Characterization of Defatted Products Obtained from the Parmigiano–Reggiano Manufacturing Chain: Determination of Peptides and Amino Acids Content and Study of the Digestibility and Bioactive Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Cheese Defatting
2.2. Fatty Acids Determination by GasChromatrography( GC-FID)
2.3. Cholesterol Analysis by GC-FID
2.4. Total Nitrogen and Fat Content Determination
2.5. Isolation of the Peptide and Amino Acid Fractions
2.6. Simulated Gastrointestinal Digestion
2.7. LC-MS Amino Acids Quantification
2.8. LC–MS Characterization of Peptides and Proteins
2.9. Antioxidant Capacity of the Digested Samples
2.10. ACE Inhibition Capacity of the Digested Samples
2.11. Data Analysis
3. Results and Discussion
3.1. Defatted Products
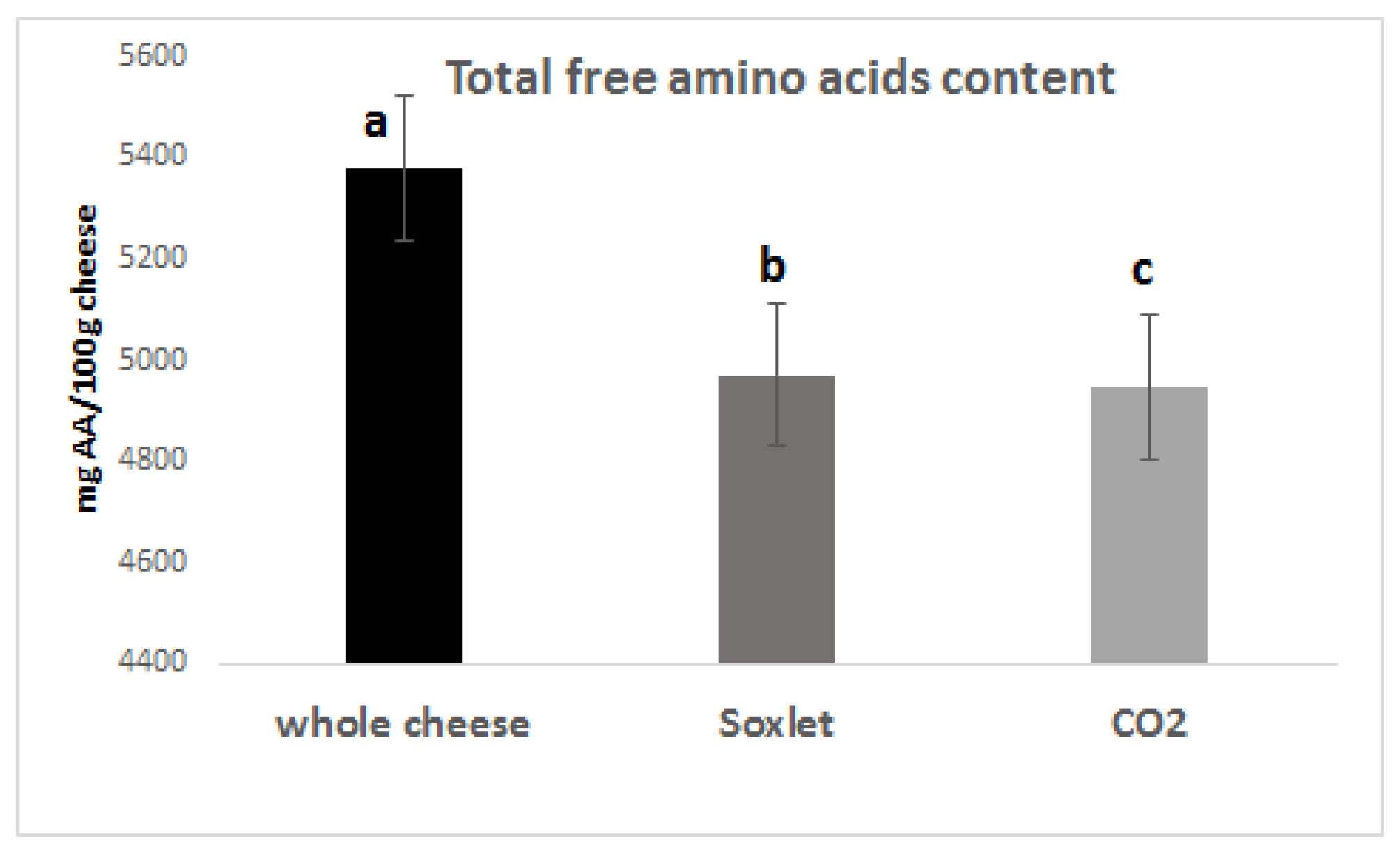
3.2. Free Amino Acids in the PR Samples
3.3. Characterization of Peptides and Proteins in the Aqueous Extracts
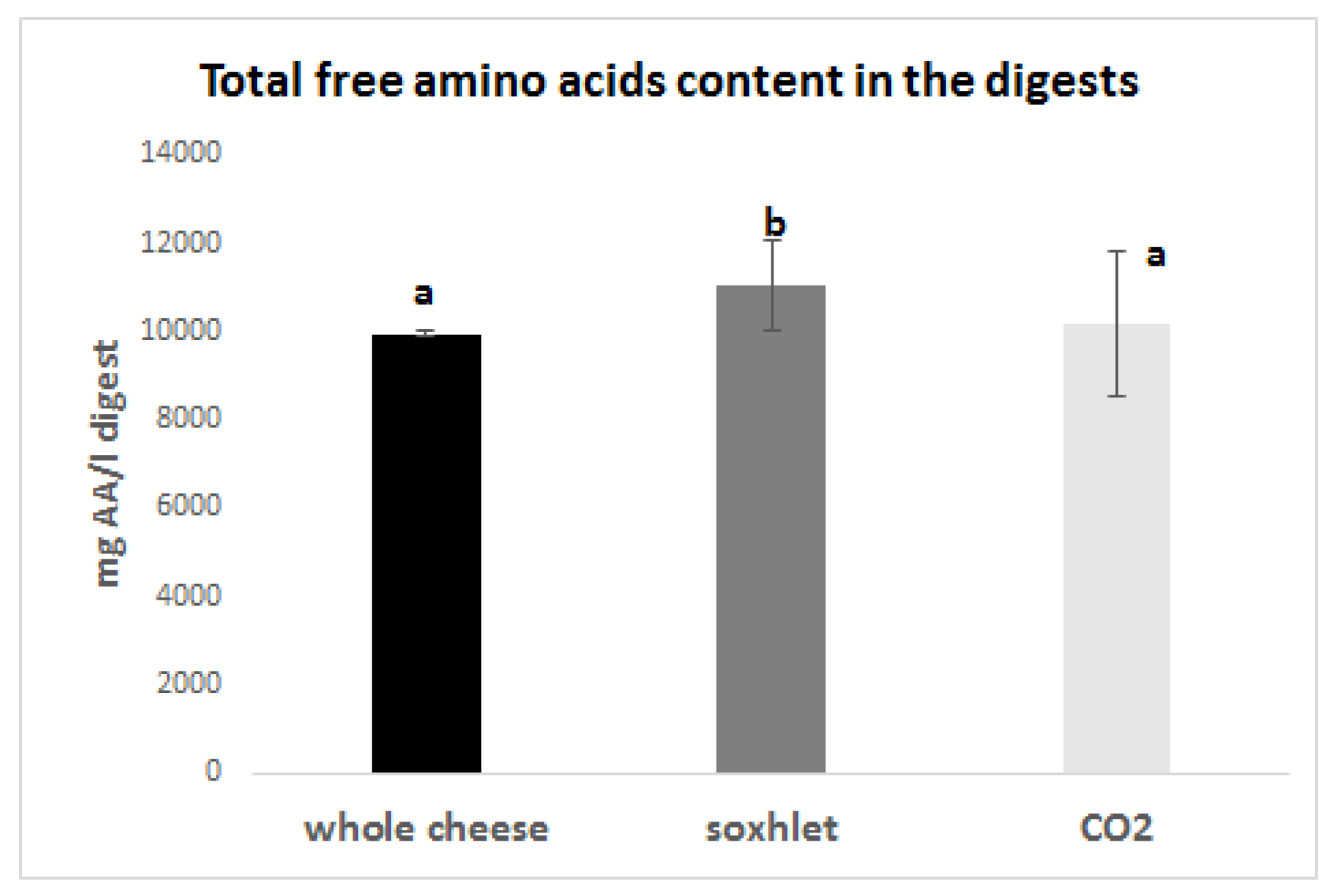
3.4. Simulated Gastrointestinal Digestion
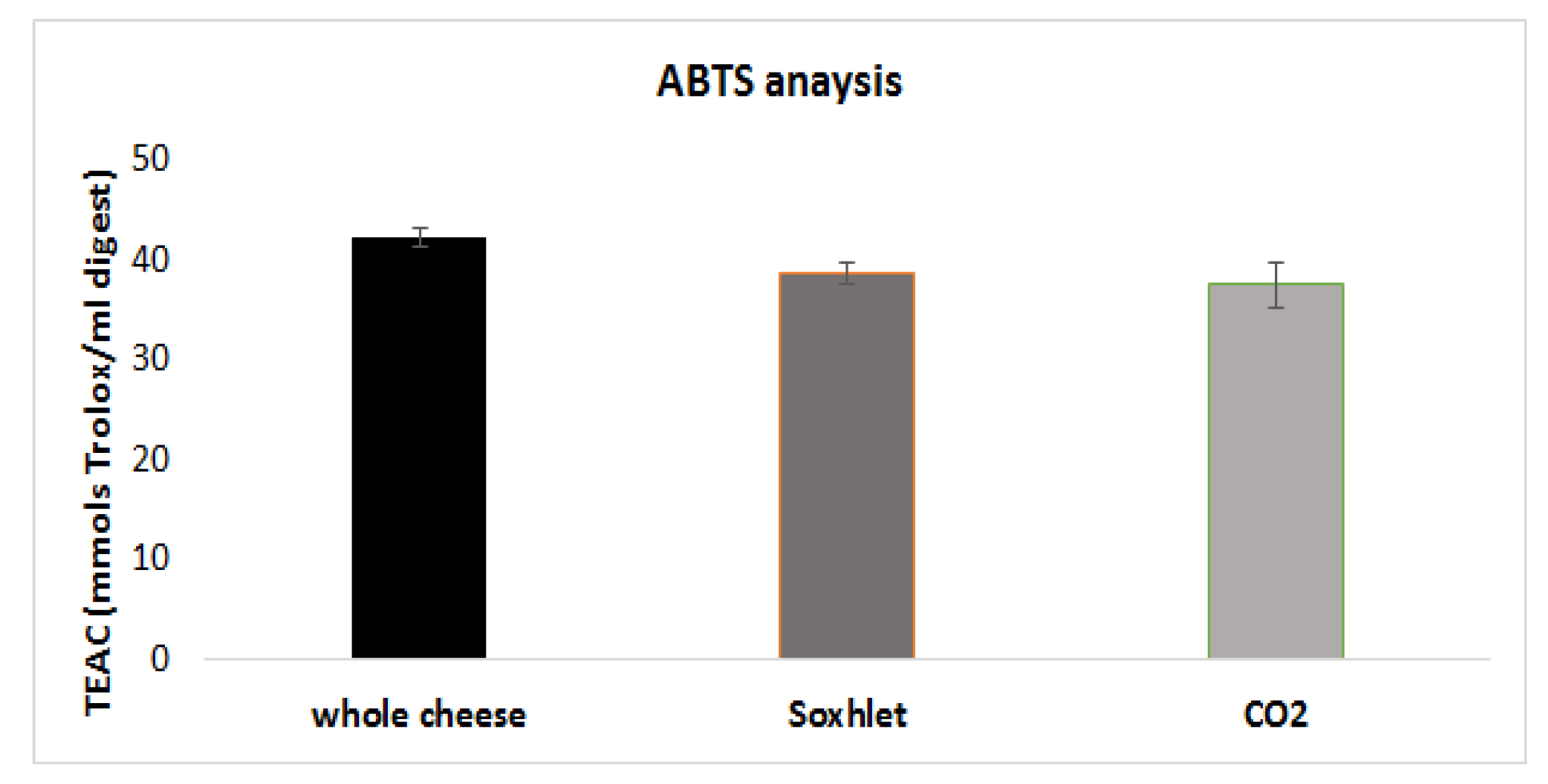
3.5. Antioxidant Activity of the Digested Samples
3.6. ACE inhibition Capacity of the Digested Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Parmigiano-Reggiano Single Document. Available online: https://www.parmigianoreggiano.com/ (accessed on 20 December 2019).
- Parmigiano Reggiano Official Website. Available online: https://www.parmigianoreggiano.com/made/nutritional_features/default.aspx (accessed on 20 December 2019).
- MacGibbon, A.H.K.; Taylor, M.W. Composition and structure of bovine milk lipids. In Advanced Dairy Chemistry; Fox, P.F., McSweeney, P.L.H., Eds.; Springer: New York, NY, USA, 2006; pp. 1–42. [Google Scholar]
- Gordon, T. The diet-heart idea: Outline of a history. Am. J. Epidemiol. 1988, 127, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Mensink, R.P.; Katan, M.B. Effect of dietary fatty acids on serum lipids and lipoproteins. A meta-analysis of 27 trials. Arterioscler. Thromb. 1992, 12, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-T.; Yen, S.T. Knowledge of Dietary Fats among US Consumers. J. Am. Diet. Assoc. 2010, 110, 613–618. [Google Scholar]
- Undeland, I.; Härröd, M.; Lingnert, H. Comparison between methods using low-toxicity solvents for the extraction of lipids from herring (Clupea harengus). Food Chem. 1998, 61, 355–365. [Google Scholar] [CrossRef]
- Biondo, P.B.F.; dos Santos, V.J.; Montanher, P.F.; de OS Junior, O.; Matsushita, M.; Almeida, V.C.; Visentainer, J.V. A new method for lipid extraction using low toxicity solvents developed for canola (Brassica napus L.) and soybean (Glycine max L. Merrill) seeds. Anal. Methods 2015, 7, 9773–9778. [Google Scholar] [CrossRef]
- Ashraf Gaber, M. Low-Fat Cheese: A Modern Demand. Int. J. Dairy Sci. 2015, 10, 249–265. [Google Scholar]
- Carunchia Whetstine, M.E.; Drake, M.A.; Nelson, B.K.; Barbano, D.M. Flavor profiles of full-fat and reduced-fat cheese and cheese fat made from aged cheddar with the fat removed using a novel process. J. Dairy Sci. 2006, 89, 505–517. [Google Scholar] [CrossRef]
- Yee, J.L.; Khalil, H.; Jimenez -Flores, R. Flavor partition and fat reduction in cheese by supercritical fluid extraction: processing variables. Lait 2007, 87, 269–285. [Google Scholar] [CrossRef]
- Manirakiza, P.; Covaci, A.; Schepens, P. Comparative Study on Total Lipid Determination using Soxhlet, Roese-Gottlieb, Bligh & Dyer, and Modified Bligh & Dyer Extraction Methods. J. Food Compost. Anal. 2001, 14, 93–100. [Google Scholar]
- Perretti, G.; Marconi, O.; Montanari, L.; Fantozzi, P. Rapid determination of total fats and fat-soluble vitamins in Parmigiano cheese and salami by SFE. LWT 2004, 37, 87–92. [Google Scholar] [CrossRef]
- Verardo, V.; Gómez-Caravaca, A.M.; Gori, A.; Losi, G.; Caboni, M.F. Bioactive lipids in the butter production chain from Parmigiano Reggiano cheese area. J. Sci. Food Agric. 2013, 93, 3625–3633. [Google Scholar] [CrossRef] [PubMed]
- Christie, W.W. A simple procedure for rapid transmethylation of glycerolipids and cholesteryl esters. J. Lipid Res. 1982, 23, 1072–1075. [Google Scholar] [PubMed]
- Guerra, E.; Downey, E.; O’Mahony, J.A.; Caboni, M.F.; O’Shea, C.A.; Ryan, A.C.; Kelly, A.L. Influence of duration of gestation on fatty acid profiles of human milk. Eur. J. Lipid Sci. Technol. 2016, 118, 1775–1787. [Google Scholar] [CrossRef]
- AOAC International Website. Available online: https://www.aoac.org (accessed on 3 March 2020).
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn; Bourlieu, C.; Carriere, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardized static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant capacity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Cushman, D.W.; Cheung, H.S. Spectrophotometric assay properties of the angiotensin-converting enzyme of rabbit lung. Biochem. Pharmacol. 1971, 20, 1637–1648. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Okubo, A.; Yamazaki, S.; Takano, T. Purification and characterization of angiotensin I-converting enzyme inhibitors from sour milk. J. Dairy Sci. 1995, 78, 777–783. [Google Scholar] [CrossRef]
- Vibha, D.; Shabina, K. Comparative study of different extraction processes for hemp (Cannabis sativa) seed oil considering physical, chemical and industrial-scale economic aspects. J. Clean. Prod. 2019, 207, 645–657. [Google Scholar]
- Kyo-Yeon, L.M.; Shafiur, R.; Ah-Na, K.; Khalid, G.; Sung-Won, K.; Jiyeon, C.; William, L.K.; Sung-Gil, C. Quality characteristics and storage stability of low-fat tofu prepared with defatted soy flours treated by supercritical-CO2 and hexane. LWT 2019, 100, 237–243. [Google Scholar]
- Rahman, M.S.; Seo, J.K.; Choi, S.G.; Gul, K.; Yang, H.S. Physicochemical characteristics and microbial safety of defatted bovine heart and its lipid extracted with supercritical-CO2 and solvent extraction. LWT 2018, 95, 355–371. [Google Scholar] [CrossRef]
- Istituto Nazionale di Ricerca per gli Alimenti e la Nutrizione (INRAN) Dossier Il Parmigiano Reggiano un Prodotto Naturalmente Funzionale; National Research Institute on Food and Nutrition: Rome, Italy, 2008.
- Hillmann, H.; Hofmann, T. Quantitation of Key Tastants and Reengineering the Taste of Parmesan Cheese. J. Agric. Food Chem. 2016, 64, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Virgili, R.; Parolari, G.; Bolzoni, L.; Barbieri, G.; Mangia, A.; Careri, M.; Spagnoli, S.; Panari, G.; Zannoni, M. Sensory-Chemical Relationships in Parmigiano-Reggiano Cheese. Food Sci. Technol. 1994, 27, 491–495. [Google Scholar] [CrossRef]
- Careri, M.; Spagnoli, S.; Panari, G.; Zannoni, M.; Barbieri, G. Chemical parameters of the non-volatile fraction of ripened Parmigiano-Reggiano cheese. Int. Dairy J. 1996, 6, 147–155. [Google Scholar] [CrossRef]
- Indpep Tool. Available online: http://web.expasy.org/findpept/?_ga=1.4745075.1565211071.1480347985 (accessed on 20 December 2019).
- Proteomics Toolkit. Available online: http://db.systemsbiology.net:8080/proteomicsToolkit/FragIonServlet.html (accessed on 20 December 2019).
- Sforza, S.; Cavatorta, V.; Lambertini, F.; Galaverna, G.; Dossena, A.; Marchelli, R. Cheese peptidomics: a detailed study on the evolution of the oligopeptide fraction in Parmigiano-Reggiano cheese from curd to 24 months of aging. J. Dairy Sci. 2012, 95, 3514–3526. [Google Scholar] [CrossRef]
- Sforza, S.; Ferroni, L.; Galaverna, G.; Dossena, A.; Marchelli, R. Extraction, semi-quantification, and fast on-line identification of oligopeptides in Grana Padano cheese by HPLC-MS. J. Agric. Food Chem. 2003, 51, 2130–2135. [Google Scholar] [CrossRef]
- Mackie, A.; Macierzanka, A. Colloidal aspects of protein digestion. Curr. Opin. Colloid. Interface Sci. 2010, 15, 102–108. [Google Scholar] [CrossRef]
- Korhonen, H. Milk-derived bioactive peptides: from science to applications. J. Funct. Foods 2009, 1, 177–187. [Google Scholar] [CrossRef]
- Bottari, B.; Quartieri, A.; Prandi, B.; Raimondi, S.; Leonardi, A.; Rossi, M.; Ulrici, A.; Gatti, M.; Sforza, S.; Nocetti, M.; et al. Characterization of the peptide fraction from digested Parmigiano Reggiano cheese and its effect on growth of lactobacilli and bifidobacteria. Int. J. Food Microbiol. 2017, 16, 32–41. [Google Scholar] [CrossRef]
- Bottesini, C.; Paolella, S.; Lambertini, F.; Galaverna, G.; Tedeschi, T.; Dossena, A.; Marchelli, R.; Sforza, S. Antioxidant capacity of water soluble extracts from Parmigiano-Reggiano cheese. Int. J. Food Sci. Nutr. 2013, 64, 953–958. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef]
- Kitts, D.D.; Weiler, K. Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery. Curr. Pharm. Des. 2003, 9, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- BIOPEP Database. Available online: http://www.uwm.edu.pl/biochemia/index.php/pl/biopep, (accessed on 20 December 2019).
- Lu, Y.; Govindasamy-Lucey, S.; Lucey, J.A. Angiotensin-I-converting enzyme-inhibitory peptides in commercial Wisconsin Cheddar cheeses of different ages. J. Dairy Sci. 2016, 99, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Nakamura, T.; Kitazawa, H.; Kawai, Y.; Itoh, T. Isolation and Structural Analysis of Antihypertensive Peptides That Exist Naturally in Gouda Cheese. J. Dairy Sci. 2000, 83, 1434–1440. [Google Scholar] [CrossRef]


| Sample | Protein (%) | Fat (%) |
|---|---|---|
| Whole Cheese | 35.7 ± 0.1 | 37.7 ± 0.4 |
| Soxhlet-defatted | 50.7 ± 2.2 | 2.6 ± 0.2 |
| CO2-defatted | 46.2 ± 0.9 | 4.6 ± 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buhler, S.; Riciputi, Y.; Perretti, G.; Caboni, M.F.; Dossena, A.; Sforza, S.; Tedeschi, T. Characterization of Defatted Products Obtained from the Parmigiano–Reggiano Manufacturing Chain: Determination of Peptides and Amino Acids Content and Study of the Digestibility and Bioactive Properties. Foods 2020, 9, 310. https://doi.org/10.3390/foods9030310
Buhler S, Riciputi Y, Perretti G, Caboni MF, Dossena A, Sforza S, Tedeschi T. Characterization of Defatted Products Obtained from the Parmigiano–Reggiano Manufacturing Chain: Determination of Peptides and Amino Acids Content and Study of the Digestibility and Bioactive Properties. Foods. 2020; 9(3):310. https://doi.org/10.3390/foods9030310
Chicago/Turabian StyleBuhler, Sofie, Ylenia Riciputi, Giuseppe Perretti, Maria Fiorenza Caboni, Arnaldo Dossena, Stefano Sforza, and Tullia Tedeschi. 2020. "Characterization of Defatted Products Obtained from the Parmigiano–Reggiano Manufacturing Chain: Determination of Peptides and Amino Acids Content and Study of the Digestibility and Bioactive Properties" Foods 9, no. 3: 310. https://doi.org/10.3390/foods9030310
APA StyleBuhler, S., Riciputi, Y., Perretti, G., Caboni, M. F., Dossena, A., Sforza, S., & Tedeschi, T. (2020). Characterization of Defatted Products Obtained from the Parmigiano–Reggiano Manufacturing Chain: Determination of Peptides and Amino Acids Content and Study of the Digestibility and Bioactive Properties. Foods, 9(3), 310. https://doi.org/10.3390/foods9030310






