Effect of Non-Thermal Atmospheric Plasma on Food-Borne Bacterial Pathogens on Ready-to Eat Foods: Morphological and Physico-Chemical Changes Occurring on the Cellular Envelopes
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Preparation and Iinoculation of Samples
2.3. NTAP Treatments
- Ncontrol = number of microorganisms in untreated control samples (CFU)
- Ntreated = number of microorganisms in NTAP-treated samples (CFU)
2.4. Fourier Transform Infrared (FTIR) Spectroscopic Analyses
2.5. Transmission Electron Microscopy (TEM) Analyses
2.6. Membrane Integrity Tests
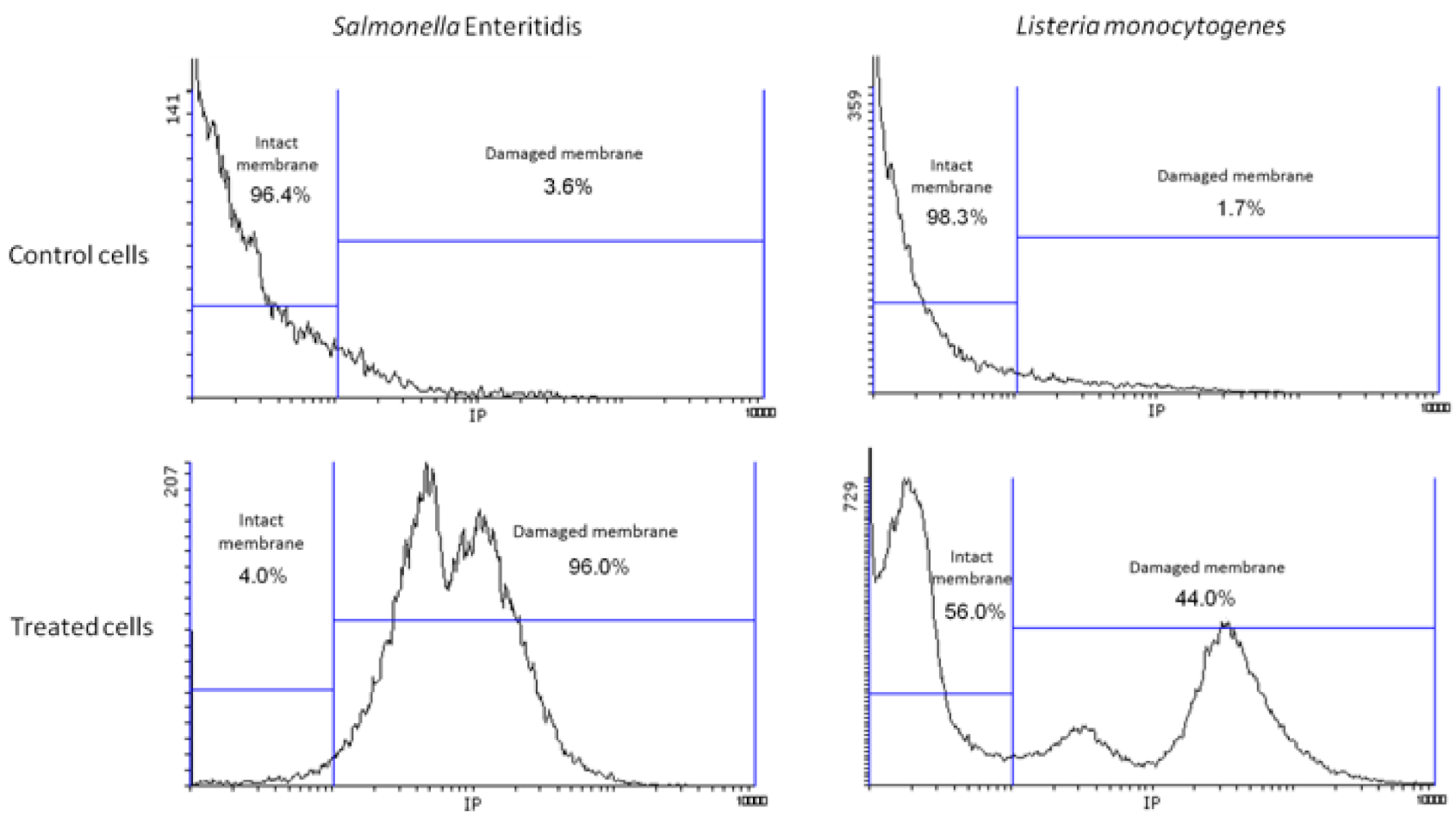
2.6.1. Assessment of Propidium Iodide (PI) Uptake
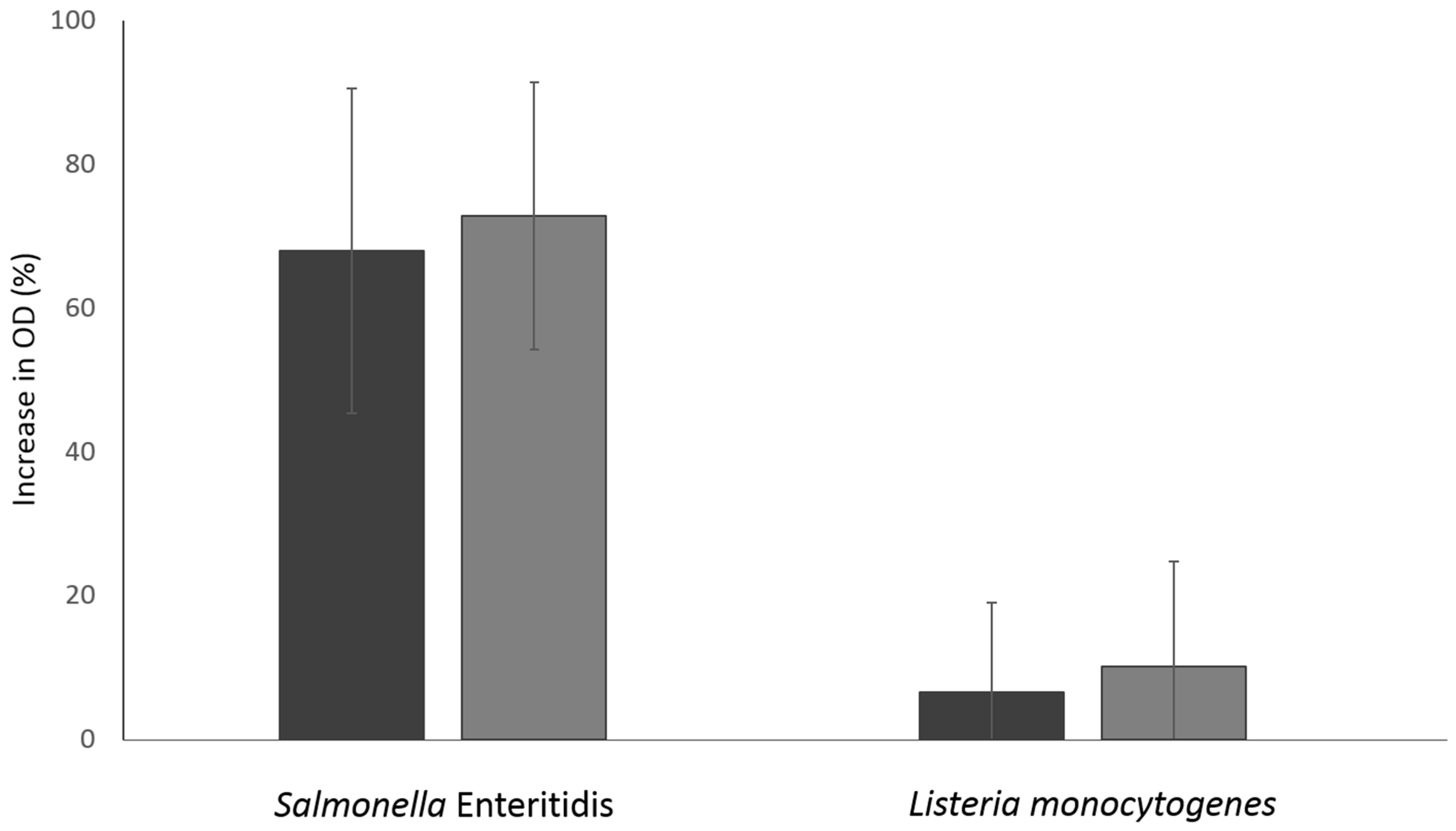
2.6.2. Measurement of the Leakage of Intracellular Nucleic Acids and Proteins
2.7. Statistical Analysis
3. Results and Discussion
3.1. Effectiveness of NTAP on Bacterial Inactivation
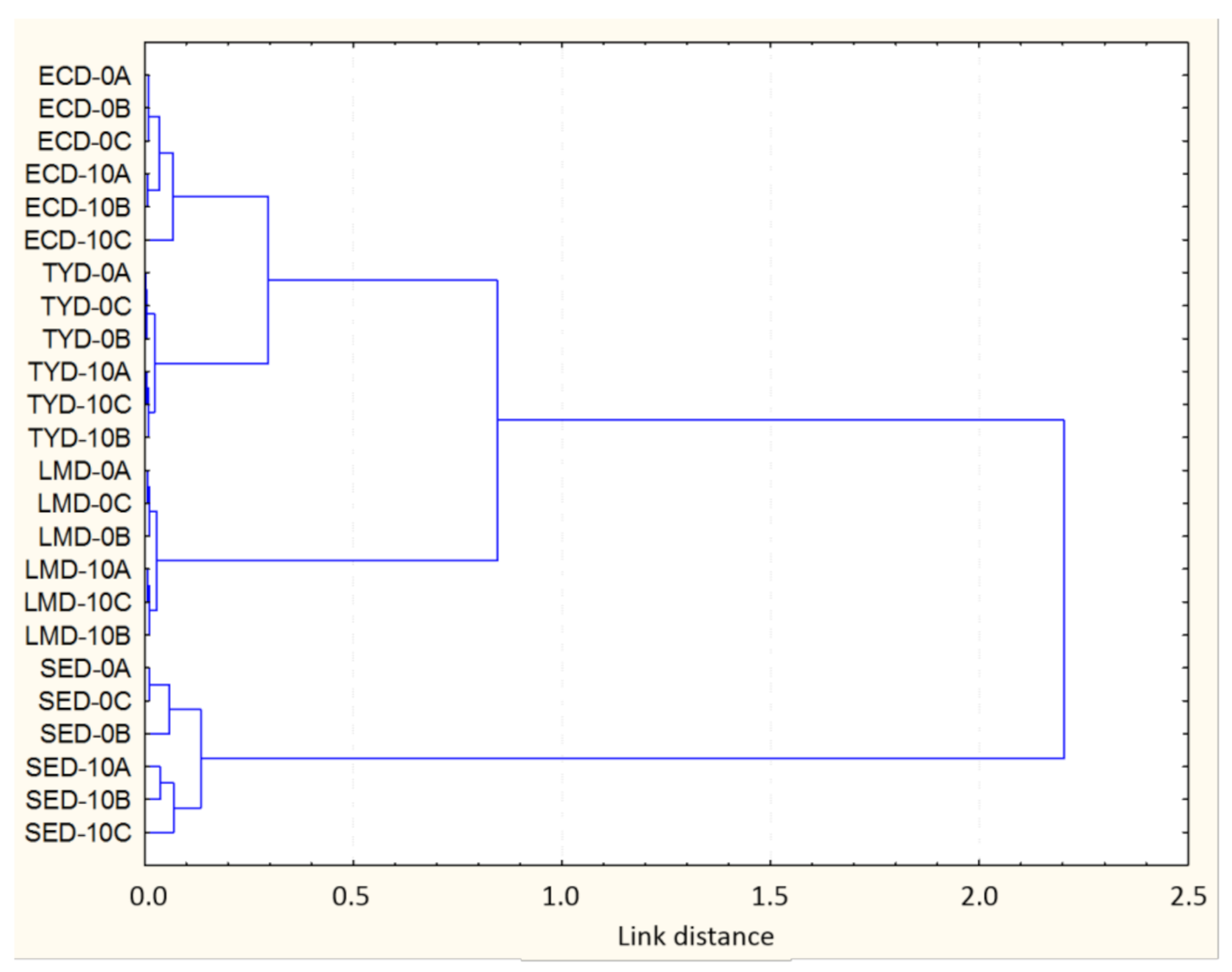
3.2. Bacterial Chemical Changes Induced by NTAP
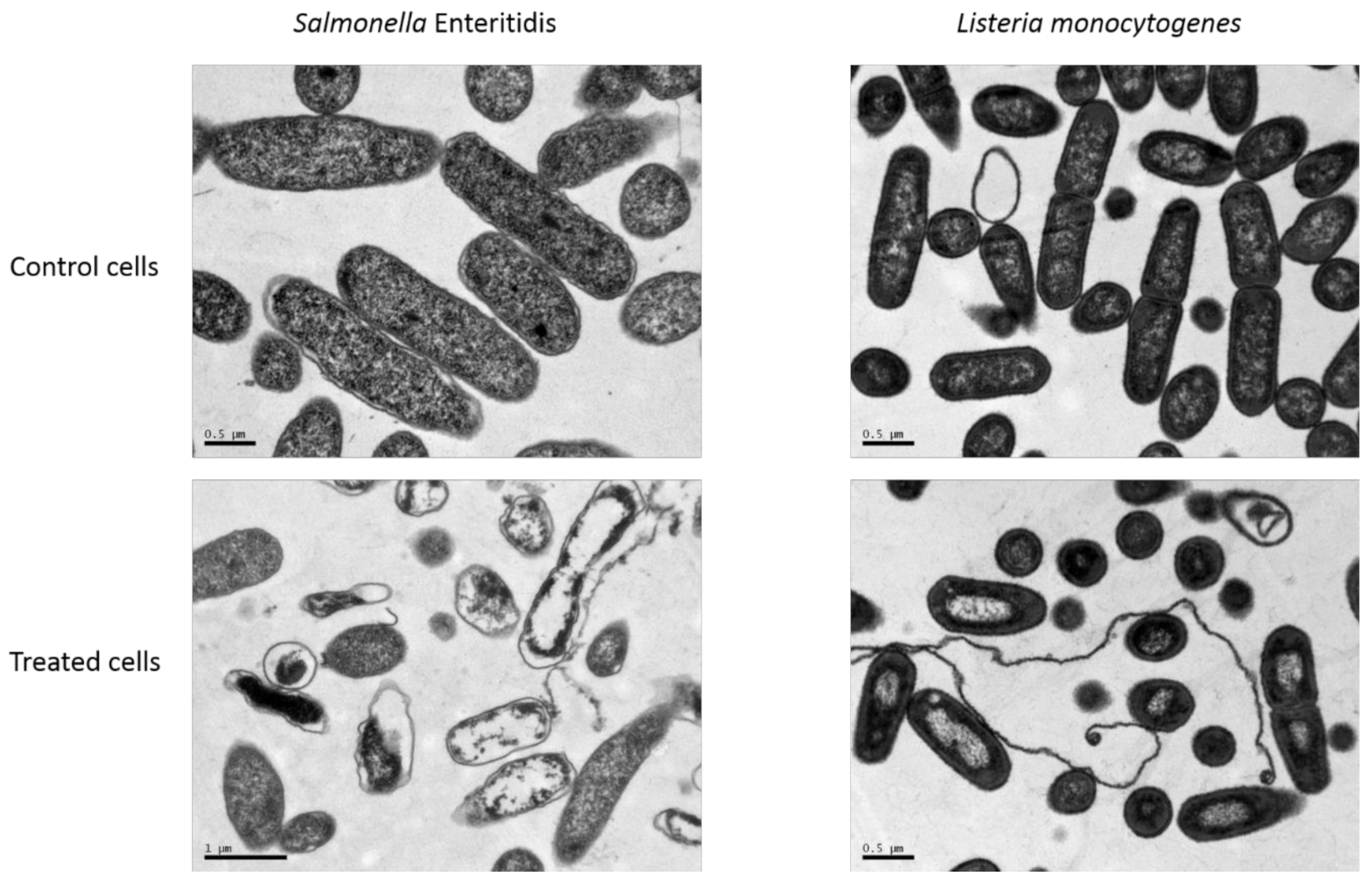
3.3. Bacterial Morphological Changes Induced by NTAP
3.4. Damage in Bacterial Membrane Integrity Induced by NTAP
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- EFSA. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- Lambertz, S.T.; Nilsson, C.; Brådenmark, A.; Sylvén, S.; Johansson, A.; Jansson, L.M.; Lindblad, M. Prevalence and level of Listeria monocytogenes in ready-to-eat foods in Sweden 2010. Int. J. Food Microbiol. 2012, 160, 24–31. [Google Scholar] [CrossRef]
- Xavier, C.; Gonzales-Barron, U.; Paula, V.; Estevinho, L.; Cadavez, V. Meta-analysis of the incidence of foodborne pathogens in Portuguese meats and their products. Food Res. Int. 2014, 55, 311–323. [Google Scholar] [CrossRef]
- Soderqvist, K.; Lambertz, S.T.; Vagsholm, I.; Boqvist, S. Foodborne bacterial pathogens in retail prepacked ready-to-eat mixed ingredient salads. J. Food Prot. 2016, 79, 978–985. [Google Scholar] [CrossRef]
- Silva, B.N.; Cadavez, V.; Teixeira, J.A.; Gonzales-Barron, U. Meta-analysis of the incidence of foodborne pathogens in vegetables and fruits from retail establishments in Europe. Curr. Opin. Food Sci. 2017, 18, 21–28. [Google Scholar] [CrossRef]
- Alkawareek, M.Y.; Gorman, S.P.; Gram, W.G.; Gilmore, B.F. Potencial celular targets and antibacterial efficacy of atmospheric pressure non-thermal plasma. Int. J. Antimicrob. Agents 2014, 43, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Paek, K.; Ju, W.T.; Lee, Y. Sterilization of bacteria; yeast and bacterial endospores by atmospheric-pressure cold plasma using helium and oxygen. J. Microbiol. 2006, 44, 269–275. [Google Scholar] [PubMed]
- Niemira, B.A.; Sites, J. Cold plasma inactivates Salmonella Stanley and Escherichia coli O157:H7 inoculated on golden delicious apples. J. Food Prot. 2008, 71, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Song, H.P.; Jung, H.S.; Choe, W.H.; Ham, J.S.; Lee, J.H.; Jo, C.R. Effect of atmospheric pressure plasma jet on inactivation of Listeria monocytogenes; quality and genotoxicity of cooked egg white and yolk. Korean J. Food Sci. Anim. Resour. 2012, 32, 561–570. [Google Scholar] [CrossRef]
- Baier, M.; Görgen, M.; Ehlbeck, J.; Knorr, D.; Herppich, W.B.; Schlüte, O. Non-thermal atmospheric pressure plasma: Screening for gentle process conditions and antibacterial efficiency on perishable fresh produce. Innov. Food Sci. Emerg. Technol. 2014, 22, 147–157. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, E.J.; Choi, E.H.; Kim, Y.J. Inactivation of Staphylococcus aureus on the beef jerky by radio-frequency atmospheric pressure plasma discharge treatment. Innov. Food Sci. Emerg. Technol. 2014, 22, 124–130. [Google Scholar] [CrossRef]
- Choi, S.; Puligundla, P.; Mok, C. Effect of corona discharge plasma on microbial decontamination of dried squid shreds including physico-chemical and sensory evaluation. LWT Food Sci. Technol. 2017, 75, 323–328. [Google Scholar] [CrossRef]
- Puligundla, P.; Kim, J.W.; Mok, C. Effect of corona discharge plasma jet treatment on decontamination and sprouting of rapeseed (Brassica napus L.) seeds. Food Control. 2017, 71, 376–382. [Google Scholar] [CrossRef]
- Joshi, S.G.; Cooper, M.; Yost, A.; Paff, M.; Ercan, U.K.; Fridman, G.; Friedman, G.; Fridman, A.; Brooks, A.D. Nonthermal dielectric-barrier discharge plasma-induced inactivation involves oxidative DNA damage and membrane lipid peroxidation in Escherichia coli. Antimicrob. Agents Chemother. 2011, 55, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Reineke, K.; Langer, K.; Hertwig, C.; Ehlbeck, J.; Schlüter; O. The impact of different process gas compositions on the inactivation effect of an atmospheric pressure plasma jet on Bacillus spores. Innov. Food Sci. Emerg. Technol. 2015, 30, 112–118. [Google Scholar] [CrossRef]
- Song, H.P.; Kim, B.; Choe, J.M.; Jung, S.; Moon, S.Y.; Choe, W.; Jo, C. Evaluation of atmospheric pressure plasma to improve the safety of slice cheese and ham inoculated by 3-strain cocktail Listeria monocytogenes. Food Microbiol. 2009, 26, 432–436. [Google Scholar] [CrossRef]
- Colagar, A.H.; Memariani, H.; Sohbatzadeh, F.; Omran, A.V. Nonthermal atmospheric argon plasma jet effects on Escherichia coli biomacromolecules. Appl. Biochem. Biotechnol. 2013, 171, 1617–1629. [Google Scholar] [CrossRef]
- Surowsky, B.; Fröhling, A.; Gottschalk, N.; Schlüter, O.; Knorr, D. Impact of cold plasma on Citrobacter freundii in apple juice: Inactivation kinetics and mechanisms. Int. J. Food Microbiol. 2014, 174, 63–71. [Google Scholar] [CrossRef]
- Yost, A.D.; Joshi, S.G. Atmospheric nonthermal plasma-treated PBS inactivates Escherichia coli by oxidative DNA damage. PLoS ONE 2015, 10, e0139903. [Google Scholar] [CrossRef]
- Itooka, K.; Takahashi, K.; Kimata, Y.; Izawa, S. Cold atmospheric pressure plasma causes protein denaturation and endoplasmic reticulum stress in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2018, 102, 2279–2288. [Google Scholar] [CrossRef]
- Xu, H.; Ma, R.; Zhu, Y.; Du, M.; Zhang, H.; Jiao, Z. A systematic study of the antimicrobial mechanisms of cold atmospheric-pressure plasma for water disinfection. Sci. Total Environ. 2020, 703, 134965. [Google Scholar] [CrossRef] [PubMed]
- Laroussi, M.; Mendis, D.A.; Rosenberg, M. Plasma interaction with microbes. New J. Phys. 2003, 5, 41.1–41.10. [Google Scholar] [CrossRef]
- Muranyi, P.; Wunderlich, J.; Langowski, H.C. Modification of bacterial structures by a low-temperature gas plasma and influence on packaging material. J. Appl. Microbiol. 2010, 109, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Yun, G. The sterilization of Escherichia coli by dielectric-barrier discharge plasma at atmospheric pressure. Appl. Surf. Sci. 2011, 257, 7065–7070. [Google Scholar] [CrossRef]
- Tseng, S.; Abramzon, N.; Jackson, J.O.; Lin, W.-J. Gas discharge plasmas are effective in inactivating Bacillus and Clostridium spores. Appl. Microbiol. Biotechnol. 2012, 93, 2563–2570. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, R.; Zhang, Q.; Feng, H.; Liang, Y.; Zhang, J.; Fang, J. Assessment of the physicochemical properties and biological effects of water activated by non-thermal plasma above and beneath the water surface. Plasma Process. Polym. 2015, 12, 439–449. [Google Scholar] [CrossRef]
- Butscher, D.; Zimmermann, D.; Schuppler, M.; von Rohr, P.R. Plasma inactivation of bacterial endospores on wheat grains and polymeric model substrates in a dielectric barrier discharge. Food Control. 2016, 60, 636–645. [Google Scholar] [CrossRef]
- Laroussi, M.; Leipold, F. Evaluation of the roles of reactive species, heat, and UV radiation in the inactivation of bacterial cells by air plasmas at atmospheric pressure. Int. J. Mass Spectrom. 2004, 233, 81–86. [Google Scholar] [CrossRef]
- Calvo, T.; Álvarez-Ordóñez, A.; Prieto, M.; González-Raurich, M.; López, M. Influence of processing parameters and stress adaptation on the inactivation of Listeria monocytogenes by Non-Thermal Atmospheric Plasma (NTAP). Food Res. Int. 2016, 89, 631–637. [Google Scholar] [CrossRef]
- Mastwijk, H.C.; Wichers, H.J.; van Dijk, C.; Schuten, H.J. Observation of free hole gasses at ambient conditions. arXiv 2009, arXiv:0711.4737v2. [Google Scholar]
- Mols, M.; Mastwijk, H.; Nierop Groot, M.; Abee, T. Physiological and transcriptional response of Bacillus cereus treated with low-temperature nitrogen gas plasma. J. Appl. Microbiol. 2013, 115, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.; Shearer, N.; Wilson, D.R.; Thompson, A. Effect of microbial loading on the efficiency of cold atmospheric gas plasma inactivation of Salmonella enterica serovar Typhimurium. Int. J. Food Microbiol. 2012, 152, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Ordoñez, A.; Mouwen, D.J.M.; Lopez, M.; Prieto, M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J. Micrbiol. Meth. 2011, 84, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Mouwen, D.J.M.; Capita, R.; Alonso-Calleja, C.; Weijtens, M.J.B.M.; Prieto, M. Discrimination of Enterobacterial Repetitive Intergenic Consensus PCR Types of Campylobacter coli and Campylobacter jejuni by Fourier Transform Infrared Spectroscopy. Appl. Environ. Microbiol. 2005, 71, 4318–4324. [Google Scholar] [CrossRef]
- Lis, K.A.; Kehrenberg, C.; Boulaaba, A.; von Köckritz-Blickwede, M.; Binder, S.; Li, Y.; Zimmermann, J.L.; Pfeifer, Y.; Ahlfeld, B. Inactivation of multidrug-resistant pathogens and Yersinia enterocolitica with cold atmospheric-pressure plasma on stainless-steel surfaces. Int. J. Antimicrob. Agents 2018, 52, 811–818. [Google Scholar] [CrossRef]
- Colejo, S.; Alvarez-Ordóñez, A.; Prieto, M.; González-Raurich, M.; López, M. Evaluation of ultraviolet light (UV); non-thermal atmospheric plasma (NTAP) and their combination for the control of foodborne pathogens in smoked salmon and their effect on quality attributes. Innov. Food Sci. Emerg. Technol. 2018, 50, 84–93. [Google Scholar] [CrossRef]
- Fernández, A.; Noriega, E.; Thompson, A. Inactivation of Salmonella enterica serovar Typhimurium on fresh produce by cold atmospheric gas plasma technology. Food Microbiol. 2013, 33, 24–29. [Google Scholar] [CrossRef]
- Bermúdez-Aguirre, D.; Wemlinger, E.; Pedrow, P.; Barbosa-Cánovas, G.; Garcia-Perez, M. Effect of atmospheric pressure cold plasma (APCP) on the inactivation of Escherichia coli in fresh produce. Food Control. 2013, 34, 149–157. [Google Scholar] [CrossRef]
- Ziuzina, D.; Patil, S.; Cullen, P.J.; Keener, K.M.; Bourke, P. Atmospheric cold plasma inactivation of Escherichia coli; Salmonella enterica serovar Typhimurium and Listeria monocytogenes inoculated on fresh produce. Food Microbiol. 2014, 42, 109–116. [Google Scholar] [CrossRef]
- Baier, M.; Ehlbeck, J.; Knorr, D.; Herppich, W.B.; Schlüter, O. Impact of plasma processed air (PPA) on quality parameters of fresh produce. Postharvest Biol. Technol. 2015, 100, 120–126. [Google Scholar] [CrossRef]
- Lee, H.J.; Jung, H.; Choe, W.; Ham, J.S.; Lee, J.H.; Jo, C. Inactivation of Listeria monocytogenes on agar and processed meat surfaces by atmospheric pressure plasma jets. Food Microbiol. 2011, 28, 1468–1471. [Google Scholar] [CrossRef] [PubMed]
- Klämpfl, T.G.; Isbary, G.; Shimizu, T.; Li, Y.F.; Zimmermann, J.L.; Stolz, W.; Schlegel, J.; Morfill, G.E.; Schmidt, H.U. Cold atmospheric air plasma sterilization against spores and other microorganisms of clinical interest. Appl. Environ. Microbiol. 2012, 78, 5077–5082. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, T.; Uehara, K.; Sasaki, Y.; Hidekazu, M.; Matsumura, Y.; Iwasawa, A.; Ito, N.; Kohno, M.; Azuma, T.; Okino, A. Microbial inactivation in the liquid phase induced by multigas plasma jet. PLoS ONE 2015, 10, e0132381. [Google Scholar] [CrossRef]
- Gabriel, A.A.; Aba, R.P.M.; Tayamora, D.J.L.; Colambo, J.C.R.; Siringan, M.A.T.; Rosario, L.M.D.; Tumlos, R.Y.; Ramos, H.J. Reference organism selection for microwave atmospheric pressure plasma jet treatment of young coconut liquid endosperm. Food Control. 2016, 69, 74–82. [Google Scholar] [CrossRef]
- Marsili, L.; Espie, S.; Anderson, J.G.; MacGregor, S.J. Plasma inactivation of food-related microorganisms in liquids. Radiat. Phys. Chem. 2002, 65, 507–513. [Google Scholar] [CrossRef]
- Nishime, T.M.C.; Borges, A.C.; Koga-Ito, C.Y.; Machida, M.; Hein, L.R.O.; Kostov, K.G. Non-thermal atmospheric pressure plasma jet applied to inactivation of different microorganisms. Surf. Coat. Technol. 2017, 312, 19–24. [Google Scholar] [CrossRef]
- Curk, M.C.; Peledan, F.; Hubert, J.C. Fourier transform infrared (FTIR) spectroscopy for identifying Lactobacillus species. FEMS Microbiol. Lett. 1994, 123, 241–248. [Google Scholar] [CrossRef]
- Álvarez-Ordoñez, A.; Prieto, M. Changes in ultrastructure and Fourier Transform Infrared spectrum of Salmonella enterica serovar Typhimurium cells after exposure to stress conditions. Appl. Environ. Microbiol. 2010, 76, 7598–7607. [Google Scholar] [CrossRef]
- Alvarez-Ordoñez, A.; Prieto, M. FT-IR Spectroscopy in Food Microbiology; Springer: New York, NY, USA, 2012; ISBN 978-1-4614-3812-0. [Google Scholar]
- Al-Holy, M.A.; Lin, M.; Cavinato, A.G.; Rasco, B.A. The use of Fourier transform infrared spectroscopy to differentiate Escherichia coli O157:H7 from other bacteria inoculated into apple juice. Food Microbiol. 2006, 23, 162–168. [Google Scholar] [CrossRef]
- Prieto-Calvo, M.A.; Prieto, M.; López, M.; Álvarez-Ordoñez, A. Effects of high hydrostatic pressure on Escherichia coli ultrastructure, membrane integrity and molecular composition as assessed by FTIR spectroscopy and microscopic imaging techniques. Molecules 2014, 19, 21310–21323. [Google Scholar] [CrossRef]
- Calvo, T.; Alvarez-Ordóñez, A.; Prieto, M.; Bernardo, A.; López, M. Stress adaptation has a minor impact on the effectivity of Non-Thermal Atmospheric Plasma (NTAP) against Salmonella spp. Food Res. Int. 2017, 102, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Patil, S.; Boehm, D.; Milosavljević, V.; Cullen, P.J.; Bourke, P. Mechanism of inactivation by high voltage atmospheric cold plasma differs between Escherichia coli and Staphylococcus aureus. Appl. Environ. Microbiol. 2015, 82, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Ziuzina, D.; Han, L.; Cullen, P.J.; Bourke, P. Cold plasma inactivation of internalised bacteria and biofilms for Salmonella enterica serovar Typhimurium, Listeria monocytogenes and Escherichia coli. Int. J. Food Microbiol. 2015, 210, 53–61. [Google Scholar] [CrossRef] [PubMed]

| Treatment Time | Food | Microorganisms | |||
|---|---|---|---|---|---|
| S. Enteritidis | S. Typhimurium | E. coli O157:H7 | L. monocytogenes | ||
| 4 min | “chorizo” | 0.58 ± 0.05 c 1,2 | 0.81 ± 0.10 d 2 | 0.36 ± 0.07 a 1 | 0.37 ± 0.13 a 1 |
| salami | 0.37 ± 0.01 b 1 | 0.62 ± 0.13 cd 2 | 0.32 ± 0.08 a 1 | 0.32 ± 0.08 a 1 | |
| bacon | 0.30 ± 0.04 ab 1 | 0.34 ± 0.07 a 1 | 0.33 ± 0.04 a 1 | 0.38 ± 0.10 a 1 | |
| smoked salmon | 0.40 ± 0.05 bc 1 | 0.39 ± 0.06 abc 1 | 0.30 ± 0.05 a 1 | 0.29 ± 0.01 a 1 | |
| tofu | 0.16 ± 0.06 a 1 | 0.36 ± 0.04 ab 1 | 0.41 ± 0.0 a 2 | 0.34 ± 0.06 a 2 | |
| apple | 0.59 ± 0.15 c 1 | 0.60 ± 0.12 bcd 1 | 1.31 ± 0.18 b 2 | 0.89 ± 0.08 b 1 | |
| 8 min | “chorizo” | 0.77 ± 0.13 bc 1,2 | 1.03 ± 0.07 d 2 | 0.55 ± 0.08 a 1 | 0.71 ± 0.14 a 1 |
| salami | 0.58 ± 0.18 b 1 | 0.75 ± 0.15 bcd 1 | 0.48 ± 0.04 a 1 | 0.45 ± 0.09 a 1 | |
| bacon | 0.51 ± 0.09 b 1 | 0.53 ± 0.15 ab 1 | 0.48 ± 0.04 a 1 | 0.56 ± 0.12 a 1 | |
| smoked salmon | 0.70 ± 0.07 bc 2 | 0.71 ± 0.09 bc 2 | 0.45 ± 0.04 a 1 | 0.49 ± 0.01 a 1 | |
| tofu | 0.20 ± 0.07 a 1 | 0.39 ± 0.04 a 2 | 0.50 ± 0.07 a 2 | 0.40 ± 0.06 a 2 | |
| apple | 0.96 ± 0.10 c 1 | 0.87 ± 0.11 cd 1 | 1.50 ± 0.11 b 2 | 1.12 ± 0.26 b 1,2 | |
| 15 min | “chorizo” | 1.01 ± 0.10 bc 1 | 1.20 ± 0.12 b 1 | 0.89 ± 0.18 a 1 | 1.04 ± 0.17 bc 1 |
| salami | 1.09 ± 0.10 bc 1 | 1.10 ± 0.24 b 1 | 0.65 ± 0.13 a 1 | 0.80 ± 0.25 ab 1 | |
| bacon | 0.82 ± 0.24 b 1 | 1.00 ± 0.35 ab 1 | 0.64 ± 0.14 a 1 | 1.00 ± 0.20 bc 1 | |
| smoked salmon | 0.86 ± 0.14 b 1,2 | 1.02 ± 0.16 b 2 | 0.57 ± 0.08 a 1 | 0.57 ± 0.10 ab 1 | |
| Tofu | 0.22 ± 0.03 a 1 | 0.46 ± 0.03 a 2 | 0.57 ± 0.06 a 2 | 0.42 ± 0.10 a 2 | |
| apple | 1.30 ± 0.20 c 1 | 1.34 ± 0.11 b 1 | 1.79 ± 0.06 b 2 | 1.47 ± 0.19 c 1,2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvo, T.; Prieto, M.; Alvarez-Ordóñez, A.; López, M. Effect of Non-Thermal Atmospheric Plasma on Food-Borne Bacterial Pathogens on Ready-to Eat Foods: Morphological and Physico-Chemical Changes Occurring on the Cellular Envelopes. Foods 2020, 9, 1865. https://doi.org/10.3390/foods9121865
Calvo T, Prieto M, Alvarez-Ordóñez A, López M. Effect of Non-Thermal Atmospheric Plasma on Food-Borne Bacterial Pathogens on Ready-to Eat Foods: Morphological and Physico-Chemical Changes Occurring on the Cellular Envelopes. Foods. 2020; 9(12):1865. https://doi.org/10.3390/foods9121865
Chicago/Turabian StyleCalvo, Tamara, Miguel Prieto, Avelino Alvarez-Ordóñez, and Mercedes López. 2020. "Effect of Non-Thermal Atmospheric Plasma on Food-Borne Bacterial Pathogens on Ready-to Eat Foods: Morphological and Physico-Chemical Changes Occurring on the Cellular Envelopes" Foods 9, no. 12: 1865. https://doi.org/10.3390/foods9121865
APA StyleCalvo, T., Prieto, M., Alvarez-Ordóñez, A., & López, M. (2020). Effect of Non-Thermal Atmospheric Plasma on Food-Borne Bacterial Pathogens on Ready-to Eat Foods: Morphological and Physico-Chemical Changes Occurring on the Cellular Envelopes. Foods, 9(12), 1865. https://doi.org/10.3390/foods9121865







