Control of Penicillium glabrum by Indigenous Antagonistic Yeast from Vineyards
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling, Microbial Counts, and Isolation of Molds and Yeast
2.2. Identification of Isolates
2.3. Screening for Antagonistic Activity by Direct Confrontation
2.4. Characterization of the Mechanism of Antagonism
2.4.1. Production of Antifungal Volatile Organic Compounds
2.4.2. Effect of Antagonistic Yeasts on Spore Germination of Pathogenic Molds
2.4.3. Parasitism of P. glabrum hyphae
2.4.4. Production of Antifungal Extracellular Substances
Antibiosis
Production of Lytic Enzymes
2.4.5. In Vitro Biofilm Formation
2.4.6. Competition for Nutrients
2.4.7. In Vivo Assay and Competition for Nutrients Determination
2.4.8. Analysis of Results
3. Results
3.1. Microbial Counts and Species Identification of Yeasts and Molds
3.2. Screening of Antagonistic Activity by Direct Confrontation
3.3. Characterization of the Mechanism of Antagonistic Activity against P. glabrum
3.4. In Vivo Assay on Wounded Grapes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Setati, M.E.; Jacobson, D.; Andong, U.-C.; Bauer, F.F. The Vineyard Yeast Microbiome, a Mixed Model Microbial Map. PLoS ONE 2012, 7, e52609. [Google Scholar] [CrossRef] [PubMed]
- Sage, L.; Garon, D.; Seigle-Murandi, F. Fungal Microflora and Ochratoxin A Risk in French Vineyards. J. Agric. Food Chem. 2004, 52, 5764–5768. [Google Scholar] [CrossRef]
- Schmid, F.; Moser, G.; Müller, H.; Berg, G. Functional and Structural Microbial Diversity in Organic and Conventional Viticulture: Organic Farming Benefits Natural Biocontrol Agents. Appl. Environ. Microbiol. 2011, 77, 2188–2191. [Google Scholar] [CrossRef] [PubMed]
- Diguta, C.; Vincent, B.; Guilloux-Benatier, M.; Alexandre, H.; Rousseaux, S. PCR ITS-RFLP: A useful method for identifying filamentous fungi isolates on grapes. Food Microbiol. 2011, 28, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, S.; Diguta, C.F.; Radoï-Matei, F.; Alexandre, H.; Guilloux-Benatier, M. Non-Botrytis grape-rotting fungi responsible for earthy and moldy off-flavors and mycotoxins. Food Microbiol. 2014, 38, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Rabosto, X.; Carrau, M.; Paz, A.; Boido, E.; Dellacassa, E.; Carrau, F.M. Grapes and vineyard soils as sources of microorganisms for biological control of Botrytis cinerea. Am. J. Enol. Vitic. 2006, 57, 332–338. [Google Scholar]
- Wang, X.; Glawe, D.A.; Kramer, E.; Weller, D.M.; Okubara, P.A. Biological Control of Botrytis cinerea: Interactions with Native Vineyard Yeasts from Washington State. Phytopathology 2018, 108, 691–701. [Google Scholar] [CrossRef]
- Jacometti, M.; Wratten, S.D.; Walter, M. Review: Alternatives to synthetic fungicides for Botrytis cinerea management in vineyards. Aust. J. Grape Wine Res. 2010, 16, 154–172. [Google Scholar] [CrossRef]
- Ky, I.; Lorrain, B.; Jourdes, M.; Pasquier, G.; Fermaud, M.; Gény, L.; Rey, P.; Doneche, B.; Teissedre, P.-L. Assessment of grey mould (Botrytis cinerea) impact on phenolic and sensory quality of Bordeaux grapes, musts and wines for two consecutive vintages. Aust. J. Grape Wine Res. 2012, 18, 215–226. [Google Scholar] [CrossRef]
- Morales, H.; Silva, L.; Paterson, R.R.M.; Venâncio, A.; Lima, N. Effects of the origins of Botrytis cinerea on earthy aromas from grape broth media further inoculated with Penicillium expansum. Food Microbiol. 2011, 28, 1048–1053. [Google Scholar] [CrossRef]
- Tjamos, S.E.; Antoniou, P.P.; Kazantzidou, A.; Antonopoulos, D.F.; Papageorgiou, I.; Tjamos, E.C. Aspergillus niger and Aspergillus carbonarius in Corinth Raisin and Wine-Producing Vineyards in Greece: Population Composition, Ochratoxin A Production and Chemical Control. J. Phytopathol. 2004, 152, 250–255. [Google Scholar] [CrossRef]
- Garcia-Cela, E.; Crespo-Sempere, A.; Ramos, A.; Sanchis, V.; Marin, S. Ecophysiological characterization of Aspergillus carbonarius, Aspergillus tubingensis and Aspergillus niger isolated from grapes in Spanish vineyards. Int. J. Food Microbiol. 2014, 173, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Covarelli, L.; Beccari, G.; Marini, A.; Tosi, L. A review on the occurrence and control of ochratoxigenic fungal species and ochratoxin A in dehydrated grapes, non-fortified dessert wines and dried vine fruit in the Mediterranean area. Food Control 2012, 26, 347–356. [Google Scholar] [CrossRef]
- Jiang, C.; Shi, J.; Zhu, C. Fruit spoilage and ochratoxin a production by Aspergillus carbonarius in the berries of different grape cultivars. Food Control 2013, 30, 93–100. [Google Scholar] [CrossRef]
- Dachery, B.; Hernandes, K.C.; Veras, F.F.; Schmidt, L.; Augusti, P.R.; Manfroi, V.; Zini, C.A.; Welke, J.E. Effect of Aspergillus carbonarius on ochratoxin a levels, volatile profile and antioxidant activity of the grapes and respective wines. Food Res. Int. 2019, 126, 108687. [Google Scholar] [CrossRef] [PubMed]
- Ostry, V.; Malir, F.; Cumova, M.; Kyrova, V.; Toman, J.; Grosse, Y.; Pospichalova, M.; Ruprich, J. Investigation of patulin and citrinin in grape must and wine from grapes naturally contaminated by strains of Penicillium expansum. Food Chem. Toxicol. 2018, 118, 805–811. [Google Scholar] [CrossRef]
- Abdullah, Q.; Mahmoud, A.; Al-harethi, A. Isolation and identification of fungal post-harvest rot of some fruits in Yemen. PSM Microbiol. 2016, 1, 36–44. [Google Scholar]
- Lorenzini, M.; Simonato, B.; Favati, F.; Bernardi, P.; Sbarbati, A.; Zapparoli, G. Filamentous fungi associated with natural infection of noble rot on withered grapes. Int. J. Food Microbiol. 2018, 272, 83–86. [Google Scholar] [CrossRef]
- Bau, M.; Bragulat, M.R.; Abarca, M.L.; Minguez, S.; Cabañes, F.J. Ochratoxigenic species from Spanish wine grapes. Int. J. Food Microbiol. 2005, 98, 125–130. [Google Scholar] [CrossRef]
- Freire, L.; Passamani, F.R.F.; Thomas, A.B.; Nassur, R.D.C.M.R.; Silva, L.M.; Paschoal, F.N.; Pereira, G.E.; Prado, G.; Batista, L.R. Influence of physical and chemical characteristics of wine grapes on the incidence of Penicillium and Aspergillus fungi in grapes and ochratoxin A in wines. Int. J. Food Microbiol. 2017, 241, 181–190. [Google Scholar] [CrossRef]
- Serra, R.; Lourenço, A.M.G.; Alípio, P.; Venâncio, A. Influence of the region of origin on the mycobiota of grapes with emphasis on Aspergillus and Penicillium species. Mycol. Res. 2006, 110, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Börjesson, T.S.; Stöllman, U.M.; Schnürer, J.L. Off-odorous compounds produced by molds on oatmeal agar: Identification and relation to other growth characteristics. J. Agric. Food Chem. 1993, 41, 2104–2111. [Google Scholar] [CrossRef]
- Börjesson, T.; Stöllman, U.; Schnürer, J. Volatile metabolites produced by six fungal species compared with other indicators of fungal growth on cereal grains. Appl. Environ. Microbiol. 1992, 58, 2599–2605. [Google Scholar] [CrossRef] [PubMed]
- Bardas, G.A.; Tzelepis, G.D.; Lotos, L.; Karaoglanidis, G.S. First report of Penicillium glabrum causing fruit rot of pomegranate (Punica granatum) in Greece. Plant Dis. 2009, 93, 1347. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, D.; Amatulli, M.T.; Garibaldi, A.; Gullino, M.L. First Report of Penicillium glabrum Causing a Postharvest Fruit Rot of Pomegranate (Punica granatum) in the Piedmont Region of Italy. Plant Dis. 2010, 94, 1066. [Google Scholar] [CrossRef] [PubMed]
- Duduk, N.; Lazarević, M.; Žebeljan, A.; Vasić, M.; Vico, I. Blue mould decay of stored onion bulbs caused by Penicillium polonicum, P. glabrum and P. expansum. J. Phytopathol. 2017, 165, 662–669. [Google Scholar] [CrossRef]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Hahn, M. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 2014, 7, 133–141. [Google Scholar] [CrossRef]
- Li, H.X.; Xiao, C.L. Characterization of Fludioxonil-Resistant and Pyrimethanil-Resistant Phenotypes of Penicillium expansum from Apple. Phytopathology 2008, 98, 427–435. [Google Scholar] [CrossRef]
- Bowyer, P.; Denning, D.W. Environmental fungicides and triazole resistance in Aspergillus. Pest Manag. Sci. 2014, 70, 173–178. [Google Scholar] [CrossRef]
- Tripathi, P.; Dubey, N. Exploitation of natural products as an alternative strategy to control postharvest fungal rotting of fruit and vegetables. Postharvest Biol. Technol. 2004, 32, 235–245. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]
- Gasser, F.; Berg, G.; Cardinale, M.; Schildberger, B. Biocontrol of Botrytis cinerea by successful introduction of Pantoea ananatis in the grapevine phyllosphere. Int. J. Wine Res. 2012, 4, 53. [Google Scholar] [CrossRef]
- Magnin-Robert, M.; Quantinet, D.; Couderchet, M.; Aziz, A.; Trotel-Aziz, P. Differential induction of grapevine resistance and defense reactions against Botrytis cinerea by bacterial mixtures in vineyards. BioControl 2012, 58, 117–131. [Google Scholar] [CrossRef]
- Lemos, W.J.; Bovo, B.; Nadai, C.; Crosato, G.; Carlot, M.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol Ability and Action Mechanism of Starmerella bacillaris (Synonym Candida zemplinina) Isolated from Wine Musts against Gray Mold Disease Agent Botrytis cinerea on Grape and Their Effects on Alcoholic Fermentation. Front. Microbiol. 2016, 7, 1249. [Google Scholar] [CrossRef]
- Pertot, I.; Giovannini, O.; Benanchi, M.; Caffi, T.; Rossi, V.; Mugnai, L. Combining biocontrol agents with different mechanisms of action in a strategy to control Botrytis cinerea on grapevine. Crop. Prot. 2017, 97, 85–93. [Google Scholar] [CrossRef]
- Arfaoui, M.; Vallance, J.; Bruez, E.; Rezgui, A.; Melki, I.; Chebil, S.; Sadfi-Zouaouia, N.; Rey, P. Isolation, identification and in vitro characterization of grapevine rhizobacteria to control ochratoxigenic Aspergillus spp. on grapes. Biol. Control 2019, 129, 201–211. [Google Scholar] [CrossRef]
- Lappa, I.K.; Mparampouti, S.; Lanza, B.; Panagou, E.Z. Control of Aspergillus carbonarius in grape berries by Lactobacillus plantarum: A phenotypic and gene transcription study. Int. J. Food Microbiol. 2018, 275, 56–65. [Google Scholar] [CrossRef]
- Apaliya, M.T.; Zhang, H.; Zheng, X.; Yang, Q.; Mahunu, G.K.; Kwaw, E. Exogenous trehalose enhanced the biocontrol efficacy of Hanseniaspora uvarum against grape berry rots caused by Aspergillus tubingensis and Penicillium commune. J. Sci. Food Agric. 2018, 98, 4665–4672. [Google Scholar] [CrossRef]
- De Curtis, F.; De Felice, D.; Ianiri, G.; De Cicco, V.; Castoria, R. Environmental factors affect the activity of biocontrol agents against ochratoxigenic Aspergillus carbonarius on wine grape. Int. J. Food Microbiol. 2012, 159, 17–24. [Google Scholar] [CrossRef]
- Dimakopoulou, M.; Tjamos, S.E.; Antoniou, P.P.; Pietri, A.; Battilani, P.; Avramidis, N.; Markakis, E.A.; Tjamos, E.C. Phyllosphere grapevine yeast Aureobasidium pullulans reduces Aspergillus carbonarius (sour rot) incidence in wine-producing vineyards in Greece. Biol. Control 2008, 46, 158–165. [Google Scholar] [CrossRef]
- Ponsone, M.L.; Chiotta, M.L.; Combina, M.; Dalcero, A.; Chulze, S. Biocontrol as a strategy to reduce the impact of ochratoxin A and Aspergillus section Nigri in grapes. Int. J. Food Microbiol. 2011, 151, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Zahavi, T.; Cohen, L.; Weiss, B.; Schena, L.; Daus, A.; Kaplunov, T.; Zutkhi, J.; Ben-Arie, R.; Droby, S. Biological control of Botrytis, Aspergillus and Rhizopus rots on table and wine grapes in Israel. Postharvest Biol. Technol. 2000, 20, 115–124. [Google Scholar] [CrossRef]
- Assaf, L.R.; Pedrozo, L.P.; Nally, M.C.; Pesce, V.M.; Toro, M.E.; de Figueroa, L.C.; Vazquez, F. Use of yeasts from different environments for the control of Penicillium expansum on table grapes at storage temperature. Int. J. Food Microbiol. 2020, 320, 108520. [Google Scholar] [CrossRef] [PubMed]
- Diguța, C.F.; Matei, F.; Cornea, C.P. Biocontrol perspectives of Aspergillus carbonarius, Botrytis cinerea and Pencillium expansum on grapes using epiphytic bacteria isolated from Romanian vineyards. Rom. Biotechnol. Lett. 2016, 21, 11126–11132. [Google Scholar]
- Godana, E.A.; Yang, Q.; Wang, K.; Zhang, H.; Zhang, X.; Zhao, L.; Abdelhai, M.H.; Legrand, N.N.G. Bio-control activity of Pichia anomala supplemented with chitosan against Penicillium expansum in postharvest grapes and its possible inhibition mechanism. LWT 2020, 124, 109188. [Google Scholar] [CrossRef]
- Droby, S.; Vinokur, V.; Weiss, B.; Cohen, L.; Daus, A.; Goldschmidt, E.E.; Porat, R. Induction of Resistance to Penicillium digitatum in Grapefruit by the Yeast Biocontrol Agent Candida oleophila. Phytopathology 2002, 92, 393–399. [Google Scholar] [CrossRef]
- Gallardo, G.; Ruiz-Moyano, S.; Hernández, A.; Benito, M.J.; Córdoba, J.J.; Pérez-Nevado, F.; Martín, A. Application of ISSR-PCR for rapid strain typing of Debaryomyces hansenii isolated from dry-cured Iberian ham. Food Microbiol. 2014, 42, 205–211. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.; Taylor, J. 38-Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Hong, S.-B.; Go, S.-J.; Shin, H.-D.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycology 2005, 97, 1316–1329. [Google Scholar] [CrossRef]
- Ruiz-Moyano, S.; Hernández, A.; Galvan, A.I.; Córdoba, M.G.; Casquete, R.; Serradilla, M.J.; Martín, A. Selection and application of antifungal VOCs-producing yeasts as biocontrol agents of grey mould in fruits. Food Microbiol. 2020, 92, 103556. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Moyano, S.; Martín, A.; Villalobos, M.; Calle, A.; Serradilla, M.J.; Córdoba, J.J.; Hernández, A. Yeasts isolated from figs (Ficus carica L.) as biocontrol agents of postharvest fruit diseases. Food Microbiol. 2016, 57, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Bueso, G.; Mangieri, N.; Maghradze, D.; Foschino, R.; Valdetara, F.; Cantoral, J.M.; Vigentini, I. Wild Grape-Associated Yeasts as Promising Biocontrol Agents against Vitis vinifera Fungal Pathogens. Front. Microbiol. 2017, 8, 2025. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.; Wingfield, M.J.; Guarro, J.; Hernández-Restrepo, M.; Sutton, D.A.; Acharya, K.; Barber, P.A.; Boekhout, T.; Dimitrov, R.A.; Dueñas, M.; et al. Fungal Planet description sheets: 320–370. Persoonia—Mol. Phylogeny Evol. Fungi 2015, 34, 167–266. [Google Scholar] [CrossRef] [PubMed]
- Settanni, L.; Sannino, C.; Francesca, N.; Guarcello, R.; Moschetti, G. Yeast ecology of vineyards within Marsala wine area (western Sicily) in two consecutive vintages and selection of autochthonous Saccharomyces cerevisiae strains. J. Biosci. Bioeng. 2012, 114, 606–614. [Google Scholar] [CrossRef]
- Bougreau, M.; Ascencio, K.; Bugarel, M.; Nightingale, K.; Loneragan, G. Yeast species isolated from Texas High Plains vineyards and dynamics during spontaneous fermentations of Tempranillo grapes. PLoS ONE 2019, 14, e0216246. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Lianou, A.; Nychas, G.J.E. Yeast Populations Residing on Healthy or Botrytis-Infected Grapes from a Vineyard in Attica, Greece. Appl. Environ. Microbiol. 2007, 73, 2765–2768. [Google Scholar] [CrossRef]
- Guzzon, R.; Franciosi, E.; Larcher, R. A new resource from traditional wines: Characterisation of the microbiota of “Vino Santo” grapes as a biocontrol agent against Botrytis cinerea. Eur. Food Res. Technol. 2014, 239, 117–126. [Google Scholar] [CrossRef]
- Drumonde-Neves, J.; Franco-Duarte, R.; Lima, T.; Schuller, D.; Pais, C. Yeast Biodiversity in Vineyard Environments Is Increased by Human Intervention. PLoS ONE 2016, 11, e0160579. [Google Scholar] [CrossRef]
- Castrillo, D.; Crego, E.R.; González, N.N.; Blanco, P. Yeast diversity on grapes from Galicia, NW Spain: Biogeographical patterns and the influence of the farming system. OENO One 2019, 53. [Google Scholar] [CrossRef]
- Longo, E.; Cansado, J.; Agrelo, D.; Villa, T.G. Effect of climatic conditions on yeast diversity in grape musts from northwest Spain. Am. J. Enol. Vitic. 1991, 42, 141–144. [Google Scholar]
- García-Cela, E.; Crespo-Sempere, A.; Gil-Serna, J.; Porqueres, A.; Marin, S. Fungal diversity, incidence and mycotoxin contamination in grapes from two agro-climatic Spanish regions with emphasis on Aspergillus species. J. Sci. Food Agric. 2015, 95, 1716–1729. [Google Scholar] [CrossRef] [PubMed]
- Rotolo, C.; Angelini, R.M.D.M.; Dongiovanni, C.; Pollastro, S.; Fumarola, G.; Di Carolo, M.; Perrelli, D.; Natale, P.; Faretra, F. Use of biocontrol agents and botanicals in integrated management of Botrytis cinerea in table grape vineyards. Pest Manag. Sci. 2018, 74, 715–725. [Google Scholar] [CrossRef]
- Prendes, L.P.; Merín, M.G.; Fontana, A.R.; Bottini, R.A.; Ramirez, M.L.; De Ambrosini, V.I.M. Isolation, identification and selection of antagonistic yeast against Alternaria alternata infection and tenuazonic acid production in wine grapes from Argentina. Int. J. Food Microbiol. 2018, 266, 14–20. [Google Scholar] [CrossRef]
- Bleve, G.; Grieco, F.; Cozzi, G.; Logrieco, A.; Visconti, A. Isolation of epiphytic yeasts with potential for biocontrol of Aspergillus carbonarius and A. niger on grape. Int. J. Food Microbiol. 2006, 108, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Rathnayake, R.M.S.P.; Savocchia, S.; Schmidtke, L.M.; Steel, C.C. Characterisation of Aureobasidium pullulans isolates from Vitis vinifera and potential biocontrol activity for the management of bitter rot of grapes. Eur. J. Plant Pathol. 2018, 151, 593–611. [Google Scholar] [CrossRef]
- Choińska, R.; Piasecka-Jóźwiak, K.; Chabłowska, B.; Dumka, J.; Łukaszewicz, A. Biocontrol ability and volatile organic compounds production as a putative mode of action of yeast strains isolated from organic grapes and rye grains. Antonie van Leeuwenhoek 2020, 113, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Arrebola, E.; Sivakumar, D.; Korsten, L. Effect of volatile compounds produced by Bacillus strains on postharvest decay in citrus. Biol. Control 2010, 53, 122–128. [Google Scholar] [CrossRef]
- Spadaro, D.; Gullino, M.L. State of the art and future prospects of the biological control of postharvest fruit diseases. Int. J. Food Microbiol. 2004, 91, 185–194. [Google Scholar] [CrossRef]
- Lutz, M.C.; Lopes, C.A.; Rodriguez, M.E.; Sosa, M.C.; Sangorrín, M. Efficacy and putative mode of action of native and commercial antagonistic yeasts against postharvest pathogens of pear. Int. J. Food Microbiol. 2013, 164, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Hernández, S.; Reyes-Pérez, J.J.; Chiquito-Contreras, R.G.; Rincon-Enriquez, G.; Cerdan-Cabrera, C.R.; Hernandez-Montiel, L.G. Biocontrol of postharvest fruit fungal diseases by bacterial antagonists: A review. Agronomy 2019, 9, 121. [Google Scholar] [CrossRef]
- Jamalizadeh, M.; Etebarian, H.R.; Aminian, H.; Alizadeh, A. A review of mechanisms of action of biological control organisms against post-harvest fruit spoilage. EPPO Bull. 2011, 41, 65–71. [Google Scholar] [CrossRef]
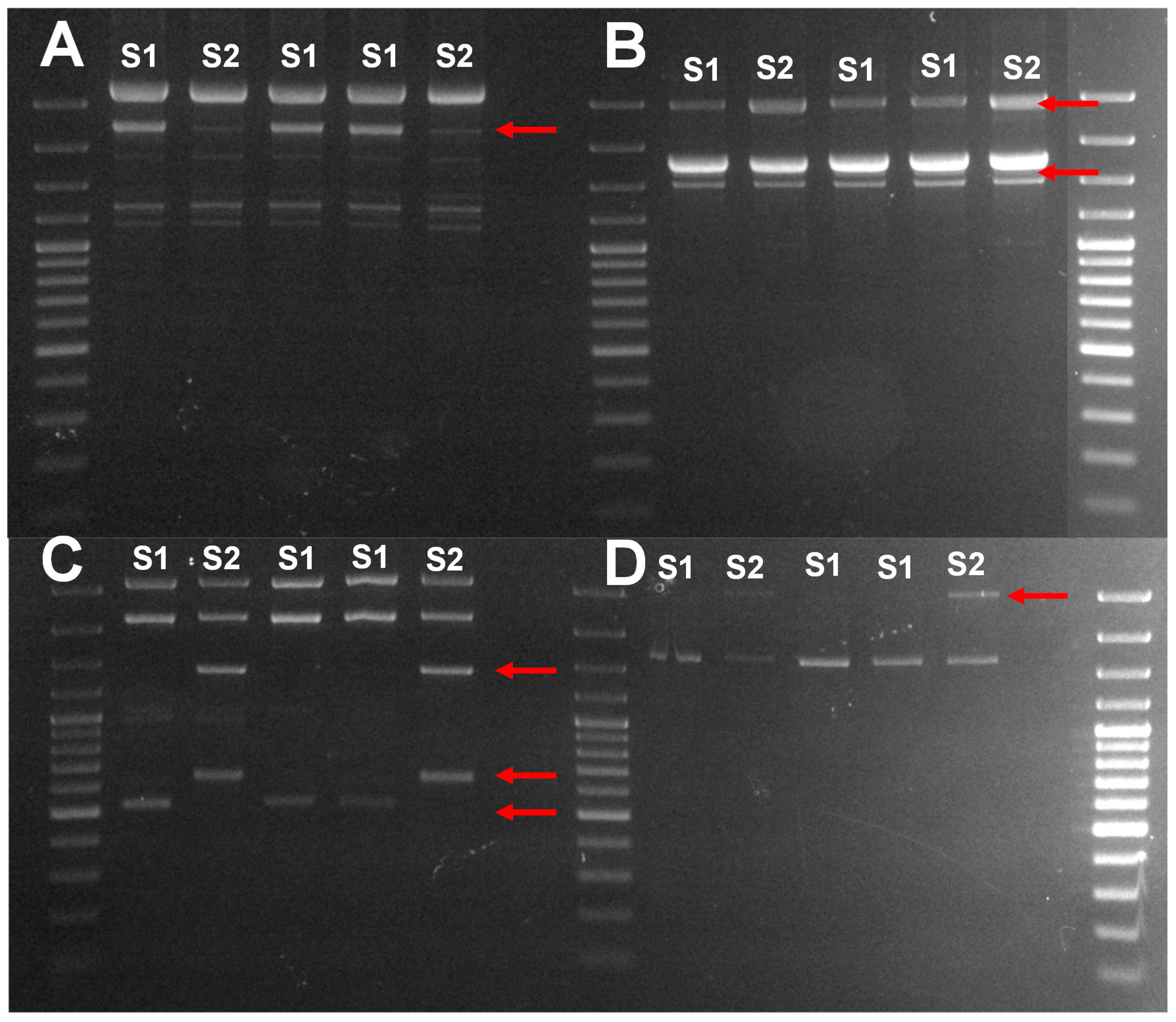
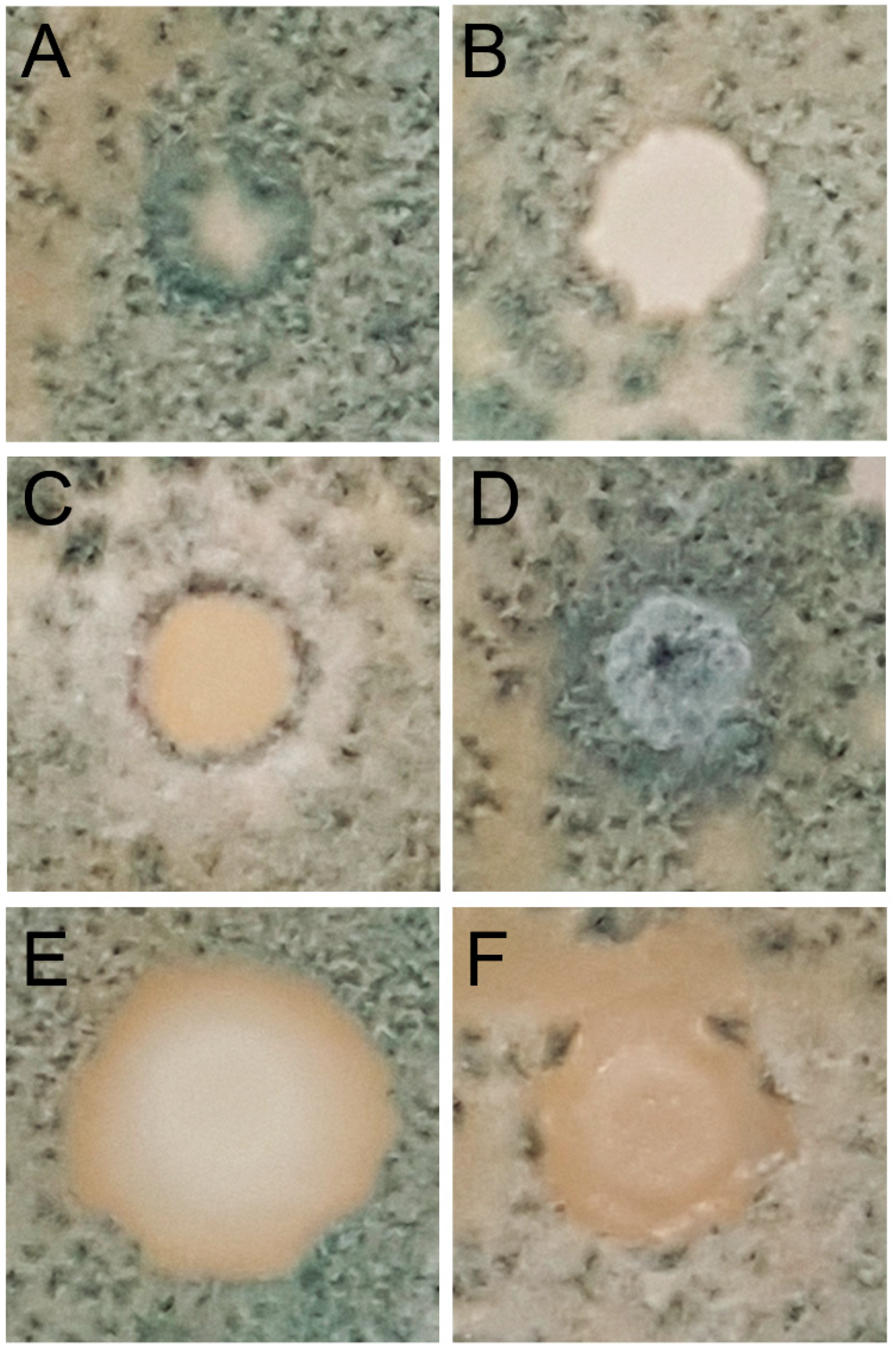
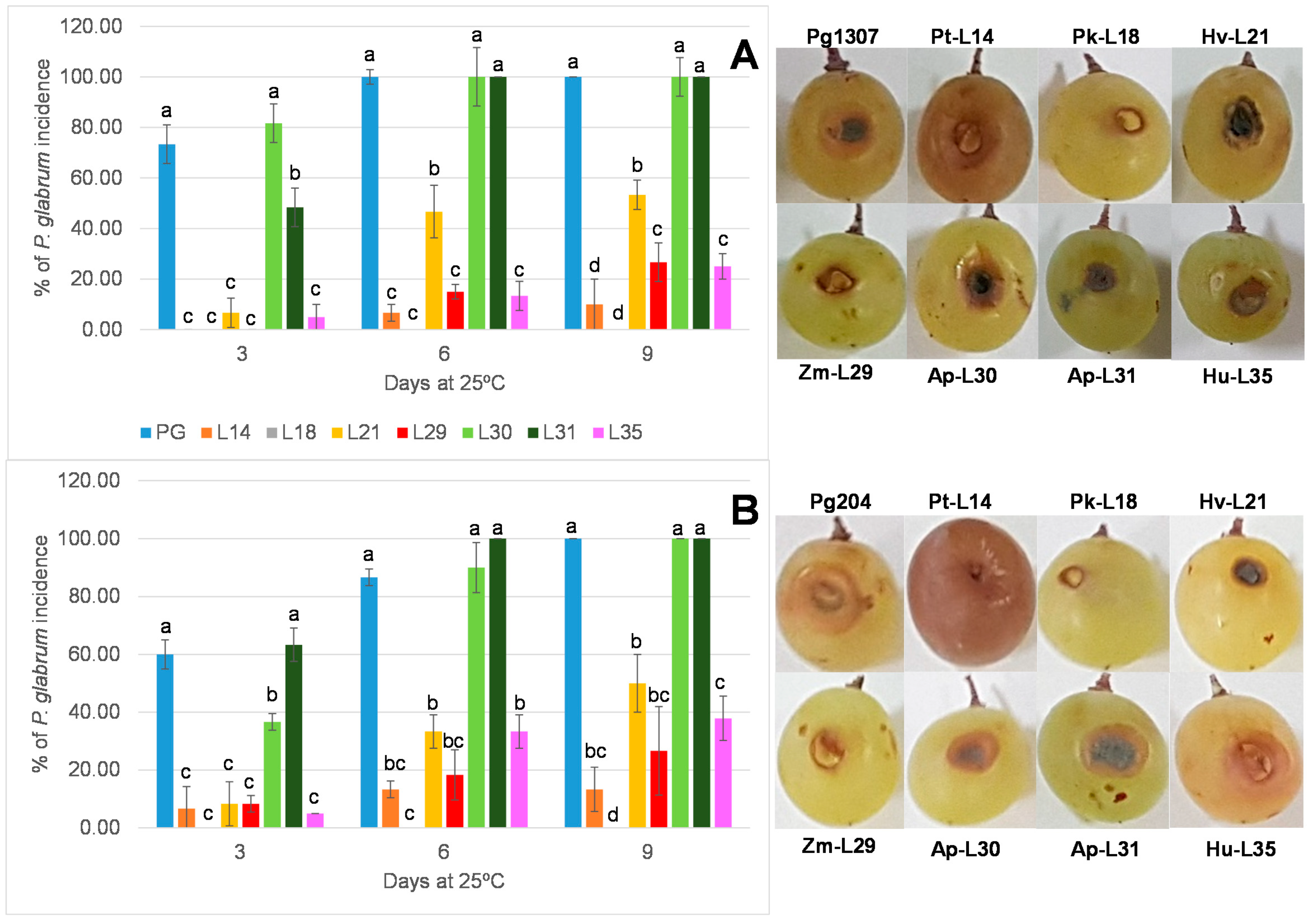
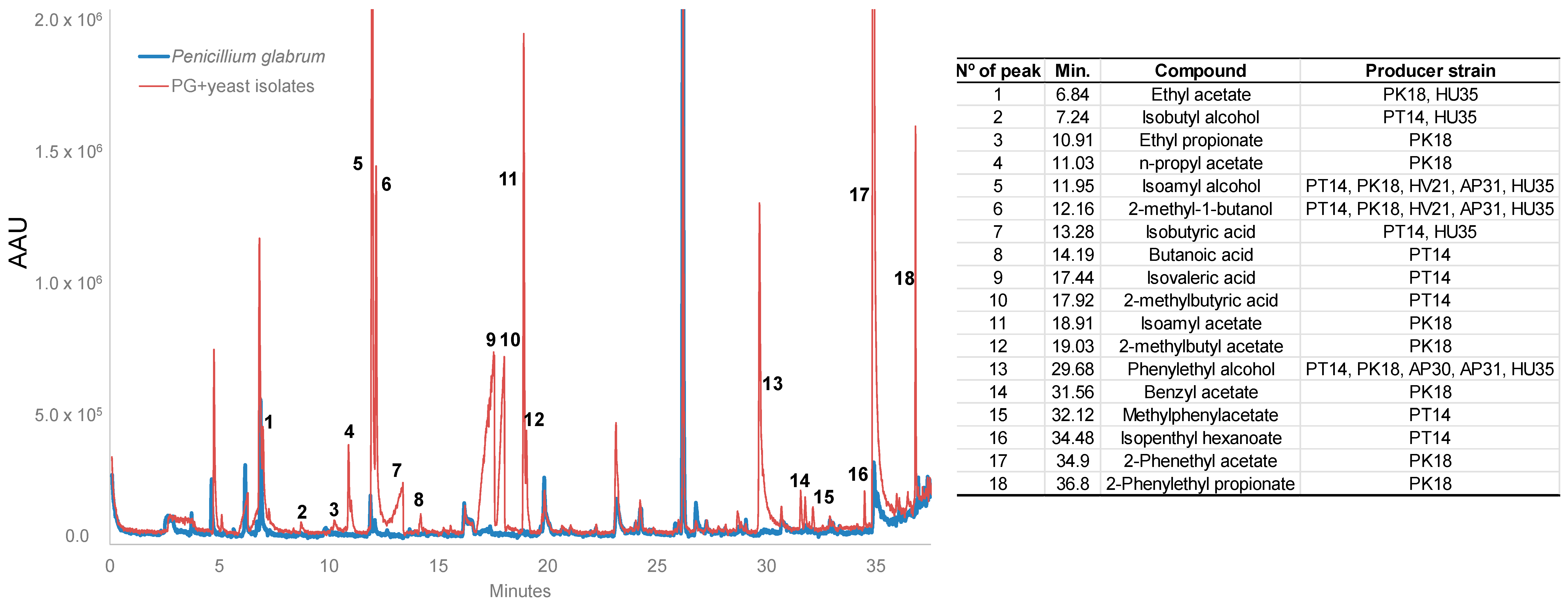
| N° Isolates | Presence Per Ten Bunches | Species Identification | Percentage of Identity | Accession Number |
|---|---|---|---|---|
| Yeasts | ||||
| 13 | 10 | Pichia terricola | 100 | NR153994 |
| 8 | 6 | Pichia kudriavzevii | 100 | KY104583 |
| 17 | 10 | Aureobasidium pullulans | 100 | KX869960 |
| 8 | 4 | Hanseniaspora uvarum | 99.86 | KY103573 |
| 6 | 2 | Hormonema viticola | 99.82 | NR137620 |
| 2 | 1 | Zygoascus meyerae | 100 | KY106012 |
| Filamentous fungi | ||||
| 18 | 10 | Penicillium glabrum | 100 | AY373915 |
| 1 | 1 | Cladosporium cladosporioides | 100 | HQ832794 |
| 11 | 4 | Alternaria alternata | 100 | MG733652 |
| 3 | 2 | Aspergillus nigricans | 100 | KP124341 |
| Confrontation | (mm Day−1) | Lag Phase (Day−1) | % Reduction in Size of Mycelia (Day 4) | |||
|---|---|---|---|---|---|---|
| Media | SD 1 | Media | SD | Media | SD | |
| Penicillium glabrum 1307 | ||||||
| Control | 5.45 d,e,f,1 | 0.07 | 0.86 e,f | 0.01 | - | - |
| Pichia terricola L14 | 5.77 a,b,c,d | 0.29 | 1.52 a,b | 0.19 | 9.17 b | 4.00 |
| Pichia kudriavzevii L18 | 4.73 g | 0.25 | 1.65 a | 0.12 | 31.48 a | 2.50 |
| Hormonema viticola L21 | 4.94 f,g | 0.07 | 0.87 e,f | 0.03 | 6.06 b,c,d | 3.61 |
| Zygoascus meyerae L29 | 5.77 a,b,c,d | 0.14 | 1.11 c,d,e | 0.09 | −0.10 d,e | 5.73 |
| Aureobasidium pullulans L30 | 5.72 b,c,d | 0.16 | 1.03 d,e | 0.04 | −1.65 e | 3.75 |
| Aureobasidium pullulans L31 | 5.59 c,d,e | 0.09 | 1.00 d,e | 0.08 | −2.38 e | 2.60 |
| Hanseniaspora uvarum L35 | 5.51 d,e | 0.08 | 1.02 d,e | 0.09 | 1.38 c,d,e | 6.02 |
| Penicillium glabrum 204 | ||||||
| Control | 5.51 e,f | 0.16 | 0.61 e | 0.09 | - | - |
| Pichia terricola L14 | 5.30 e,f,g | 0.04 | 1.02 d,e | 0.03 | 9.842 b | 3.34 |
| Pichia kudriavzevii L18 | 5.10 f,g,h | 0.24 | 1.35 a,b | 0.11 | 23.91 a | 1.68 |
| Hormonema viticola L21 | 6.18 a,b,c | 0.05 | 1.22 b,c | 0.04 | 11.23 b | 1.96 |
| Zygoascus meyerae L29 | 5.85 d,e | 0.14 | 0.77 e | 0.08 | 2.69 c,d,e | 3.05 |
| Aureobasidium pullulans L30 | 6.27 a,b | 0.26 | 1.24 a,b,c | 0.11 | 2.08 c,d,e | 4.44 |
| Aureobasidium pullulans L31 | 6.08 a,b,c,d | 0.31 | 1.08 b,c,d,e | 0.21 | 7.00 b,c | 2.05 |
| Hanseniaspora uvarum L35 | 6.41 a | 0.04 | 1.33 a,b | 0.02 | 7.37 b,c | 1.95 |
| p | <0.001 | <0.001 | <0.001 | |||
| Confrontation | % Spores Germinated | Size of Germ Tube (μM) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 105 Cell mL−1 | 106 Cell mL−1 | 107 Cell mL−1 | 105 Cell mL−1 | 106 Cell mL−1 | 107 Cell mL−1 | ||||||||
| Media | SD 1 | Media | SD | Media | SD | Media | SD | Media | SD | Media | SD | ||
| Penicillium glabrum 1307 | |||||||||||||
| Control | 88.78 | 4.03 | 71.61 | 30.50 | |||||||||
| Pichia terricola L14 | 77.45 * | 5.32 | 65.97 * | 8.68 | 64.88 * | 5.25 | 35.13 * | 12.42 | 26.06 * | 9.088 | 27.93 * | 7.16 | |
| Pichia kudriavzevii L18 | 77.31 | 10.60 | 81.72 | 3.91 | 81.04 | 6.46 | 38.26 * | 11.31 | 34.20 * | 17.61 | 35.13 * | 14.70 | |
| Hormonema viticola L21 | 91.28 | 6.58 | 93.44 | 5.10 | 92.52 | 5.01 | 74.23 | 32.42 | 85.73 | 37.72 | 68.46 | 37.55 | |
| Zygoascus meyerae L29 | 93.22 | 4.36 | 93.87 | 4.10 | 94.36 | 3.75 | 77.13 | 28.73 | 73.53 | 36.32 | 73.82 | 36.33 | |
| Aureobasidium pullulans L30 | 90.93 | 3.89 | 92.49 | 4.90 | 91.56 | 5.78 | 84.20 | 38.48 | 73.46 | 33.67 | 80.27 | 37.05 | |
| Aureobasidium pullulans L31 | 93.61 | 1.83 | 93.59 | 4.23 | 87.53 | 3.92 | 67.60 | 24.58 | 66.26 | 15.10 | 61.66 | 22.46 | |
| Hanseniaspora uvarum L35 | 94.13 | 0.61 | 91.48 | 4.88 | 92.18 | 5.53 | 72.40 | 28.96 | 77.49 | 27.99 | 76.46 | 18.67 | |
| Penicillium glabrum 204 | |||||||||||||
| Control | 93.74 | 3.93 | 87.02 | 42.20 | |||||||||
| Pichia terricola L14 | 75.50 * | 3.42 | 70.84 * | 2.82 | 63.61 * | 7.28 | 36.46 * | 12.40 | 29.46 * | 17.53 | 29.13 * | 15.45 | |
| Pichia kudriavzevii L18 | 87.02 | 2.68 | 88.25 | 3.16 | 90.54 | 6.12 | 48.40 * | 20.20 | 42.40 * | 24.23 | 37.33 * | 18.18 | |
| Hormonema viticola L21 | 93.44 | 4.87 | 90.68 | 3.24 | 89.65 | 3.84 | 76.46 | 24.73 | 84.02 | 30.53 | 62.93 | 29.32 | |
| Zygoascus meyerae L29 | 90.84 | 4.46 | 89.56 | 5.91 | 89.69 | 3.84 | 63.66 | 27.15 | 66.06 | 22.90 | 66.61 | 17.12 | |
| Aureobasidium pullulans L30 | 86.71 | 5.87 | 90.67 | 4.70 | 88.58 | 4.75 | 101.2 | 15.86 | 72.26 | 26.35 | 64.13 | 20.53 | |
| Aureobasidium pullulans L31 | 92.74 | 7.21 | 88.28 | 4.58 | 84.35 | 10.50 | 65.66 | 25.76 | 69.01 | 28.62 | 80.66 | 24.07 | |
| Hanseniaspora uvarum L35 | 97.89 | 2.88 | 95.69 | 4.23 | 92.08 | 5.96 | 71.13 | 29.59 | 62.42 | 22.52 | 64.06 | 15.91 | |
| p | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |||||||
| Strain. | Parasitism | Antibiosis | Enzymatic Activities | Biofilm Formation | Competition for Nutrients | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chitinase | β-Glucanase | Pectinase | Protease | Iron Depletion | In Vivo Competition | ||||
| Pichia terricola L14 | − | − | − | + | − | − | + | − | + |
| Pichia kudriavzevii L18 | − | − | − | − | − | − | − | − | − |
| Hormonema viticola L21 | − | − | + | + | + | − | − | − | − |
| Zygoascus meyerae L29 | − | − | + | + | + | − | + | − | − |
| Aureobasidium pullulans L30 | − | − | + | + | + | + | − | − | − |
| Aureobasidium pullulans L31 | − | − | + | + | + | + | − | − | − |
| Hanseniaspora uvarum L35 | − | − | − | − | − | − | + | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabañas, C.M.; Hernández, A.; Martínez, A.; Tejero, P.; Vázquez-Hernández, M.; Martín, A.; Ruiz-Moyano, S. Control of Penicillium glabrum by Indigenous Antagonistic Yeast from Vineyards. Foods 2020, 9, 1864. https://doi.org/10.3390/foods9121864
Cabañas CM, Hernández A, Martínez A, Tejero P, Vázquez-Hernández M, Martín A, Ruiz-Moyano S. Control of Penicillium glabrum by Indigenous Antagonistic Yeast from Vineyards. Foods. 2020; 9(12):1864. https://doi.org/10.3390/foods9121864
Chicago/Turabian StyleCabañas, Catalina M., Alejandro Hernández, Ana Martínez, Paula Tejero, María Vázquez-Hernández, Alberto Martín, and Santiago Ruiz-Moyano. 2020. "Control of Penicillium glabrum by Indigenous Antagonistic Yeast from Vineyards" Foods 9, no. 12: 1864. https://doi.org/10.3390/foods9121864
APA StyleCabañas, C. M., Hernández, A., Martínez, A., Tejero, P., Vázquez-Hernández, M., Martín, A., & Ruiz-Moyano, S. (2020). Control of Penicillium glabrum by Indigenous Antagonistic Yeast from Vineyards. Foods, 9(12), 1864. https://doi.org/10.3390/foods9121864





