The Role of an Acidic Peptide in Controlling the Oxidation Process of Walnut Oil
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. The Mechanism of Action of Acidic Peptide in Oil
2.2.1. Preparation of Oil Samples
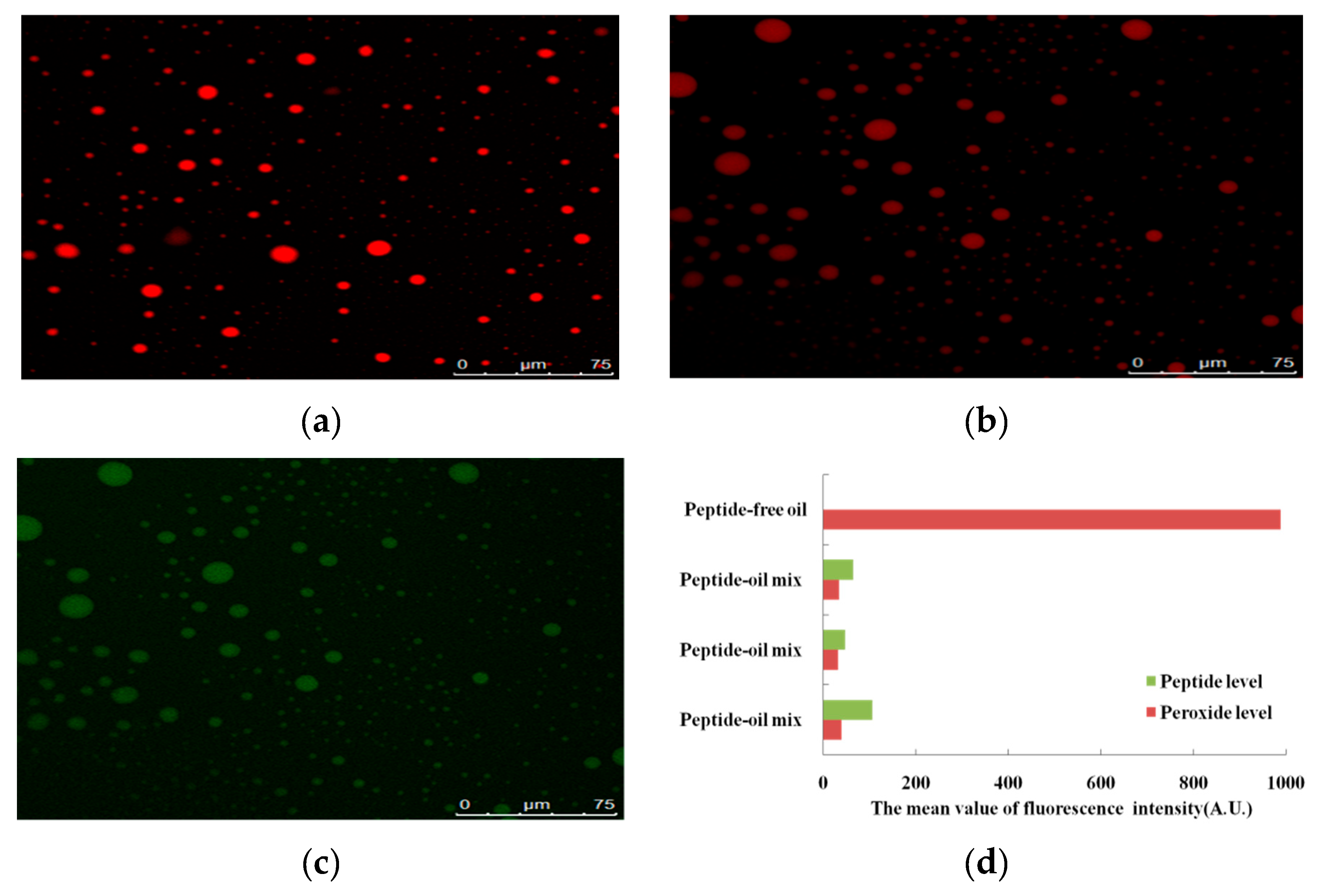
2.2.2. Observation of Lipid Peroxide
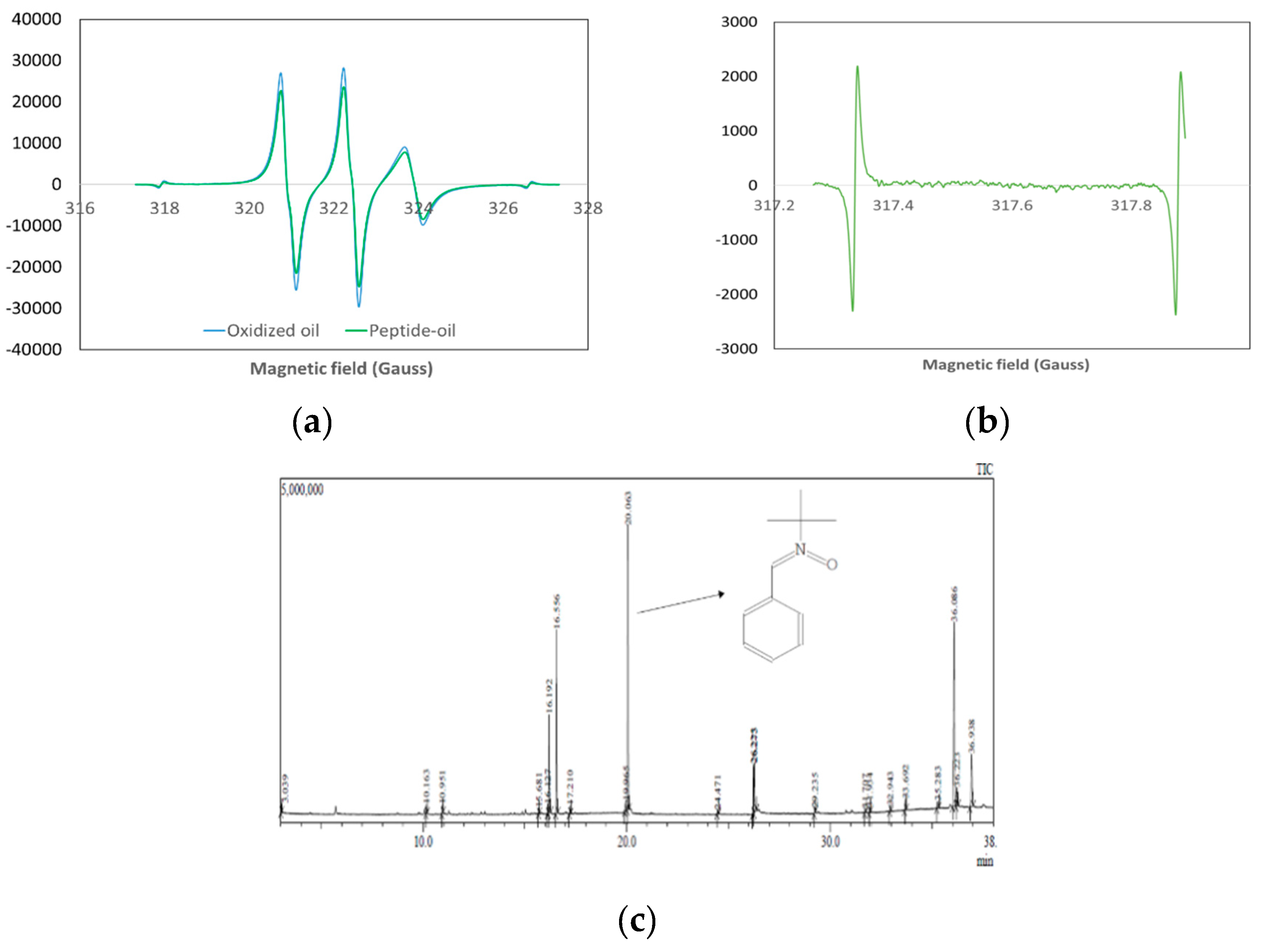
2.2.3. Evaluation of Lipid-Derived Radical Production
2.2.4. Examination of the Lipid-Derived Radical
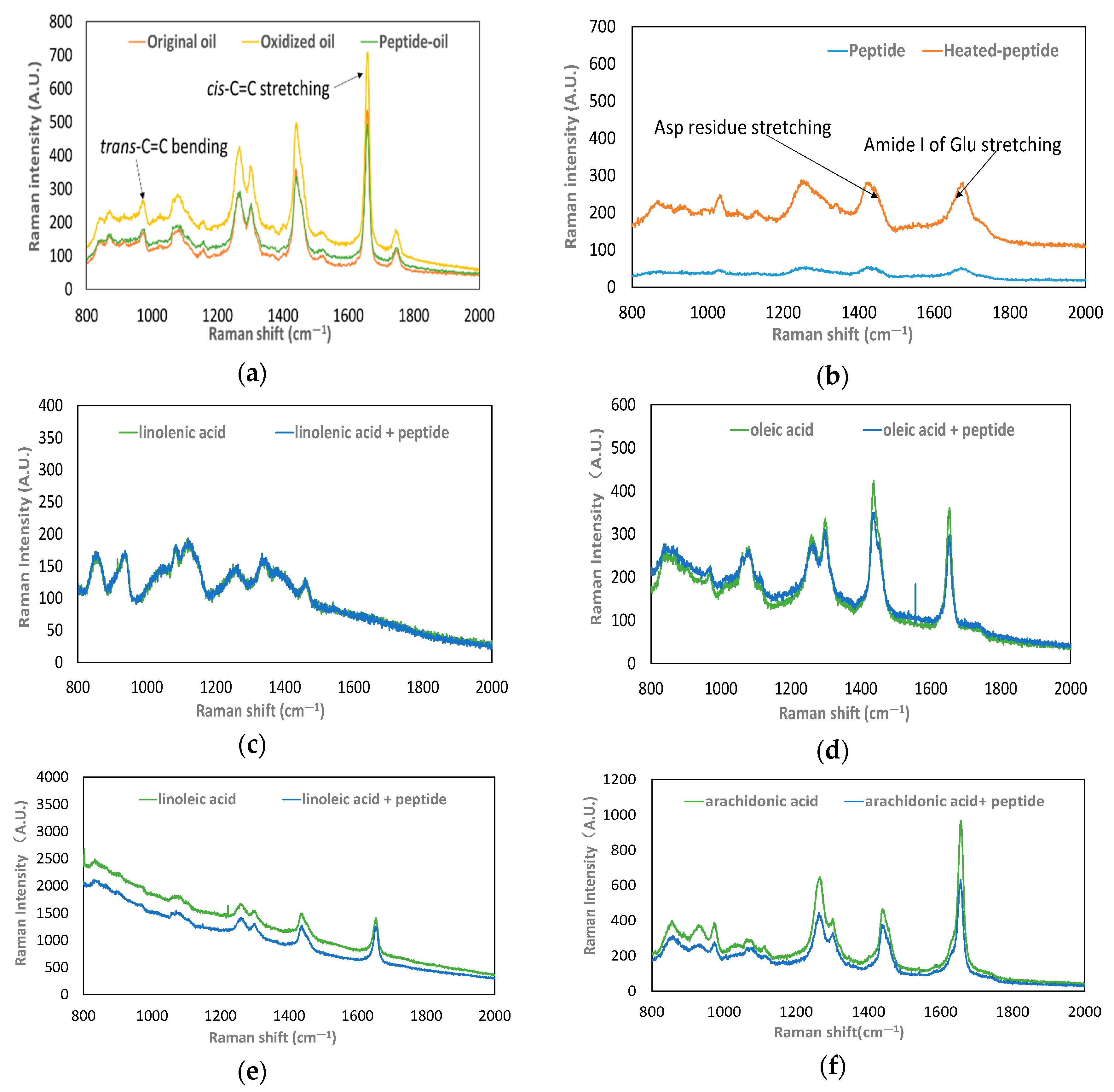
2.2.5. Investigation of the Raman Shift of the Lipid
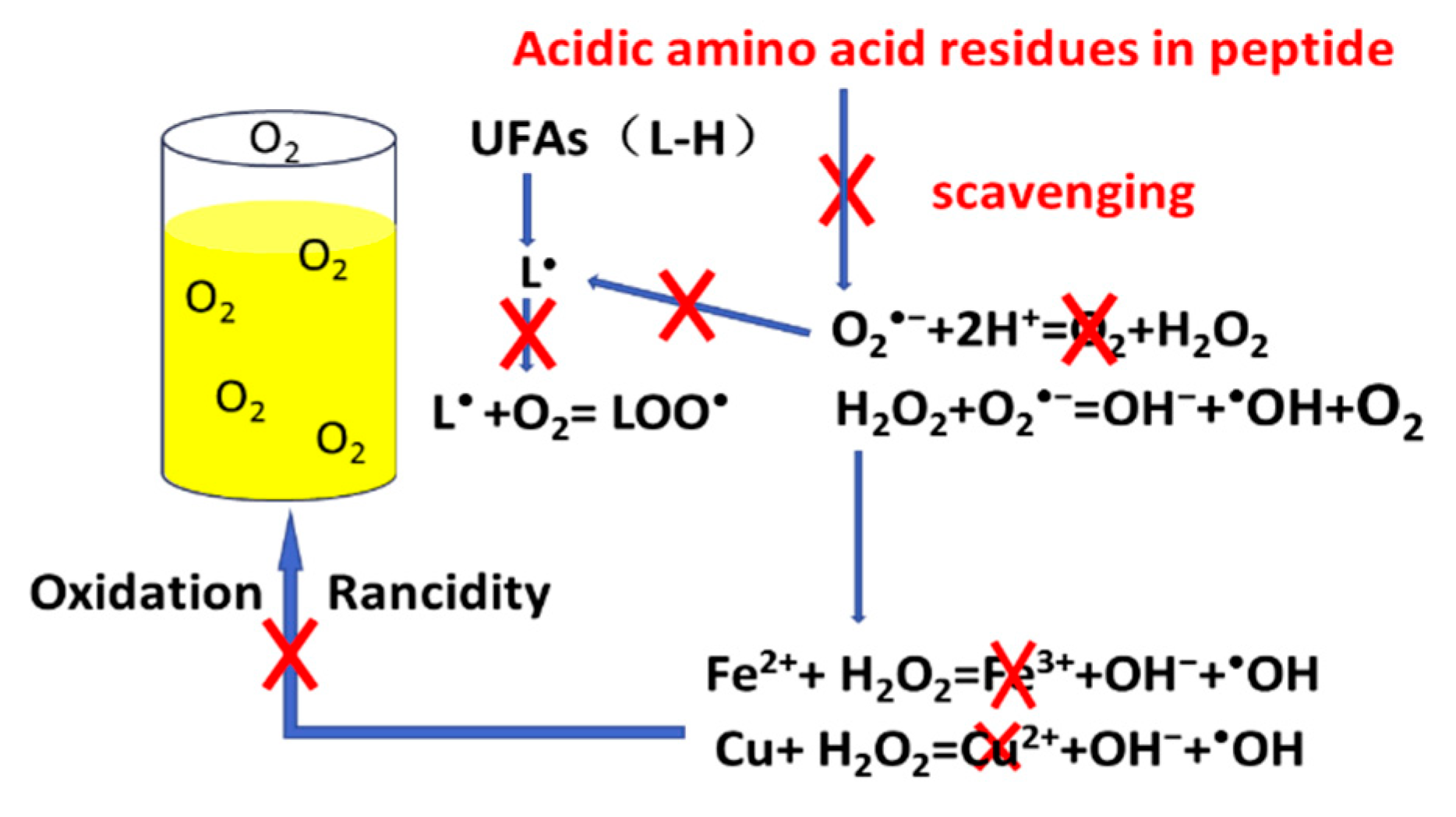
2.3. Effect of Acidic Amino Acid Residues in Anti-Oxidative Peptides
2.3.1. Superoxide Radical Scavenging Assay
2.3.2. Linoleic Acid Peroxidation Inhibition Assay
2.4. Statistical Analysis
3. Results
3.1. The Mechanism of Action of an Acidic Peptide in Oil
3.1.1. Reduction of Lipid Peroxide
3.1.2. Inhibition of Lipid-Derived Radical Production
3.1.3. Peptide Binding with UFAs
3.2. Effect of Acidic Amino Acid Residues in Peptides
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Afonso, C.B.; Sousa, B.C.; Pitt, A.R.; Spickett, C.M. A mass spectrometry approach for the identification and localization of small aldehyde modifications of proteins. Arch. Biochem. Biophys. 2018, 646, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Kanavouras, A.; Coutelieris, F.A. Systematic transition from description to prediction for the oxidation in packaged olive oil. Food Chem. 2017, 229, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, C.; Westberg, M.; Breitenbach, T.; Bregnhøj, M.; Ogilby, P.R. Monitoring Interfacial Lipid Oxidation in Oil-in-Water Emulsions Using Spatially Resolved Optical Techniques. Anal. Chem. 2017, 89, 6239–6247. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, J.; Grypa, R.; Ellingworth, J.; Duodu, K.G.; Kock, H.L.D. The effects of antioxidants and shelf life conditions on oxidation markers in a sunflower oil salad dressing emulsion (SOSDE). Food Chem. 2016, 213, 230–237. [Google Scholar] [CrossRef]
- Xu, L.; Yu, X.; Liu, L.; Zhang, R. A novel method for qualitative analysis of edible oil oxidation using an electronic nose. Food Chem. 2016, 202, 229–235. [Google Scholar] [CrossRef]
- Qi, B.; Zhang, Q.; Sui, X.; Wang, Z.; Li, Y.; Jiang, L. Differential scanning calorimetry study—Assessing the influence of composition of vegetable oils on oxidation. Food Chem. 2016, 194, 601–607. [Google Scholar] [CrossRef]
- Porter, N.A.; Caldwell, S.E.; Mills, K.A. Mechanisms of free radical oxidation of unsaturated lipids. Lipids 1995, 30, 277. [Google Scholar] [CrossRef]
- Elias, R.J.; Kellerby, S.S.; Decker, E.A. Antioxidant activity of proteins and peptides. Crit. Rev. Food Sci. 2008, 48, 430. [Google Scholar] [CrossRef]
- Guo, H.; Yoshiaki, K.; Masami, Y. Structures and properties of antioxidative peptides derived from royal jelly protein. Food Chem. 2009, 113, 238–245. [Google Scholar] [CrossRef]
- Park, E.Y.; Murakami, H.; Mori, T.; Matsumura, Y. Effects of protein and peptide addition on lipid oxidation in powder model system. J. Agric. Food Chem. 2005, 53, 137–144. [Google Scholar] [CrossRef]
- Beermann, C.; Euler, M.; Herzberg, J.; Stahl, B. Anti-oxidative capacity of enzymatically released peptides from soybean protein isolate. Eur. Food Res. Technol. 2009, 229, 637–644. [Google Scholar] [CrossRef]
- Saiga, A.; Tanabe, S.; Nishimura, T. Antioxidant activity of peptides obtained from porcine myofibrillar proteins by protease treatment. J. Agric. Food Chem. 2003, 51, 3661–3667. [Google Scholar] [CrossRef] [PubMed]
- Jie, Y.; Zhao, H.F.; Sun, X.Q.; Lv, X.R.; Zhang, Z.; Zhang, B.L. Isolation of antioxidative peptide from Caragana ambigua seeds’ protein hydrolysate and its mechanism in retarding lipid auto-oxidation. J. Sci. food Agric. 2019, 99, 3078–3085. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, V.P.; Kapila, S.; Sonfack, T.K.; Kapila, R. Antioxidative peptide derived from enzymatic digestion of buffalo casein. Int. Dairy J. 2015, 42, 1–5. [Google Scholar] [CrossRef]
- Qian, S.Y.; Yue, G.H.; Tomer, K.B.; Mason, R.P. Identification of all classes of spin-trapped carbon-centered radicals in soybean lipoxygenase-dependent lipid peroxidations of omega-6 polyunsaturated fatty acids via LC/ESR, LC/MS, and tandem MS. Free Radic. Biol. Med. 2003, 34, 1017–1028. [Google Scholar] [CrossRef]
- Shamaei, S.; Seiiedlou, S.S.; Aghbashlo, M.; Tsotsas, E.; Kharaghani, A. Microencapsulation of walnut oil by spray drying: Effects of wall material and drying conditions on physicochemical properties of microcapsules. Innov. Food Sci. Emergy 2017, 39, 101–112. [Google Scholar] [CrossRef]
- Li, B.; Wang, H.; Zhao, Q.; Ouyang, J.; Wu, Y. Rapid detection of authenticity and adulteration of walnut oil by FTIR and fluorescence spectroscopy: A comparative study. Food Chem. 2015, 181, 25–30. [Google Scholar] [CrossRef]
- Li, B.; Chen, F.; Wang, X.; Ji, B.; Wu, Y. Isolation and identification of antioxidative peptides from porcine collagen hydrolysate by consecutive chromatography and electrospray ionization-mass spectrometry. Food Chem. 2007, 102, 1135–1143. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Wang, L.; Guo, X.; Wang, X.; Yao, H. Isolation and identification of antioxidative peptides from rice endosperm protein enzymatic hydrolysate by consecutive chromatography and MALDI-TOF/TOF MS/MS. Food Chem. 2010, 119, 226–234. [Google Scholar] [CrossRef]
- Tsopmo, A.; Romanowski, A.; Banda, L.; Lavoie, J.C.; Jenssen, H.; Friel, J.K. Novel anti-oxidative peptides from enzymatic digestion of human milk. Food Chem. 2011, 126, 1138–1143. [Google Scholar] [CrossRef]
- Lin, Z.; Su, G.; Ren, J.; Gu, L.; You, L.; Zhao, M. Isolation and characterization of an oxygen radical absorbance activity peptide from defatted peanut meal hydrolysate and its antioxidant properties. J. Agric. Food Chem. 2012, 60, 5431–5437. [Google Scholar] [CrossRef]
- Thomsen, M.K.; Kristensen, D.; Skibsted, L.H. Electron spin resonance spectroscopy for determination of the oxidative stability of food lipids. J. Am. Oil Chem. Soc. 2000, 77, 725–730. [Google Scholar] [CrossRef]
- Williams, H.E.; Claybourn, M.; Green, A.R. Investigating the free radical trapping ability of NXY-059, S-PBN and PBN. Free Radic. Res. 2007, 41, 6. [Google Scholar] [CrossRef] [PubMed]
- Janzen, E.G.; Towner, R.A.; Krygsman, P.H.; Haire, D.L.; Poyer, J.L. Structure identification of free radicals by EER and GC/MS of PBN spin adducts from the in vitro and in vivo rat liver metabolism of halothane. Free Radic. Res. 1990, 9, 9. [Google Scholar] [CrossRef]
- Yue, Q.S.; Tomer, K.B.; Yue, G.H.; Guo, Q.; Kadiiska, M.B.; Mason, R.P. Characterization of the initial carbon-centered pentadienyl radical and subsequent radicals in lipid peroxidation: Identification via on-line high performance liquid chromatography/electron spin resonance and mass spectrometry. Free Radic. Biol. Med. 2002, 33, 998–1009. [Google Scholar] [CrossRef]
- Polyakov, N.E.; Leshina, T.V.; Salakhutdinov, N.F.; Konovalova, T.A.; Kispert, L.D. Antioxidant and redox properties of supramolecular complexes of carotenoids with β-glycyrrhizic acid. Free Radic. Biol. Med. 2006, 40, 1804–1809. [Google Scholar] [CrossRef]
- Chen, H.; Cao, P.; Li, B.; Sun, D.; Li, J.; Liu, Y. High sensitive and efficient detection of edible oils adulterated with used frying oil by electron spin resonance. Food Control. 2017, 73, 540–545. [Google Scholar] [CrossRef]
- Buettner, G.R. Spin trapping: ESR parameters of spin adducts. Free Radic. Biol. Med. 1987, 3, 259–303. [Google Scholar] [CrossRef]
- Carmona, M.Á.; Lafont, F.; Jiménez-Sanchidrián, C.; Ruiz, J.R. Raman spectroscopy study of edible oils and determination of the oxidative stability at frying temperatures. Eur. Food Res. Technol. 2015, 116, 1451–1456. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, Y.; Xi, J.; Guo, Y.; Qian, H.; Cheng, Y.; Chen, Y.; Yao, W.R. Characterization of lipid oxidation process of beef during repeated freeze-thaw by electron spin resonance technology and Raman spectroscopy. Food Chem. 2018, 243, 58–64. [Google Scholar] [CrossRef]
- Moudache, M.; Nerín, C.; Colon, M.; Zaidi, F. Antioxidant effect of an innovative active plastic film containing olive leaves extract on fresh pork meat and its evaluation by Raman spectroscopy. Food Chem. 2017, 229, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Philippidis, A.; Papliaka, Z.E.; Anglos, D. Surface enhanced Raman and 2D-fluorescence spectroscopy for the investigation of amino acids and egg proteins. Microchem. J. 2016, 126, 230–236. [Google Scholar] [CrossRef]
- Pazderka, T.; Kopecký, V. Drop coating deposition Raman spectroscopy of proteinogenic amino acids compared with their solution and crystalline state. Spectrochim. Acta Part A 2017, 185, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Suetsuna, K. Antioxidant peptides from the protease digest of prawn (Penaeusjaponicus) muscle. Mar. Biotechnol. 2000, 2, 5–10. [Google Scholar] [CrossRef] [PubMed]
| Inhibitor No. | Sequence | Inhibition of Superoxide Anion Radical (%) | Inhibition of Linoleic Acid Oxidation (%) | Reference |
|---|---|---|---|---|
| 0 | QITEGEDGGG | 86.46 ± 0.07 a | 60.37 ± 0.16 a | [13] |
| 1 | ITEGEDGGG | 85.03 ± 0.05 a | 58.62 ± 0.39 a | |
| 2 | QITEGED | 85.00 ± 0.07 a | 55.34 ± 0.39 a | |
| 3 | QEGEDGGG | 85.54 ± 0.23 a | 53.49 ± 0.35 a | |
| 4 | QITEGGGG | 64.14 ± 0.02 b | 31.11 ± 0.17 b | |
| 5 | ED | 14.08 ± 0.05 c | 27.28 ± 0.15 c | |
| control | ||||
| 6 | HIQKEDVPSER | 87.28 ± 0.01 d | 52.53 ± 0.13 d | [14] |
| 7 | HIQKVPSER | 62.36 ± 0.02 e | 30.50 ± 0.16 e |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jie, Y.; Zhao, H.; Zhang, B. The Role of an Acidic Peptide in Controlling the Oxidation Process of Walnut Oil. Foods 2019, 8, 499. https://doi.org/10.3390/foods8100499
Jie Y, Zhao H, Zhang B. The Role of an Acidic Peptide in Controlling the Oxidation Process of Walnut Oil. Foods. 2019; 8(10):499. https://doi.org/10.3390/foods8100499
Chicago/Turabian StyleJie, Yu, Hongfei Zhao, and Bolin Zhang. 2019. "The Role of an Acidic Peptide in Controlling the Oxidation Process of Walnut Oil" Foods 8, no. 10: 499. https://doi.org/10.3390/foods8100499
APA StyleJie, Y., Zhao, H., & Zhang, B. (2019). The Role of an Acidic Peptide in Controlling the Oxidation Process of Walnut Oil. Foods, 8(10), 499. https://doi.org/10.3390/foods8100499





