol. 2008, 43, 52–61. [Google Scholar] [CrossRef]
- Airado-Rodríguez, D.; Galeano-Díaz, T.; Durán-Merás, I.; Wold, J.P. Usefulness of fluorescence excitation-emission matrices in combination with PARAFAC, as fingerprints of red wines. J. Agric. Food Chem. 2009, 57, 1711–1720. [Google Scholar] [CrossRef] [PubMed]
- Callejón, R.M.; Amigo, J.M.; Pairo, E.; Garmón, S.; Ocaña, J.A.; Morales, M.L. Classification of Sherry vinegars by combining multidimensional fluorescence, parafac and different classification approaches. Talanta 2012, 88, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Lenhardt, L.; Bro, R.; Zeković, I.; Dramićanin, T.; Dramićanin, M.D. Fluorescence spectroscopy coupled with PARAFAC and PLS DA for characterization and classification of honey. Food Chem. 2015, 175, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Muñoz de la Peña, A.; Mora Diez, N.; Mahedero García, M.C.; Bohoyo Gil, D.; Cañada-Cañada, F. A chemometric sensor for determining sulphaguanidine residues in honey samples. Talanta 2007, 73, 304–313. [Google Scholar] [CrossRef]
- Mahedero, M.C.; Diaz, N.M.; Muñoz de la Peña, A.; Espinosa Mansilla, A.; Gonzalez Gomez, D.; Bohoyo Gil, D. Strategies for solving matrix effects in the analysis of sulfathiazole in honey samples using three-way photochemically induced fluorescence data. Talanta 2005, 65, 806–813. [Google Scholar] [CrossRef]
- Zhu, S.H.; Wu, H.L.; Li, B.R.; Xia, A.L.; Han, Q.J.; Zhang, Y.; Bian, Y.C.; Yu, R.Q. Determination of pesticides in honey using excitation-emission matrix fluorescence coupled with second-order calibration and second-order standard addition methods. Anal. Chim. Acta 2008, 619, 165–172. [Google Scholar] [CrossRef]
- Włodarska, K.; Pawlak-Lemańska, K.; Khmelinskii, I.; Sikorska, E. Explorative study of apple juice fluorescence in relation to antioxidant properties. Food Chem. 2016, 210, 593–599. [Google Scholar] [CrossRef]
- Valderrama, P.; Março, P.H.; Locquet, N.; Ammari, F.; Rutledge, D.N. A procedure to facilitate the choice of the number of factors in multi-way data analysis applied to the natural samples: Application to monitoring the thermal degradation of oils using front-face fluorescence spectroscopy. Chemom. Intell. Lab. Syst. 2011, 106, 166–172. [Google Scholar] [CrossRef]
- Bro, R. Multi-Way Analysis in the Food Industry, Theory Algorithms and Applications. Ph.D. Thesis, Universiteit van Amsterdam, Amsterdam, The Netherlands, 1998. [Google Scholar]
- Bro, R.; Kiers, H.A.L. A new efficient method for determining the number of components in PARAFAC models. J. Chemom. 2003, 17, 274–286. [Google Scholar] [CrossRef]
- Borg, J. Cultivation and Diseases of Fruit Trees in the Maltese Islands; Dept. of Agriculture. Kessinger Publishing: Malta, 1922. [Google Scholar]
- Mazzitelli, O.; Calleja, A.; Sardella, D.; Farrugia, C.; Zammit-Mangion, M. Analysis of the molecular diversity of Olea europaea in the Mediterranean Island of Malta. Genet. Resour. Crop Evol. 2015, 62, 1021–1027. [Google Scholar] [CrossRef]
- Lia, F.; Farrugia, C.; Zammit-Mangion, M. Application of Elemental Analysis via Energy Dispersive X-Ray Fluorescence (ED-XRF) for the Authentication of Maltese Extra Virgin Olive Oil. Agriculture 2020, 10, 71. [Google Scholar] [CrossRef]
- Lia, F.; Farrugia, C.; Zammit-Mangion, M. A First Description of the Phenolic Profile of EVOOs from the Maltese Islands Using SPE and HPLC: Pedo-Climatic Conditions Modulate Genetic Factors. Agriculture 2019, 9, 107. [Google Scholar] [CrossRef]
- Harshman, R.A.; Lundy, M.E. Parafac-parallel factor-analysis. Comput. Stat. Data Anal. 1994, 18, 39–72. [Google Scholar] [CrossRef]
- Andersen, C.M.; Bro, R. Practical aspects of PARAFAC modeling of fluorescence excitation-emission data. J. Chemom. 2003, 17, 200–215. [Google Scholar] [CrossRef]
- Bro, R. Exploratory study of sugar production using fluorescence spectroscopy and multi-way analysis. Chemom. Intell. Lab. Syst. 1999, 4, 133–147. [Google Scholar] [CrossRef]
- Guimet, F.; Ferre, J.; Boque, R.; Rius, F.X. Application of unfold principal component analysis and parallel factor analysis to the exploratory analysis of olive oils by means of excitation-emission matrix fluorescence spectroscopy. Anal. Chim. Acta 2004, 515, 75–85. [Google Scholar] [CrossRef]
- Guimet, F.; Ferre, J.; Boque, R.; Vidal, M.; Garcia, J. Excitation-emission fluorescence spectroscopy combined with three-way methods of analysis as a complementary technique for olive oil characterization. J. Agric. Food Chem. 2005, 53, 9319–9328. [Google Scholar] [CrossRef]
- Bro, R. PARAFAC. Tutorial and applications. Chemom. Intell. Lab. Syst. 1997, 38, 149–171. [Google Scholar] [CrossRef]
- Dupuy, N.; Le Dreau, Y.; Ollivier, D.; Artaud, J.; Pinatel, C.; Kister, J. Origin of French Virgin Olive Oil Registered Designation of Origins Predicted by Chemometric Analysis of Synchronous Excitation-Emission Fluorescence Spectra. J. Agric. Food Chem. 2005, 53, 9361–9368. [Google Scholar] [CrossRef]
- Tena, N.; Aparicio, R.; García-González, D.L. Chemical changes of thermoxidized virgin olive oil determined by excitation-emission fluorescence spectroscopy (EEFS). Food Res. Int. 2012, 45, 103–108. [Google Scholar] [CrossRef]
- Zandomeneghi, M.; Zandomeneghi, G. Comment on Cluster Analysis Applied to the Exploratory Analysis of Commercial Spanish Olive Oils by Means of Excitation Emission Fluorescence Spectroscopy. J. Agric. Food Chem. 2005, 53, 5829–5830. [Google Scholar] [CrossRef] [PubMed]
- Eitenmiller, R.R.; Ye, L.; Landen, W.O., Jr. (Eds.) Ascorbic acid: Vitamin C. In Vitamin Analysis for the Health and Food Sciences, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Cheikhousman, R.; Zude, M.; Bouveresse, D.J.-R.; Léger, C.L.; Rutledge, D.N.; Birlouez-Aragon, I.; Zude-Sasse, M. Fluorescence Spectroscopy for Monitoring Deterioration of Extra Virgin Olive Oil During Heating. Anal. Bioanal. Chem. 2005, 382, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
- Wold, S. PLS for Multivariate Linear Modeling. Chemometric Methods in Molecular Design; van de Waterbeemd, H., Ed.; VCH Publishers: Weinheim, Germany, 1995; Volume 2, pp. 195–218. [Google Scholar]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikstrom, C.; Wold, S. Multi- and Megavariate Data Analysis Basic Principals and Applications; Umetrics: Umeå, Sweden, 2006. [Google Scholar]


| Core Consistency % | Explained Variation % | Residual Variation | Split-Half Similarity % | |
|---|---|---|---|---|
| PC1 | 100.00 | 62.86 | 0.952 | 98.81 |
| PC2 | 98.80 | 86.42 | 0.868 | 97.60 |
| PC3 | 90.12 | 93.12 | 0.74 | 96.20 |
| 1PC4 | 91.97 | 95.21 | 0.627 | 88.54 |
| PC5 | 83.93 | 96.47 | 0.493 | 59.18 |
| PC6 | −169.51 | 97.19 | 0.364 | 44.40 |
| PC7 | −72.12 | 97.58 | 0.261 | 0.00 |
| PC8 | −699.24 | 97.83 | 0.174 | 0.00 |
| PC9 | −11,859.88 | 98.21 | 0 | 0.00 |
| No.LV | %Variation X | %Variation Y | Overall %Variation | Training | Validation | RMSEC | RMSECV |
|---|---|---|---|---|---|---|---|
| 1 | 62.16 ± 0.57 | 57.97 ± 2.16 | 69.05 ± 9.18 | 70.21 ± 8.74 | 67.14 ± 10.98 | 0.48 ± 0.02 | 0.45 ± 0.06 |
| 2 | 73.86 ± 1.13 | 68.62 ± 2.07 | 77.14 ± 3.78 | 78.72 ± 6.49 | 74.95 ± 7.34 | 0.43 ± 0.02 | 0.44 ± 0.04 |
| 3 | 77.93 ± 1.97 | 70.60 ± 1.96 | 78.10 ± 3.60 | 78.01 ± 6.38 | 76.21 ± 6.17 | 0.41 ± 0.02 | 0.39 ± 0.06 |
| 4 | 85.18 ± 2.90 | 78.37 ± 1.74 | 80.95 ± 5.02 | 83.69 ± 5.38 | 78.87 ± 8.78 | 0.37 ± 0.02 | 0.40 ± 0.07 |
| 5 | 90.91 ± 1.48 | 81.83 ± 4.44 | 83.81 ± 4.36 | 89.36 ± 4.43 | 81.62 ± 10.82 | 0.33 ± 0.04 | 0.41 ± 0.08 |
| 6 | 93.25 ± 1.13 | 85.38 ± 3.55 | 87.62 ± 4.59 | 92.91 ± 4.58 | 85.45 ± 9.33 | 0.30 ± 0.03 | 0.40 ± 0.09 |
| 7 | 94.66 ± 1.24 | 87.29 ± 2.78 | 87.62 ± 5.95 | 93.62 ± 4.84 | 85.48 ± 12.20 | 0.28 ± 0.03 | 0.39 ± 0.08 |
| 8 | 95.13 ± 1.20 | 88.66 ± 2.14 | 88.57 ± 4.29 | 93.62 ± 3.94 | 86.78 ± 6.85 | 0.26 ± 0.02 | 0.39 ± 0.06 |
| 9 | 95.40 ± 1.26 | 89.67 ± 1.87 | 90.00 ± 3.78 | 94.33 ± 3.41 | 88.37 ± 6.85 | 0.25 ± 0.02 | 0.40 ± 0.02 |
| 10 | 95.67 ± 1.23 | 90.90 ± 1.93 | 91.90 ± 5.77 | 96.45 ± 4.29 | 90.06 ± 5.46 | 0.23 ± 0.02 | 0.40 ± 0.02 |
| 11 | 95.90 ± 1.27 | 91.98 ± 1.17 | 93.81 ± 4.36 | 98.58 ± 3.78 | 91.79 ± 5.49 | 0.22 ± 0.02 | 0.40 ± 0.02 |
| 112 | 96.62 ± 1.14 | 93.00 ± 1.15 | 94.76 ± 3.30 | 98.58 ± 2.94 | 93.18 ± 5.19 | 0.20 ± 0.02 | 0.39 ± 0.01 |
| 13 | 97.12 ± 0.64 | 94.06 ± 0.92 | 93.81 ± 2.97 | 98.58 ± 3.24 | 92.25 ± 5.19 | 0.19 ± 0.02 | 0.40 ± 0.00 |
| 14 | 97.52 ± 0.76 | 94.88 ± 0.78 | 92.86 ± 4.29 | 98.58 ± 3.71 | 91.30 ± 5.42 | 0.17 ± 0.01 | 0.43 ± 0.03 |
| 15 | 97.71 ± 0.64 | 95.77 ± 0.45 | 93.33 ± 4.36 | 99.29 ± 3.87 | 91.76 ± 5.66 | 0.16 ± 0.01 | 0.42 ± 0.05 |
| 16 | 97.86 ± 0.60 | 96.32 ± 0.46 | 92.86 ± 2.86 | 100.00 ± 4.76 | 91.11 ± 7.23 | 0.15 ± 0.01 | 0.42 ± 0.06 |
| 17 | 98.03 ± 0.62 | 96.64 ± 0.67 | 93.81 ± 2.18 | 100.00 ± 5.80 | 92.34 ± 6.14 | 0.13 ± 0.01 | 0.42 ± 0.07 |
| 18 | 98.17 ± 0.68 | 96.89 ± 0.71 | 94.76 ± 1.65 | 100.00 ± 4.39 | 93.51 ± 5.16 | 0.13 ± 0.02 | 0.41 ± 0.06 |
| 19 | 98.40 ± 0.45 | 97.47 ± 0.61 | 93.33 ± 3.60 | 100.00 ± 3.99 | 92.00 ± 7.09 | 0.11 ± 0.02 | 0.42 ± 0.03 |
| 20 | 98.56 ± 0.37 | 97.91 ± 0.46 | 61.43 ± 27.22 | 63.83 ± 27.29 | 66.89 ± 25.87 | 0.10 ± 0.01 | 0.62 ± 0.19 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lia, F.; Formosa, J.P.; Zammit-Mangion, M.; Farrugia, C. The First Identification of the Uniqueness and Authentication of Maltese Extra Virgin Olive Oil Using 3D-Fluorescence Spectroscopy Coupled with Multi-Way Data Analysis. Foods 2020, 9, 498. https://doi.org/10.3390/foods9040498
Lia F, Formosa JP, Zammit-Mangion M, Farrugia C. The First Identification of the Uniqueness and Authentication of Maltese Extra Virgin Olive Oil Using 3D-Fluorescence Spectroscopy Coupled with Multi-Way Data Analysis. Foods. 2020; 9(4):498. https://doi.org/10.3390/foods9040498
Chicago/Turabian StyleLia, Frederick, Jean Paul Formosa, Marion Zammit-Mangion, and Claude Farrugia. 2020. "The First Identification of the Uniqueness and Authentication of Maltese Extra Virgin Olive Oil Using 3D-Fluorescence Spectroscopy Coupled with Multi-Way Data Analysis" Foods 9, no. 4: 498. https://doi.org/10.3390/foods9040498
APA StyleLia, F., Formosa, J. P., Zammit-Mangion, M., & Farrugia, C. (2020). The First Identification of the Uniqueness and Authentication of Maltese Extra Virgin Olive Oil Using 3D-Fluorescence Spectroscopy Coupled with Multi-Way Data Analysis. Foods, 9(4), 498. https://doi.org/10.3390/foods9040498