Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Fermented Sausages and Sampling Procedures
2.2. Microbial Enumeration and Bulk Cell Collection
2.3. DNA Extraction from Bulk Cultures
2.4. PCR
2.5. DGGE Analysis
2.6. Sequence Analysis of DGGE Bands
2.7. Statistical Analysis
3. Results and Discussion
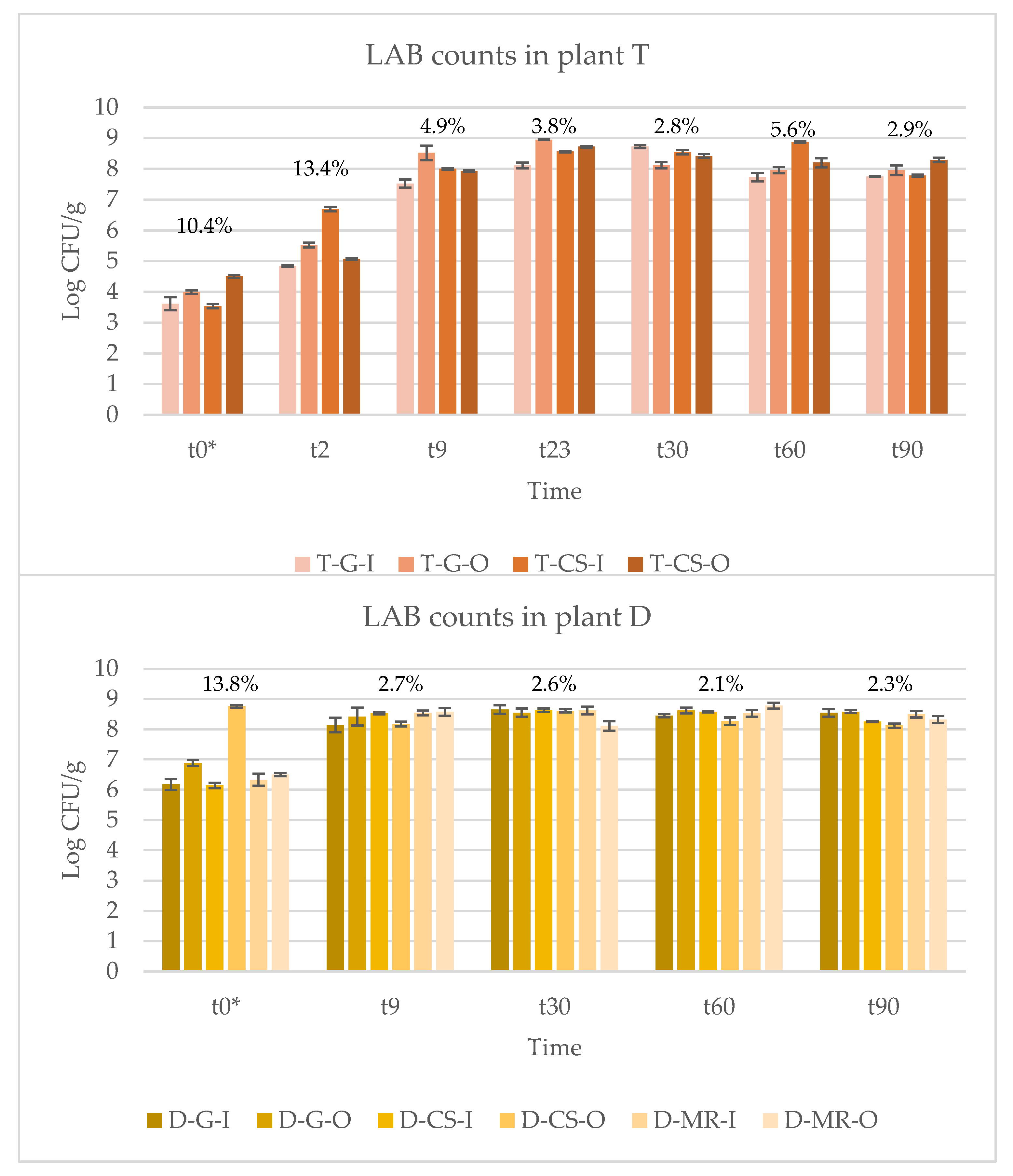
3.1. LAB Counts
3.2. Dynamics of the Detected Species
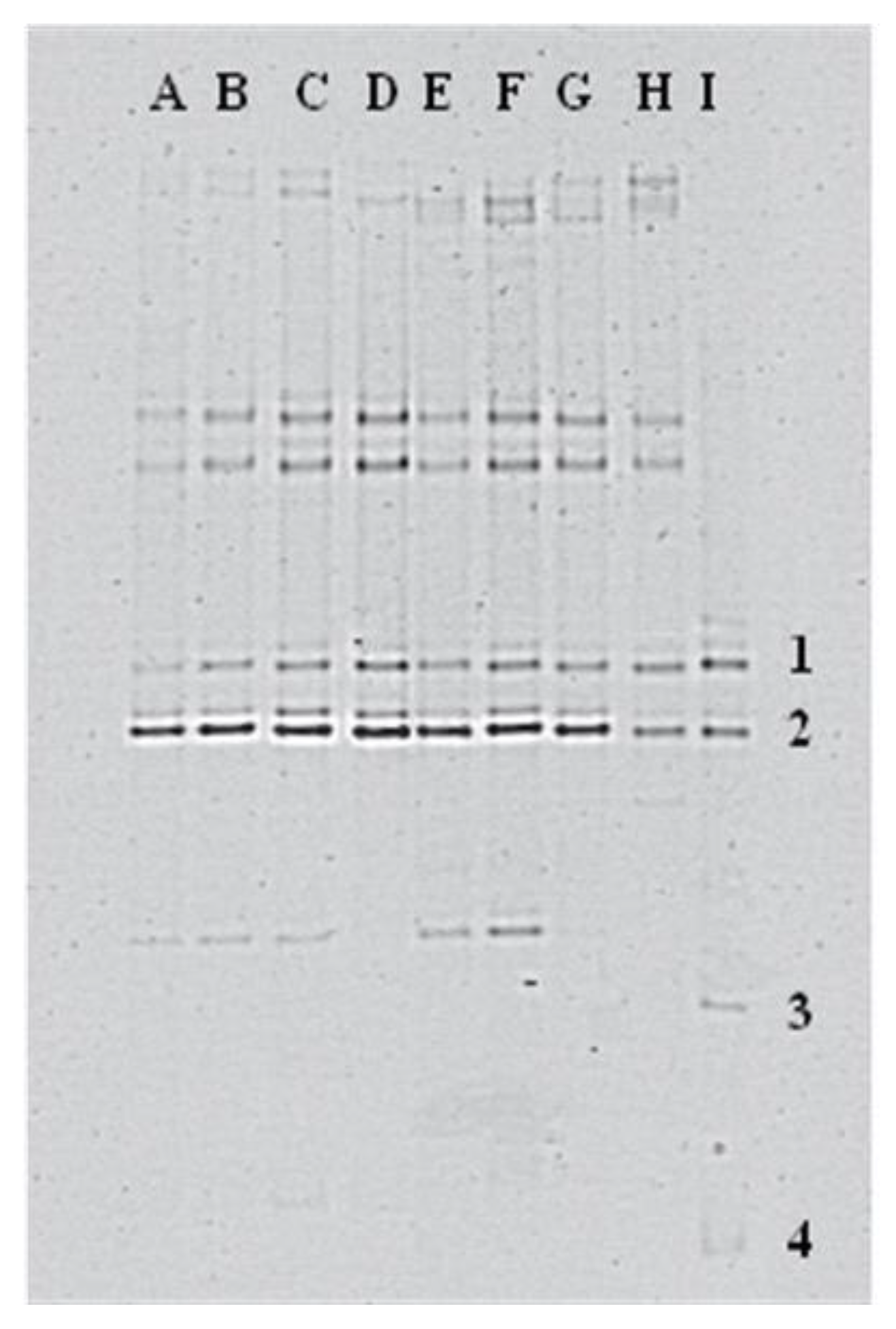
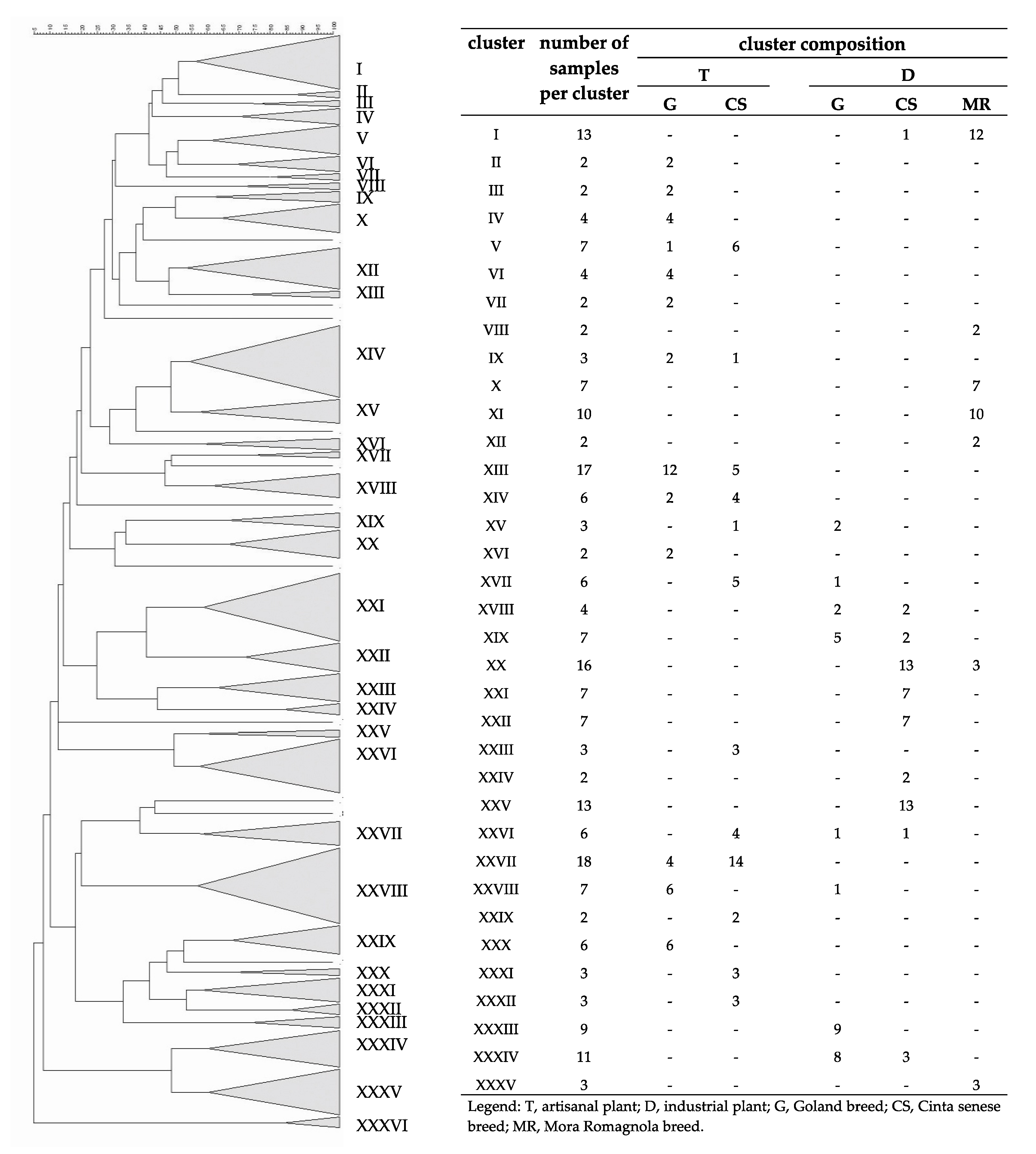
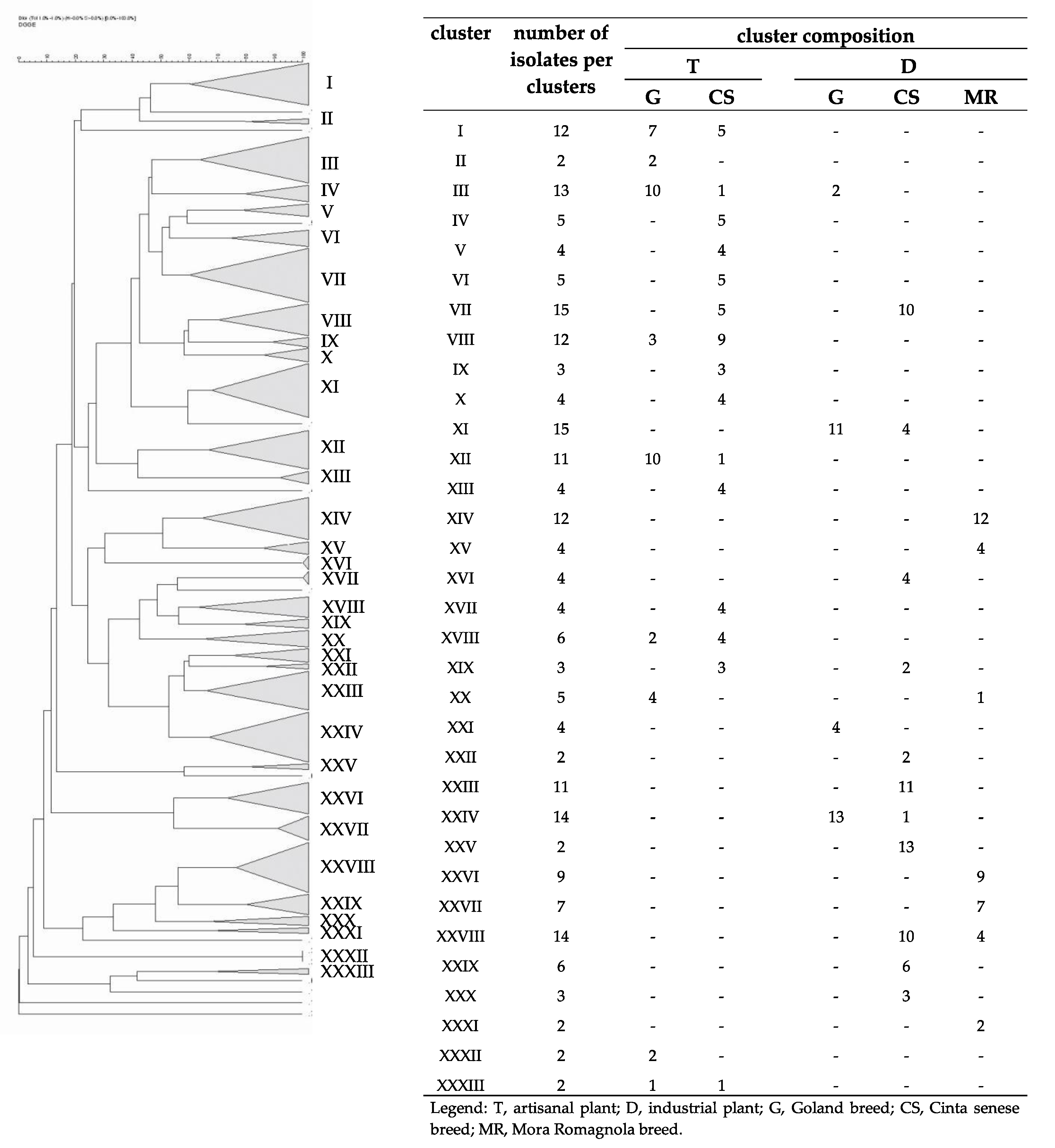
3.3. Fingerprint Analyses
Author Contributions
Funding
Conflicts of Interest
References
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Urso, R.; Cantoni, C.; Comi, G. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl. Environ. Microbiol. 2004, 70, 1883–1894. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Manzano, M.; Comi, G. Catalase positive cocci in fermented sausages: Variability due to different pork breeds, breeding system and sausage production technology. Food Microbiol. 2012, 29, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Manzano, M.; Cantoni, C.; Comi, G. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italia sausages. Appl. Environ. Microbiol. 2001, 67, 5113–6282. [Google Scholar] [CrossRef]
- Lücke, F.K. Fermented sausages. In Microbiology of Fermented Food; Wood, B.J.B., Ed.; Elsevier Applied Science: New York, NY, USA, 1985. [Google Scholar]
- Iacumin, L.; Osualdini, M.; Bovolenta, S.; Boscolo, D.; Chiesa, L.; Panseri, S.; Comi, G. Microbial, chemico-physical and volatile aromatic compounds characterization of Pitina PGI, a peculiar sausage-like product of North East Italy. Meat Sci. 2020, 162, 108081. [Google Scholar] [CrossRef]
- Coppola, R.; Marconi, E.; Rossi, F.; Dellaglio, F. Artisanal production of Naples-types salami: Chamical and microbiological aspects. Ital. J. Food Sci. 1995, 7, 57–62. [Google Scholar]
- Coppola, R.; Iorizzo, M.; Saotta, R.; Sorrentino, E.; Grazia, L. Characterization of micrococci and staphylococci isolated from sopressata molisana, a Southern Italy fermented sausage. Food Microbiol. 1997, 14, 47–53. [Google Scholar] [CrossRef]
- Iacumin, L.; Comi, G.; Cantoni, C.; Cocolin, L. Ecology and dynamics of coagulase-negative cocci isolated from naturally fermented Italian sausages. Syst. Appl. Microbiol. 2006, 29, 480–486. [Google Scholar] [CrossRef]
- Lücke, F.K.; Hechelmann, H. Starter cultures for dry sausages and raw ham. Fleischwirt 1987, 66, 1505–1508. [Google Scholar]
- Demeyer, D.I.; Verplaetse, A.; Gistelink, M. Fermentation of meat: An integrated process. Belg. J. Food Chem. Biotechnol. 1986, 41, 131–140. [Google Scholar]
- Baka, A.M.; Papavergou, E.J.; Pragalaki, T.; Bloukas, J.G.; Kotzekidou, P. Effect of selected autochthonous starter cultures on processing and quality characteristics of Greek fermented sausages. LWT Food Sci. Technol. 2011, 44, 54–61. [Google Scholar] [CrossRef]
- Geisen, R.; Lüke, F.K.; Krockel, L. Starter and protective cultures for meat and meat products. Fleischwirt 1992, 72, 894–898. [Google Scholar]
- Selgas, M.D.; Sanz, B.; Ordonez, J.A. Selected characteristics of micrococci isolated from Spanish dry fermented sausages. Food Microbiol. 1988, 5, 185–193. [Google Scholar] [CrossRef]
- Varnam, A.H.; Sutherland, J.P. Meat and Meat Products; Chapman & Hall: London, UK, 1995. [Google Scholar]
- Jones, R.J.; Hussein, H.M.; Zaigorec, M.; Brightnell, G.; Tagg, J.R. Isolation of lactic acid bacteria with inhibitory activity against pathogens and spoilage organisms associated with fresh meat. Food Microbiol. 2008, 25, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Urso, R.; Iacumin, L.; Cantoni, C.; Cattaneo, P.; Comi, G.; Cocolin, L. Culture-Dependent and -Independent Methods to Investigate the Microbial Ecology of Italian Fermented Sausages. Appl. Environ. Microbiol. 2005, 4, 1977–1986. [Google Scholar] [CrossRef]
- Comi, G.; Iacumin, L. The use of bioprotective cultures. In Strategies to Obtaining Healthier Foods; Nova Science Publishers, Inc. Hauppauge: New York, NY, USA, 2017. [Google Scholar]
- Holzapfel, R.W.; Geisen, R.; Schillinger, U. Biological preservation of foods with reference to protective cultures, becteriocins and food-grade enzymes. Int. J. Food Microbiol. 1995, 24, 343–362. [Google Scholar] [CrossRef]
- Stiles, M.E. Biopreservation by lactic acid bacteria. Antonie Von Leeuwenhoek 1996, 70, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Ben Omar, N.; Ampe, F. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Eviron. Microbiol. 2000, 66, 3664–3673. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Mauriello, G.; Aponte, M.; Moschetti, G.; Villani, F. Microbial succession during ripening of Naples-type salami, a southern Italian fermented sausage. Meat Sci. 2000, 56, 321–329. [Google Scholar] [CrossRef]
- Fontana, C.; Vignolo, G.; Cocconcelli, P.S. PCR–DGGE analysis for the identification of microbial populations from Argentinean dry fermented sausages. J. Microbiol. Methods 2005, 63, 254–263. [Google Scholar] [CrossRef]
- Iacumin, L.; Cecchini, F.; Manzano, M.; Osualdini, M.; Boscolo, D.; Orlic, S.; Comi, G. Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol. 2009, 26, 128–135. [Google Scholar] [CrossRef]
- Klijn, N.; Weerkamp, A.H.; de Vos, W.M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 1991, 57, 3390–3393. [Google Scholar] [CrossRef] [PubMed]
- Franciosa, I.; Alessandria, V.; Dolci, P.; Rantsiou, K.; Cocolin, L. Sausage fermentation and starter cultures in the era of molecular biologymethods. Int. J. Food Microbiol. 2018, 279, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Andrighetto, C.; Zampese, L.; Lombardi, A. RAPD-PCR characterization of lactobacilli isolated from artisanal meat plants and traditional fermented sausages of Veneto region. Lett. Appl. Microbiol. 2001, 33, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Papamanoli, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E.; Kotzekidou, P. Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci. 2003, 65, 859–867. [Google Scholar] [CrossRef]
- Leroy, F.; de Vuyst, L. Temperature and pH Conditions That Prevail during Fermentation of Sausages Are Optimal for Production of the Antilisterial Bacteriocin Sakacin K. Appl. Environ. Microbiol. 1999, 65, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; de Vuyst, L. The Presence of Salt and a Curing Agent Reduces Bacteriocin Production byLactobacillus sakei CTC 494, a Potential Starter Culture for Sausage Fermentation. Appl. Environ. Microbiol. 1999, 65, 5350–5356. [Google Scholar] [CrossRef]
- Martin, B.; Garriga, M.; Hugas, M.; Bover-Cid, S.; Veciana-Nogués, M.T.; Aymerich, T. Molecular, technological and safety characterization of Gram-positive catalase-positive cocci from slightly fermented sausages. Int. J. Food Microbiol. 2006, 107, 148–158. [Google Scholar] [CrossRef]
- Ravyts, F.; Steen, L.; Goemaere, O.; Paelinck, H.; De Vuyst, L.; Leroy, F. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 2010, 27, 945–954. [Google Scholar] [CrossRef]
- Fista, G.A.; Bloukas, J.G.; Siomos, A.S. Effect of leek and onion processing and quality characteristics of Greek traditional sausages. Meat Sci. 2004, 68, 163–172. [Google Scholar] [CrossRef]
- Iacumin, L.; Comi, G.; Cantoni, C.; Cocolin, L. Molecular and technological characterization of Staphylococcus xylosus isolated from naturally fermented Italian sausages by RAPD, Rep-PCR and Sau-PCR analysis. Meat Sci. 2006, 74, 281–288. [Google Scholar] [CrossRef]
- Morot-Bizot, S.; Talon, R.; Leroy-Setrin, S. Development of specific PCR primers for a rapid and accurate identification of Staphylococcus xylosus, a species used in food fermentation. J. Microbiol. Methods 2003, 55, 279–286. [Google Scholar] [CrossRef]
- Verluyten, J.; Leroy, F.; Vuyst, L. Effects of Different Spices Used in Production of Fermented Sausages on Growth of and Curvacin A Production by Lactobacillus curvatus LTH 1174. Appl. Environ. Microbiol. 2004, 70, 4807–4813. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.; Bian, G.; Su, Y.; Zhu, W. Differential Effects of Breed and Nursing on Early-Life Colonic Microbiota and Immune Status as Revealed in a Cross- Fostering Piglet. Appl. Environ. Microbiol. 2019, 85, e02510–e02518. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Huang, X.; Fang, S.; Xin, W.; Huang, L.; Chen, C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci. Rep. 2016, 6, 27427. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Yang, T.; Wang, Y.; Xing, K.; Zhang, F.; Zhao, X.; Ao, H.; Chen, S.; Liu, J.; Wanet, C. Metagenomic analysis of cecal microbiome identified microbiota and functional capacities associated with feed efficiency in landrace finishing pigs. Front. Microbiol. 2017, 8, 1546. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.R.; Ma, S.Q.; Zhu, Z.G.; Su, Y.; Zoetendal, E.G.; Mackie, R.; Liu, J.H.; Mu, C.L.; Huang, R.H.; Smidt, H.; et al. Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ. Microbiol. 2016, 18, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Monin, G.; Mejenes-Quijano, A.; Talmant, A.; Sellier, P. Influence of breed and muscle metabolic type on muscle glycolytic potential and meat pH in pigs. Meat Sci. 1987, 20, 149–158. [Google Scholar] [CrossRef]
- Zentek, J.; Ferrara, F.; Pieper, R.; Tedin, L.; Meyer, W.; Vahjen, W. Effects of dietary combinations of organic acids and medium chain fatty acids on the gastrointestinal microbial ecology and bacterial metabolites in the digestive tract of weaning piglets. J. Anim. Sci. 2013, 91, 3200–3210. [Google Scholar] [CrossRef]
- Nguyen, D.H.; Lee, K.Y.; Tran, H.N.; Kim, I.H. Effect of a protected blend of organic acids and medium-chain fatty acids on growth performance, nutrient digestibility, blood profiles, meat quality, faecal microflora, and faecal gas emission in finishing pigs. Can. J. Anim. Sci. 2019, 99, 448–455. [Google Scholar] [CrossRef]
- Albrecht, A.; Hebel, M.; Mittler, M.; Hurck, C.; Kustwan, K.; Heitkonig, B.; Bitschinski, D.; Kreyenschmidt, J. Influence of Different Production Systems on the Quality and Shelf Life of Poultry Meat: A Case Study in the German Sector. J. Food Qual. 2019, 3718057. [Google Scholar] [CrossRef]
- Gounadaki, A.S.; Skandamis, P.N. Microbial ecology of food contact surfaces and products of small-scale facilities producing traditional sausages. Food Microbiol. 2008, 25, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Duffy, E.A.; Belk, K.E.; Sofos, J.N.; Bellinger, G.R.; Pape, A.; Smith, G.C. Extent of Microbial Contamination in United States Pork Retail Products. J. Food Prot. 2001, 64, 172–178. [Google Scholar] [CrossRef]
- Saide-Albornoz, J.J.; Knipe, C.L.; Murano, E.A.; Beran, G.W. Contamination of Pork Carcasses during Slaughter, Fabrication, and Chilled Storage. J. Food Prot. 1995, 58, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Shaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- White, H. Asymptotic Theory for Econometricians; Academic Press: Orlando, FL, USA, 1980. [Google Scholar]
- Dytham, C. Choosing and Using Statistics: A Biologist’s Guide, 2nd ed.; Blackwell: Malden, MA, USA, 2003. [Google Scholar]
- Vauterin, L.; Vauterin, P. Computer-aided objective comparison of electrophoretic patterns for grouping and identification of microorganisms. Eur. Microbiol. 1992, 1, 37–41. [Google Scholar]
- Ammor, S.; Rachman, C.; Chaillou, S.; PreÅLvost, H.; Dousset, X.; Zagorec, M.; Dufour, E.; Chevallier, I. Phenotypic and genotypic identification of lactic acid bacteria isolated from a small-scale facility producing traditional dry sausages. Food Microbiol. 2005, 22, 373–382. [Google Scholar] [CrossRef]
- Comi, G.; Urso, R.; Iacumin, L.; Rantsiou, K.; Cattaneo, P.; Cantoni, C.; Cocolin, L. Characterization of naturally fermented sausages produced in North East of Italy. Meat Sci. 2005, 69, 381–392. [Google Scholar] [CrossRef]
- Greco, M.; Mazette, R.; De Santis, E.P.L.; Corona, A.; Cosseddu, A.M. Evolution and identification of lactic acid bacteria isolated during the ripening of Sardinian sausages. Meat Sci. 2005, 69, 733–739. [Google Scholar] [CrossRef]
- Talon, R.; Leroy, S.; Lebert, I. Microbial ecosystems of traditional fermented meat products: The importance of indigenous starters. Meat Sci. 2007, 77, 55–62. [Google Scholar] [CrossRef]
- Casaburi, A.; Aristoy, M.C.; Cavella, S.; Di Monaco, S.; Di Monaco, R.; Ercolini, D.; Toldra, F.; Villani, F. Biochemical and sensory characteristics of traditional fermented sausage of Vallo di Diano (Southern Italy) as affected by the use of starter cultures. Meat Sci. 2006, 76, 295–307. [Google Scholar] [CrossRef]
- Kostinek, M.; Specht, I.; Edward, V.A.; Schillinger, U.; Hertel, C.; Holzapfel, W.H.; Franz, C.M. Diversity and technological proprities of predominant lactic acid bacteria from fermented cassava used for the preparation of Gari, a traditional African food. Syst. Appl. Microbiol. 2005, 6, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Torriani, S.; Di Bucchianico, R.; Pattarini, F.; Zabeo, G.; Dell’Aglio, F. Presence and biotechnological charaterization of lactic acid bacteria and micrococcaceaea strains in Abruzzo traditional raw dry sausages. Ind. Delle Conserve 1994, 69, 3–9. [Google Scholar]
- Villani, F.; Casaburi, A.; Pennacchia, C. Microbial ecology of the soppressata of Vallo di Diano, a traditional dry fermented sausage from southern Italy, and in vitro and in situ selection of autochthonous starter cultures. Appl. Environ. Microbiol. 2007, 73, 5453–5463. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, C.; Donoso, E.; Valenzuela, Y.M.; Arizaga, R.; Salous, A.E. Effect of probiotic and prebiotic in the formulation and elaboration of sausage as an alternative of consumption. Int. J. Res. Pharm. Sci. 2019, 10, 2781–2785. [Google Scholar] [CrossRef]
- Sidira, M.; Mitropoulou, G.; Galanis, A.; Kanellaki, M.; Kourkoutas, Y. Effect of sugar content on quality characteristics and shelf-life of probiotic dry-fermented sausages produced by free or immobilized Lactobacillus casei ATCC 393. Foods 2019, 8, 219. [Google Scholar] [CrossRef]
- Mehmeti, I.; Muji, S.; Diep, D.B.; Nesa, I.F. High frequency of the potential pathogen Lactococcus garvieae in raw milk from Kosovo. Food Control. 2015, 53, 189–194. [Google Scholar] [CrossRef]
- Beck, R.; Weiss, N.; Winter, J. Lactobacillus graminis sp. nov., a New Species of Facultatively Heterofermentative Lactobacilli Surviving at Low pH in Grass Silage. Syst. Appl. Microbiol. 1988, 10, 279–283. [Google Scholar] [CrossRef]
- Lebert, I.; Leroy, S.; Giammarinaro, P.; Lebert, A.; Charcornac, J.P.; Bover-Cid, S.; Vidal-Carou, M.C.; Talon, R. Diversity of microorganisms in the environment and dry fermented sausages of small traditional French processing units. Meat Sci. 2007, 76, 112–122. [Google Scholar] [CrossRef]
| Processing Time | |||||||
|---|---|---|---|---|---|---|---|
| t0 * | t2 | t9 | t23 | t30 | t60 | t90 | |
| T-G-I | 3.61 ± 0.21 | 4.84 ± 0.03 | 7.52 ± 0.13 | 8.11 ± 0.09 | 8.72 ± 0.05 | 7.73 ± 0.14 | 7.75 ± 0.01 |
| T-G-O | 3.99 ± 0.06 | 5.52 ± 0.08 | 8.52 ± 0.24 | 8.95 ± 0.01 | 8.12 ± 0.10 | 7.96 ± 0.10 | 7.95 ± 0.16 |
| T-CS-I | 3.53 ± 0.07 | 6.69 ± 0.07 | 8.00 ± 0.02 | 8.56 ± 0.02 | 8.54 ± 0.07 | 8.87 ± 0.03 | 7.78 ± 0.03 |
| T-CS-O | 4.50 ± 0.05 | 5.07 ± 0.03 | 7.93 ± 0.03 | 8.72 ± 0.03 | 8.42 ± 0.06 | 8.20 ± 0.15 | 8.29 ± 0.07 |
| Mean | 3.91 ± 0.41 α | 5.53 ± 0.74 β | 7.99 ± 0.39 γ | 8.62 ± 0.33 δ | 8.45 ± 0.24 δ | 8.19 ± 0.46 γδ | 7.94 ± 0.23 γ |
| D-G-I | 6.17 ± 0.18 Aα | n.d. | 8.14 ± 0.24 α | 8.47 ± 0.12 β | 8.65 ± 0.14 Aβ | 8.45 ± 0.05 ABβ | 8.54 ± 0.13 ABβ |
| D-G-O | 6.88 ± 0.10 Cα | n.d. | 8.42 ± 0.30 α | n.d. | 8.55 ± 0.14 ABβ | 8.62 ± 0.10 ABβ | 8.58 ± 0.05 Bβ |
| D-CS-I | 6.14 ± 0.09 Aα | n.d. | 8.53 ± 0.04 γ | n.d. | 8.63 ± 0.06 ABγ | 8.58 ± 0.02 Bγ | 8.25 ± 0.02 Bβ |
| D-CS-O | 8.76 ± 0.04 Dα | n.d. | 8.17 ± 0.08 β | n.d. | 8.61 ± 0.05 ABγ | 8.27 ± 0.12 Aβ | 8.12 ± 0.07 Aβ |
| D-MR-I | 6.33 ± 0.20 ABα | n.d. | 8.54 ± 0.08 β | n.d. | 8.62 ± 0.13 ABβ | 8.52 ± 0.11 ABβ | 8.50 ± 0.11 ABβ |
| D-MR-O | 6.50 ± 0.05 Bα | n.d. | 8.58 ± 0.13 βγ | n.d. | 8.11 ± 0.16 Bβ | 8.78 ± 0.10 Bγ | 8.32 ± 0.12 ABβ |
| Mean | 6.80 ± 0.94 α | 8.39 ± 0.23 β | 8.47 ± 0.12 β | 8.53 ± 0.22 β | 8.53 ± 0.18 β | 8.38 ± 0.19 β | |
| Identified Species (NCBI Accession Number) | t0 | t9 | t30 | t60 | t90 |
|---|---|---|---|---|---|
| Plate Dilution at Which the Identified Species were Detected | |||||
| D-G-I | |||||
| Lactobacillus sakei (NR_113821.1) | 10−2 | n.d. | n.d. | n.d. | 10−6 |
| Lactobacillus curvatus (NR_113334.1) | n.d. | n.d. | n.d. | n.d. | 10−7 |
| Lactobacillus casei (NR_041893.1) | n.d. | n.d. | 107 | n.d. | 10−7 |
| Pediocossus pentosaceus (NR_042058.1) | 10−7 | 10−7 | 10−7 | 10−7 | 10−7 |
| D-G-O | |||||
| Lactobacillus sakei (NR_113821.1) | 10−3 | 10−6 | 10−7 | 10−7 | 10−7 |
| Lactobacillus curvatus (NR_113334.1) | n.d. | n.d. | 10−6 | 10−6 | 10−6 |
| Pediocossus pentosaceus (NR_042058.1) | 10−6 | 10−6 | 10−7 | 10−7 | 10−7 |
| D-CS-I | |||||
| Lactobacillus sakei (NR_113821.1) | 10−2 | 10−6 | 10−7 | 10−7 | 10−6 |
| Pediocossus pentosaceus (NR_042058.1) | 10−5 | 10−7 | 10−7 | 10−7 | 10−7 |
| D-CS-O | |||||
| Lactobacillus sakei (NR_113821.1) | n.d. | n.d. | n.d. | 10−5 | 10−4 |
| Lactobacillus curvatus (NR_113334.1) | n.d. | n.d. | n.d. | 10−5 | 10−5 |
| Pediocossus pentosaceus (NR_042058.1) | 10−6 | 10−7 | 10−7 | 10−6 | 10−7 |
| D-MR-I | |||||
| Lactobacillus sakei (NR_113821.1) | n.d. | n.d. | n.d. | 10−6 | 10−4 |
| Pediocossus pentosaceus (NR_042058.1) | 10−5 | 10−7 | 10−7 | 10−7 | 10−6 |
| D-MR-O | |||||
| Lactobacillus sakei (NR_113821.1) | n.d. | 10−7 | 10−7 | 10−7 | 10−7 |
| Lactobacillus curvatus (NR_113334.1) | n.d. | 10−7 | 10−7 | 10−7 | 10−7 |
| Pediocossus pentosaceus (NR_042058.1) | 10−6 | 10−7 | 10−7 | 10−7 | 10−7 |
| Identified Species (NCBI Accession Number) | t0 | t2 | t9 | t23 | t30 | t60 | t90 |
|---|---|---|---|---|---|---|---|
| Plate Dilution at Which the Identified Species were Detected | |||||||
| T-G-I | |||||||
| Lactobacillus sakei (NR_113821.1) | 10−2 | 10−5 | 10−5 | 10−5 | 10−6 | 10−6 | n.d. |
| Lactobacillus curvatus (NR_113334.1) | 10−2 | 10−4 | 10−5 | 10−5 | 10−6 | 10−6 | 10−6 |
| Lactobacillus casei (NR_041893.1) | 10−2 | 10−4 | 10−6 | n.d. | n.d. | n.d. | n.d. |
| Lactobacillus graminis (NR_042438.1) | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 10−6 |
| Lactococcus garviae (KU898985.1) | n.d. | 10−3 | 10−6 | 10−7 | n.d. | n.d. | n.d. |
| Leuconostoc mesenteroides (DQ297412.1) | n.d. | n.d. | n.d. | 10−7 | 10−6 | 10−6 | n.d. |
| Pediocossus pentosaceus (NR_042058.1) | n.d. | n.d. | n.d. | 10−5 | 10−5 | n.d. | n.d. |
| T-G-O | |||||||
| Lactobacillus sakei (NR_113821.1) | 10−2 | 10−4 | 10−7 | 10−7 | 10−6 | 10−6 | 10−6 |
| Lactobacillus curvatus (NR_113334.1) | 10−2 | 10−4 | 10−7 | 10−7 | 10−6 | n.d. | n.d. |
| Leuconostoc mesenteroides (DQ297412.1) | 102 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| T-CS-I | |||||||
| Lactobacillus sakei (NR_113821.1) | 10−2 | 10−6 | 10−6 | 10−7 | 10−7 | 10−7 | 10−7 |
| Lactobacillus curvatus (NR_113334.1) | n.d. | 10−6 | 10−6 | 10−7 | 10−7 | 10−7 | 10−7 |
| Lactococcus garviae (KU898985.1) | 10−3 | 10−4 | n.d. | n.d. | n.d. | n.d. | n.d. |
| Leuconostoc mesenteroides (DQ297412.1) | 10−2 | 10−6 | n.d. | 10−5 | 10−7 | n.d. | n.d. |
| T-CS-O | |||||||
| Lactobacillus sakei (NR_113821.1) | n.d. | 10−3 | 10−7 | 10−7 | 10−7 | 10−6 | 10−7 |
| Lactobacillus curvatus (NR_113334.1) | n.d. | 10−3 | 10−7 | 10−7 | 10−7 | 10−6 | 10−7 |
| Lactococcus garviae (KU898985.1) | n.d. | 10−3 | n.d. | n.d. | n.d. | n.d. | n.d. |
| Leuconostoc mesenteroides (DQ297412.1) | 10−3 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Comi, G.; Muzzin, A.; Corazzin, M.; Iacumin, L. Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology. Foods 2020, 9, 338. https://doi.org/10.3390/foods9030338
Comi G, Muzzin A, Corazzin M, Iacumin L. Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology. Foods. 2020; 9(3):338. https://doi.org/10.3390/foods9030338
Chicago/Turabian StyleComi, Giuseppe, Alessia Muzzin, Mirco Corazzin, and Lucilla Iacumin. 2020. "Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology" Foods 9, no. 3: 338. https://doi.org/10.3390/foods9030338
APA StyleComi, G., Muzzin, A., Corazzin, M., & Iacumin, L. (2020). Lactic Acid Bacteria: Variability Due to Different Pork Breeds, Breeding Systems and Fermented Sausage Production Technology. Foods, 9(3), 338. https://doi.org/10.3390/foods9030338







