Molecular Characterization of Microbial and Fungal Communities on Dry-Aged Beef of Hanwoo Using Metagenomic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Dry Aging of Beef and Sample Collection
2.2. DNA Preparation and Sequencing
2.3. Metagenomic Analysis
2.4. Statistical Analysis
3. Results
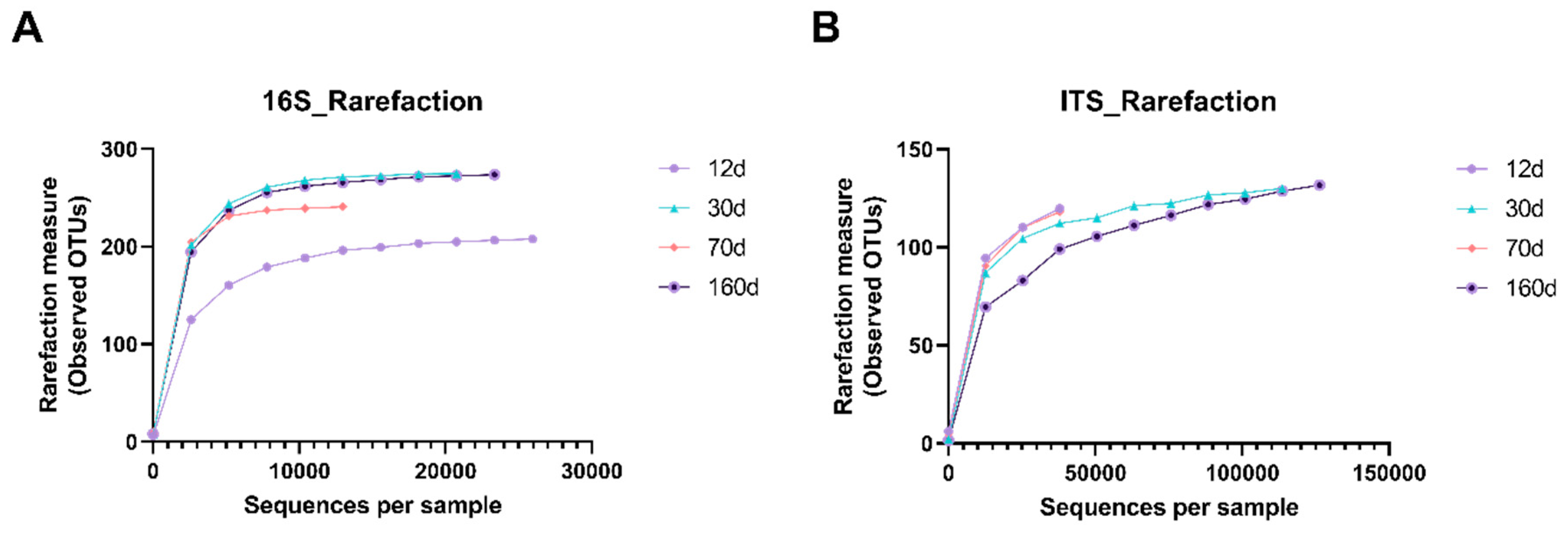
3.1. Dry Aging Leads to Changes in Species Diversity in Beef
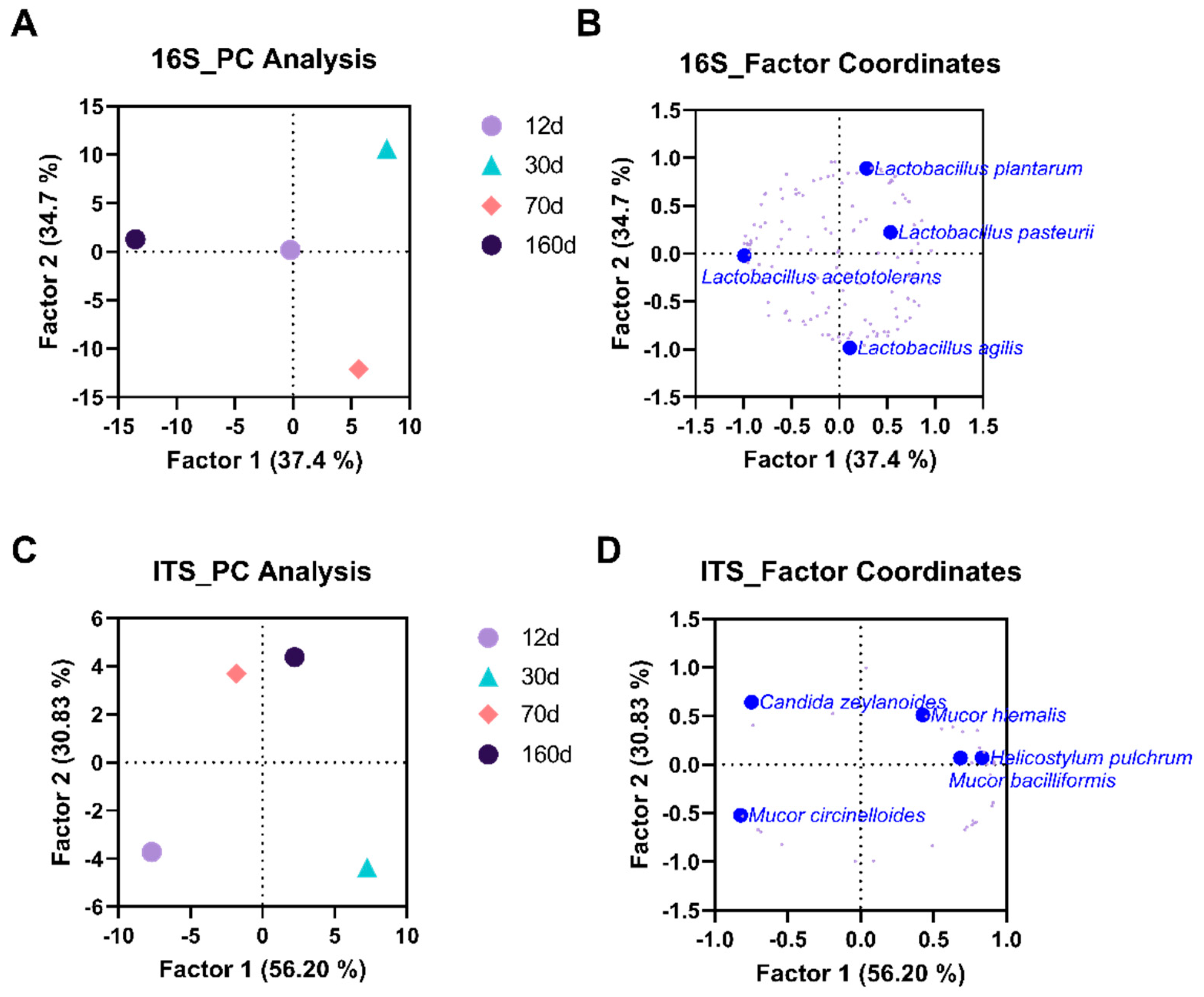
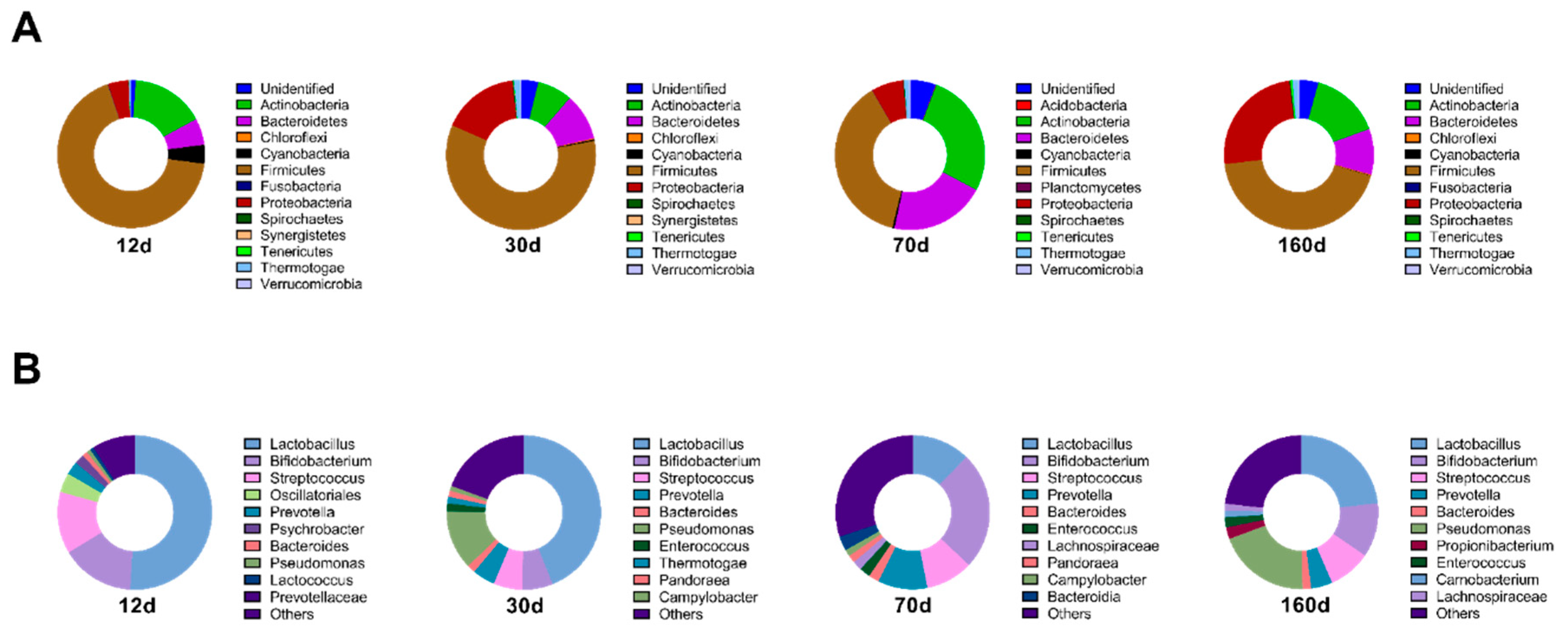
3.2. Dry Aging Alters Microbial Compositions in Beef
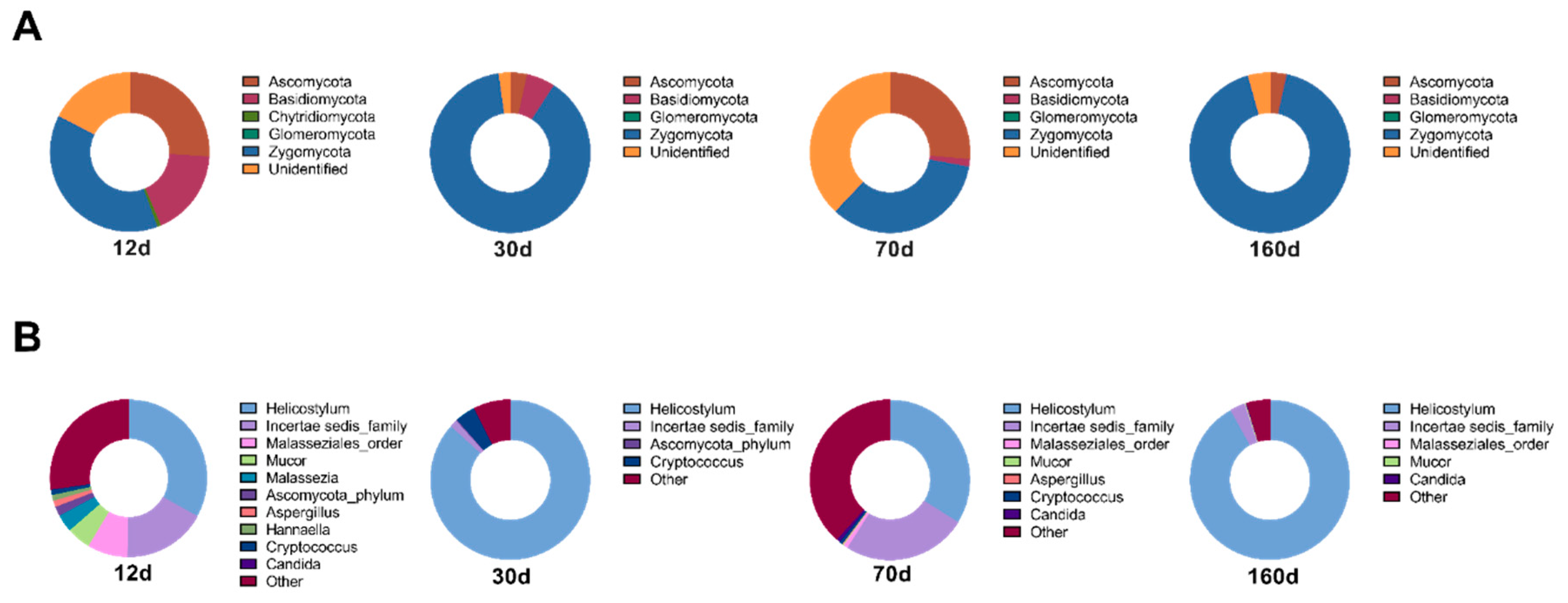
3.3. Dry Aging Alters Fungal Compositions in Beef
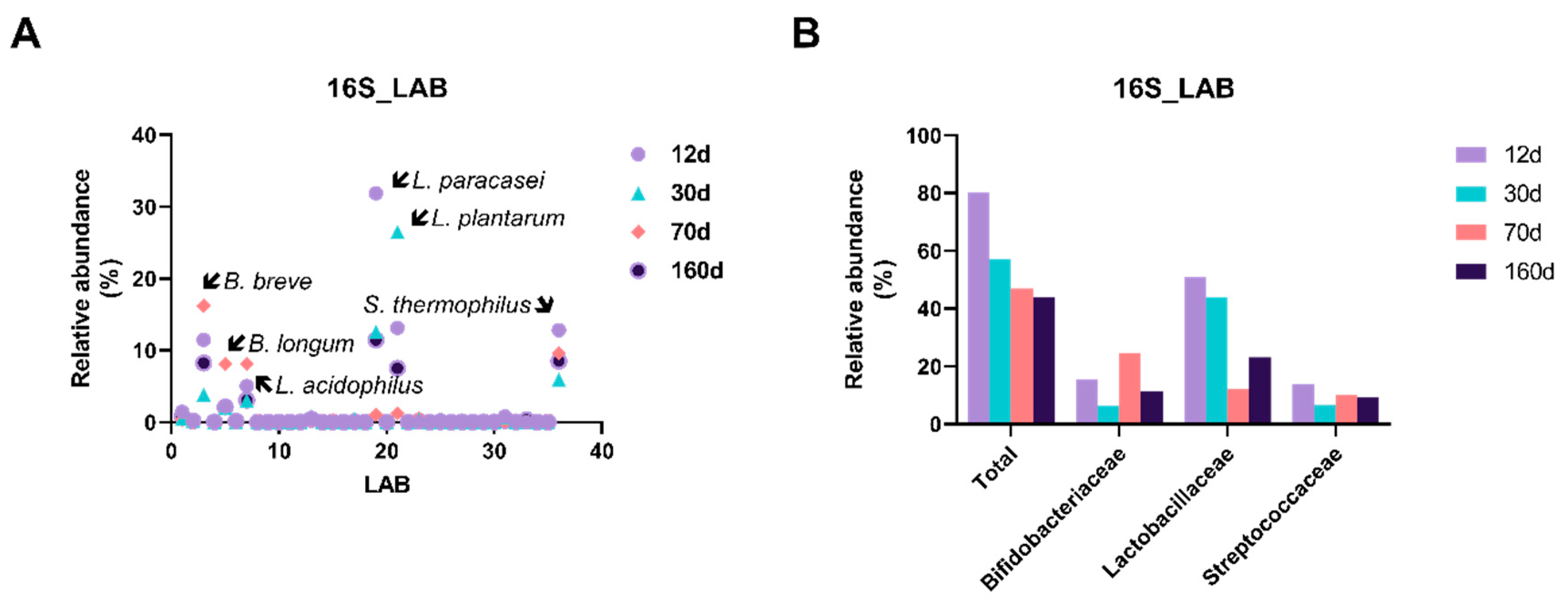
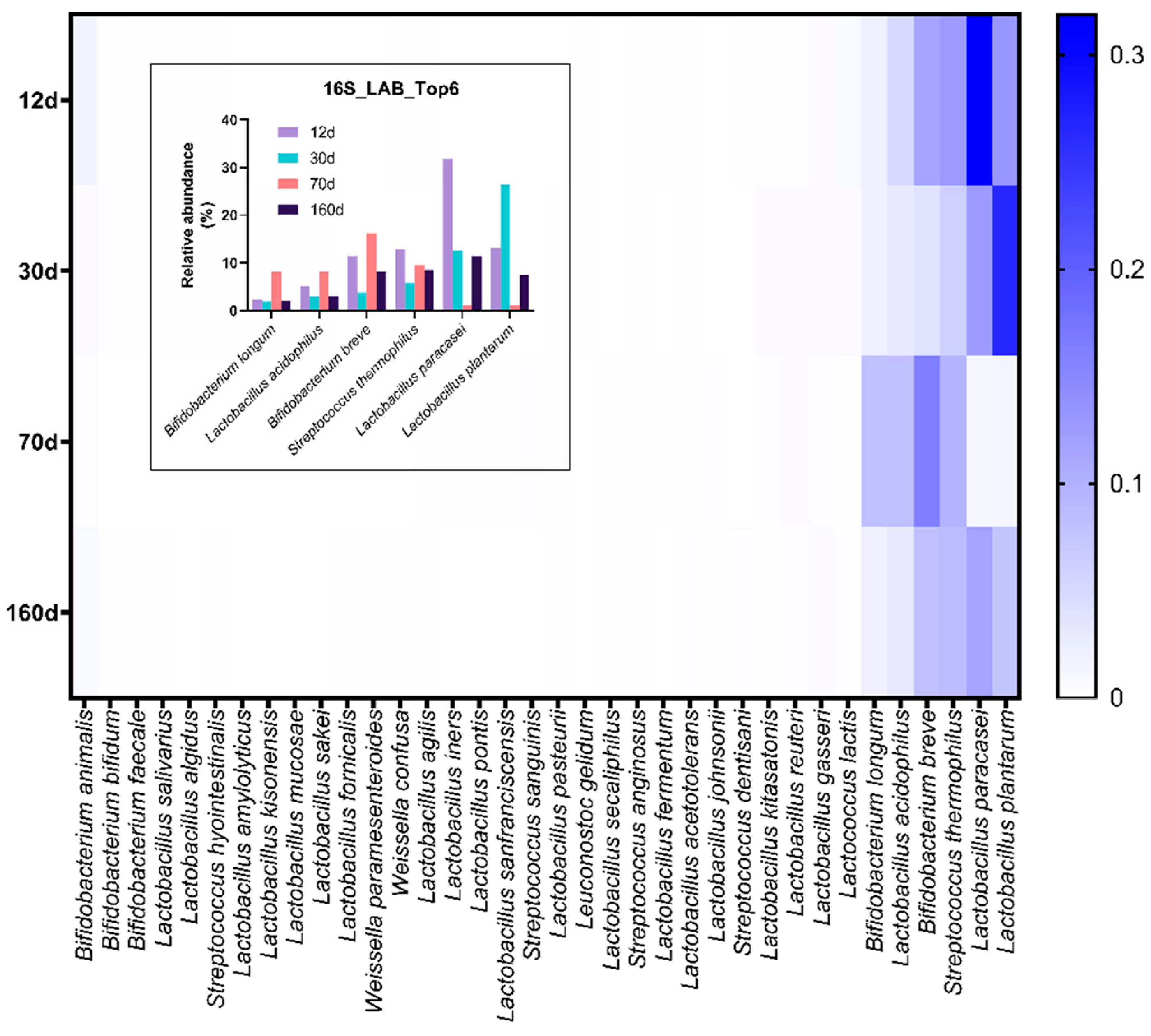
3.4. Prolonged Dry Aging Reduces the Composition of Lactic acid Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Thorslund, C.A.H.; Sandøe, P.; Aaslyng, M.D.; Lassen, J. A good taste in the meat, a good taste in the mouth–Animal welfare as an aspect of pork quality in three European countries. Livest. Sci. 2016, 193, 58–65. [Google Scholar] [CrossRef]
- Ponrajan, A.; Harrison, M.A.; Pringle, T.D.; Segers, J.R.; Lowe, B.K.; McKeith, R.O.; Stelzleni, A.M. Effect of sodium citrate plus sodium diacetate or buffered vinegar on quality attributes of enhanced beef top sirloins. Meat Sci. 2012, 91, 43–49. [Google Scholar] [CrossRef]
- Baublits, R.T.; Pohlman, F.W.; Brown, A.H.; Yancey, E.J.; Johnson, Z.B. Impact of muscle type and sodium chloride concentration on the quality, sensory, and instrumental color characteristics of solution enhanced whole-muscle beef. Meat Sci. 2006, 72, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.A.; Gray, J.I.; Rabie, S.; Lopez-Bote, C.; Booren, A.M. Polycyclic aromatic hydrocarbons in smoked food products and commercial liquid smoke flavourings. Food Addit. Contam. 1993, 10, 503–521. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Lee, H.J.; Lee, J.; Jo, C.; Yoon, Y. Identification of Microorganisms Associated with the Quality Improvement of Dry-Aged Beef Through Microbiome Analysis and DNA Sequencing, and Evaluation of Their Effects on Beef Quality. J. Food Sci. 2019, 84, 2944–2954. [Google Scholar] [CrossRef] [PubMed]
- Dashdorj, D.; Tripathi, V.K.; Cho, S.; Kim, Y.; Hwang, I. Dry aging of beef; Review. J. Anim. Sci. Technol. 2016, 58, 20. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; McGilchrist, P.; Polkinghorne, R.; Huynh, L.; Galletly, J.; Kobayashi, K.; Nishimura, T.; Bonney, S.; Kelman, K.R.; Warner, R.D. Effects of different ageing methods on colour, yield, oxidation and sensory qualities of Australian beef loins consumed in Australia and Japan. Food Res. Int. 2019, 125, 108528. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Baek, K.H.; Ko, Y.-J.; Lee, H.J.; Yim, D.-G.; Jo, C. Characteristic Metabolic Changes of the Crust from Dry-Aged Beef Using 2D NMR Spectroscopy. Molecules 2020, 25, 3087. [Google Scholar] [CrossRef]
- Kim, Y.H.B.; Kemp, R.; Samuelsson, L.M. Effects of dry-aging on meat quality attributes and metabolite profiles of beef loins. Meat Sci. 2016, 111, 168–176. [Google Scholar] [CrossRef]
- Lee, H.J.; Yoon, J.W.; Kim, M.; Oh, H.; Yoon, Y.; Jo, C. Changes in microbial composition on the crust by different air flow velocities and their effect on sensory properties of dry-aged beef. Meat Sci. 2019, 153, 152–158. [Google Scholar] [CrossRef]
- da Silva, A.C.M.; de Oliveira Pena, P.; Pflanzer, S.B.; da Silva do Nascimento, M. Effect of different dry aging temperatures on Listeria innocua as surrogate for Listeria monocytogenes. Meat Sci. 2019, 157, 107884. [Google Scholar] [CrossRef]
- Gowda, T.K.G.M.; de Zutter, L.; van Royen, G.; Houf, K.; van Damme, I. Evaluation of the microbiological quality of dry aged beef in Belgium. J. Food Process. Technol. 2016, 7, 11. [Google Scholar] [CrossRef]
- Tittor, A.W.; Tittor, M.G.; Brashears, M.M.; Brooks, J.C.; Garmyn, A.J.; Miller, M.F. Effects of simulated dry and wet chilling and aging of beef fat and lean tissues on the reduction of Escherichia coli O157:H7 and Salmonella. J. Food Prot. 2011, 74, 289–293. [Google Scholar] [CrossRef]
- Ryu, S.; Park, M.R.; Maburutse, B.E.; Lee, W.J.; Park, D.-J.; Cho, S.; Hwang, I.; Oh, S.; Kim, Y. Diversity and Characteristics of the Meat Microbiological Community on Dry Aged Beef. J. Microbiol. Biotechnol. 2018, 28, 105–108. [Google Scholar] [CrossRef]
- Katani, R.; Schilling, M.A.; Lyimo, B.; Tonui, T.; Cattadori, I.M.; Eblate, E.; Martin, A.; Estes, A.B.; Buza, T.; Rentsch, D.; et al. Microbial Diversity in Bushmeat Samples Recovered from the Serengeti Ecosystem in Tanzania. Sci. Rep. 2019, 9, 18086. [Google Scholar] [CrossRef] [PubMed]
- Maltecca, C.; Bergamaschi, M.; Tiezzi, F. The interaction between microbiome and pig efficiency: A review. J. Anim. Breed. Genet. 2020, 137, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Hugas, M.; Monfort, J.M. Bacterial starter cultures for meat fermentation. Food Chem. 1997, 59, 547–554. [Google Scholar] [CrossRef]
- Todorov, S.D.; Koep, K.S.C.; Van Reenen, C.A.; Hoffman, L.C.; Slinde, E.; Dicks, L.M.T. Production of salami from beef, horse, mutton, Blesbok (Damaliscus dorcas phillipsi) and Springbok (Antidorcas marsupialis) with bacteriocinogenic strains of Lactobacillus plantarum and Lactobacillus curvatus. Meat Sci. 2007, 77, 405–412. [Google Scholar] [CrossRef]
- Vignolo, G.M.; de Ruiz Holgado, A.P.; Oliver, G. Use of Bacterial Cultures in the Ripening of Fermented Sausages. J. Food Prot. 1989, 52, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Coman, M.M.; Cecchini, C.; Verdenelli, M.C.; Silvi, S.; Orpianesi, C.; Cresci, A. Functional foods as carriers for SYNBIO®, a probiotic bacteria combination. Int. J. Food Microbiol. 2012, 157, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Yumoto, I.; Kusano, T.; Shingyo, T.; Nodasaka, Y.; Matsuyama, H.; Okuyama, H. Assignment of Pseudomonas sp. strain E-3 to Pseudomonas psychrophila sp. nov., a new facultatively psychrophilic bacterium. Extremophiles 2001, 5, 343–349. [Google Scholar] [CrossRef]
- Matsuo, S.; Shirai, H.; Takada, Y. Isocitrate dehydrogenase isozymes from a psychrotrophic bacterium, Pseudomonas psychrophila. Arch. Microbiol. 2010, 192, 639–650. [Google Scholar] [CrossRef][Green Version]
- Dabadé, D.S.; Azokpota, P.; Nout, M.J.R.; Hounhouigan, D.J.; Zwietering, M.H.; den Besten, H.M.W. Prediction of spoilage of tropical shrimp (Penaeus notialis) under dynamic temperature regimes. Int. J. Food Microbiol. 2015, 210, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, H.; Wu, C.; Deng, W.; Wang, D.; Zhao, G.; Song, J.; Jiang, Y. Molecular analysis of dominant species in Listeria monocytogenes-positive biofilms in the drains of food processing facilities. Appl. Microbiol. Biotechnol. 2016, 100, 3165–3175. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Hong, H.; Yang, Q.; Liu, X.; Zhuang, S.; Li, Y.; Liu, J.; Luo, Y. TMT-based proteomic analysis of the fish-borne spoiler Pseudomonas psychrophila subjected to chitosan oligosaccharides in fish juice system. Food Microbiol. 2020, 90, 103494. [Google Scholar] [CrossRef] [PubMed]
- Capouya, R.; Mitchell, T.; Clark, D.I.; Clark, D.L.; Bass, P. A Survey of Microbial Communities on Dry-Aged Beef in Commercial Meat Processing Facilities. Meat Muscle Biol. 2020, 4. [Google Scholar] [CrossRef]
- Upadhyay, H.P. Helicostylum and Thamnostylum (Mucorales). Mycologia 1973, 65, 733–751. [Google Scholar] [CrossRef]
- Brilhante, R.S.N.; da Rocha, M.G.; de Guedes, G.M.; de Oliveira, J.S.; Araújo, G.D.S.; España, J.D.A.; Sales, J.A.; de Aguiar, L.; de Paiva, M.A.N.; de Cordeiro, R.A.; et al. Malassezia pachydermatis from animals: Planktonic and biofilm antifungal susceptibility and its virulence arsenal. Vet. Microbiol. 2018, 220, 47–52. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, R.; Wang, Y.; Sun, L.; Tao, S.; Li, X.; Gooneratne, R.; Zhao, J. Diversity and succession of microbial communities and chemical analysis in dried Lutianus erythropterus during storage. Int. J. Food Microbiol. 2020, 314, 108416. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, S.; Shin, M.; Cho, S.; Hwang, I.; Kim, Y.; Oh, S. Molecular Characterization of Microbial and Fungal Communities on Dry-Aged Beef of Hanwoo Using Metagenomic Analysis. Foods 2020, 9, 1571. https://doi.org/10.3390/foods9111571
Ryu S, Shin M, Cho S, Hwang I, Kim Y, Oh S. Molecular Characterization of Microbial and Fungal Communities on Dry-Aged Beef of Hanwoo Using Metagenomic Analysis. Foods. 2020; 9(11):1571. https://doi.org/10.3390/foods9111571
Chicago/Turabian StyleRyu, Sangdon, Minhye Shin, Soohyun Cho, Inho Hwang, Younghoon Kim, and Sangnam Oh. 2020. "Molecular Characterization of Microbial and Fungal Communities on Dry-Aged Beef of Hanwoo Using Metagenomic Analysis" Foods 9, no. 11: 1571. https://doi.org/10.3390/foods9111571
APA StyleRyu, S., Shin, M., Cho, S., Hwang, I., Kim, Y., & Oh, S. (2020). Molecular Characterization of Microbial and Fungal Communities on Dry-Aged Beef of Hanwoo Using Metagenomic Analysis. Foods, 9(11), 1571. https://doi.org/10.3390/foods9111571




