Compositional Shifts in the Mycobiota of ‘Shine Muscat’ Grape (Vitis labruscana Baily × V. vinifera L.) Bunches During Cold Storage at Different Temperatures
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection and Treatments
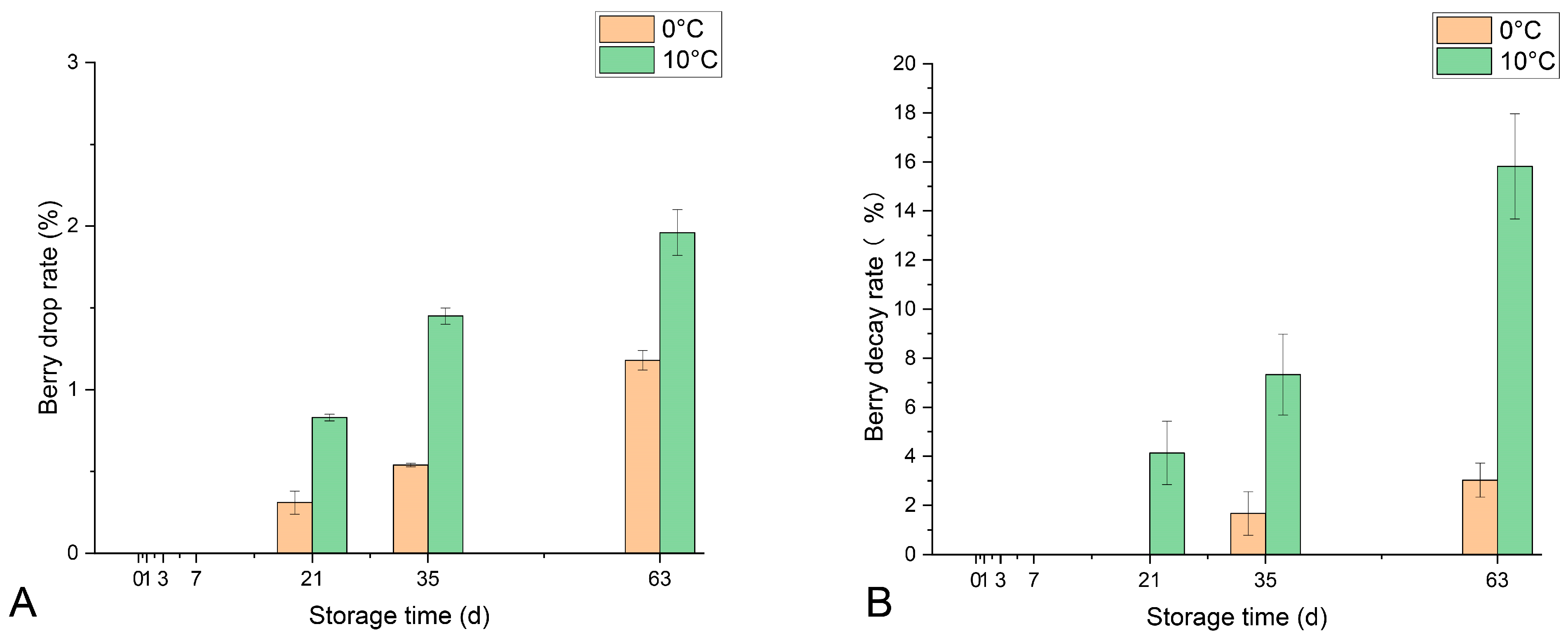
2.2. Berry Drop Rate and Decay Rate Determination
2.3. DNA Extraction, PCR, and Sequencing
2.4. Data Processing
2.5. Statistical Analysis
3. Results
3.1. Stored Berry Morphology and Physiological Indexes
3.2. Overall Analysis of Sequencing
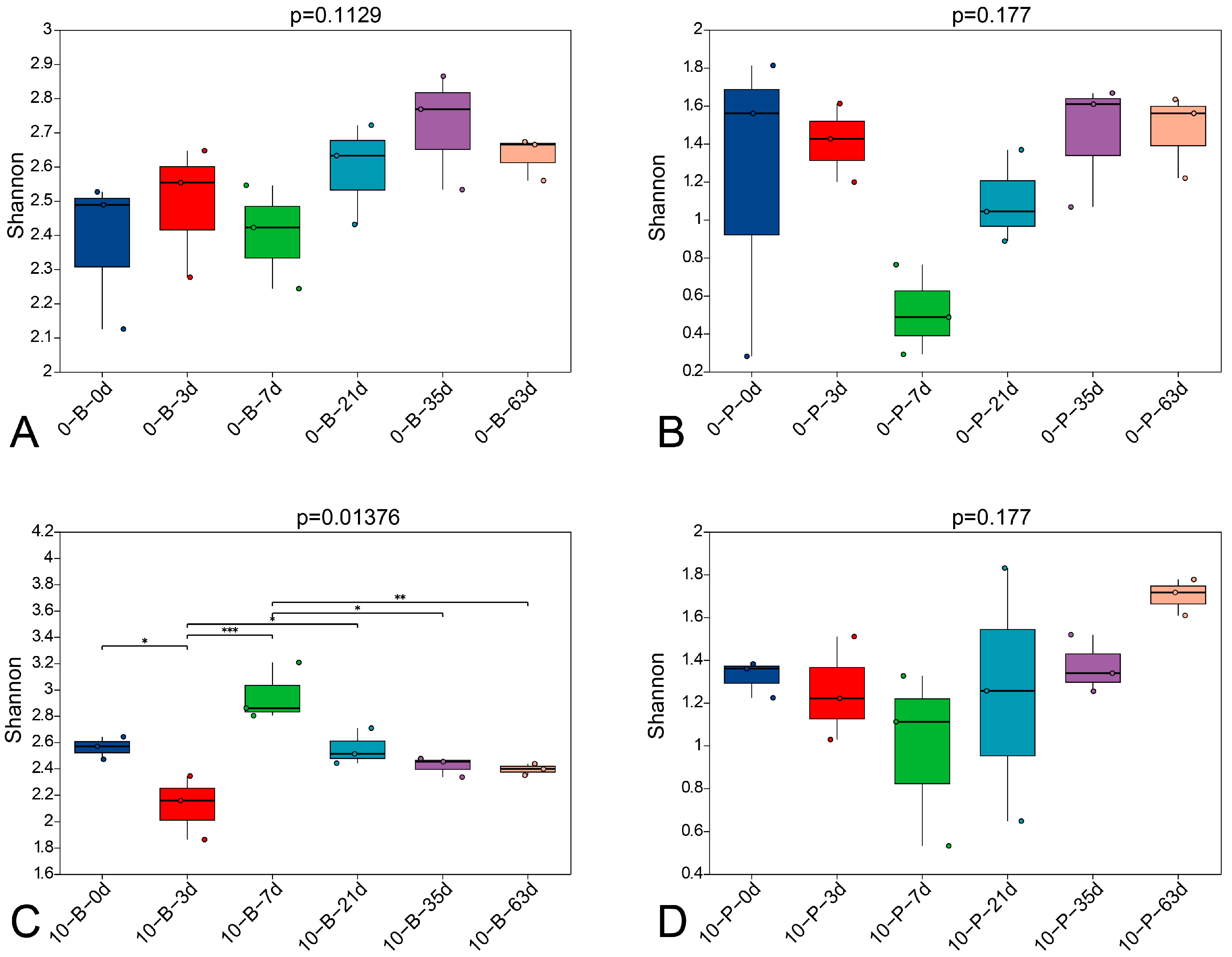
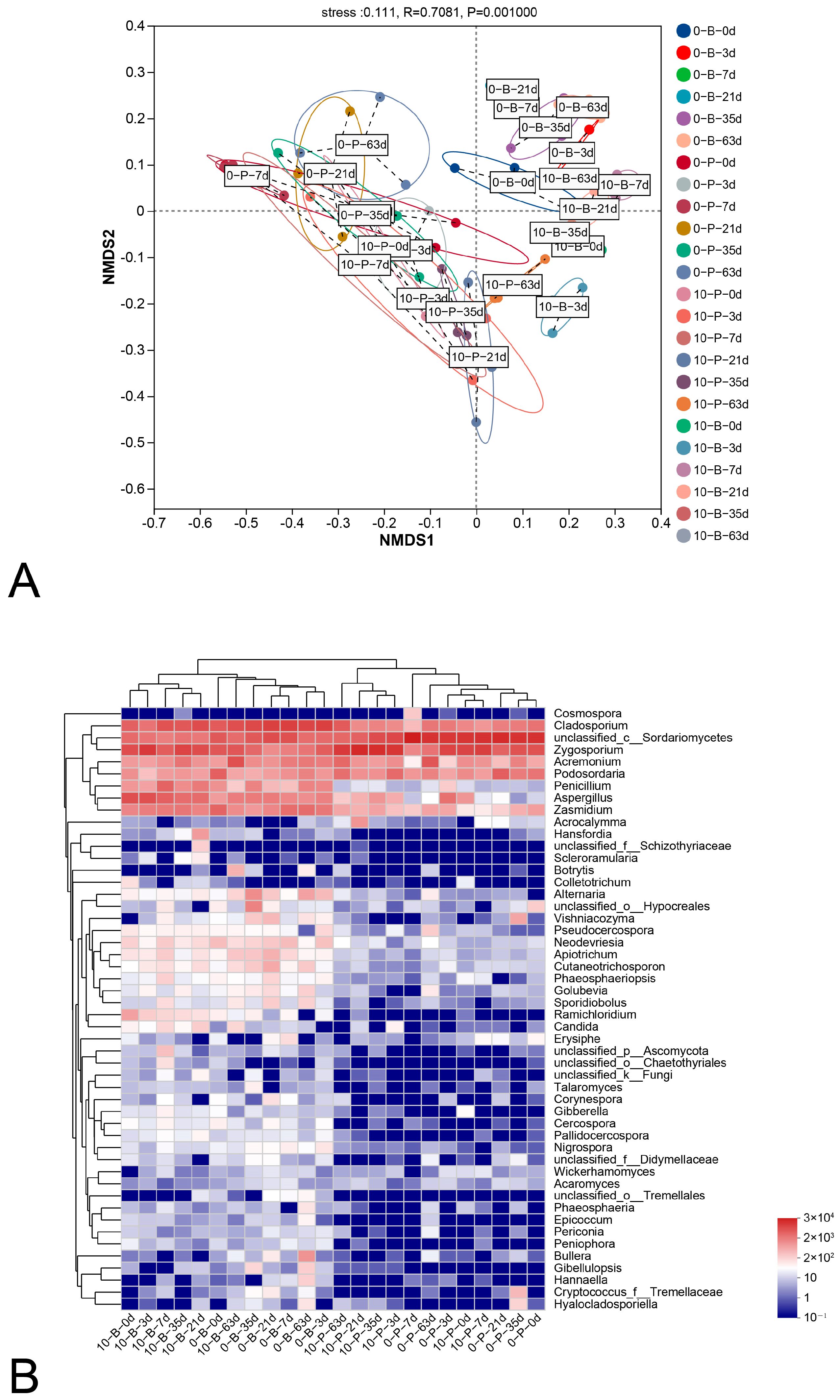
3.3. Fungal α-Diversity
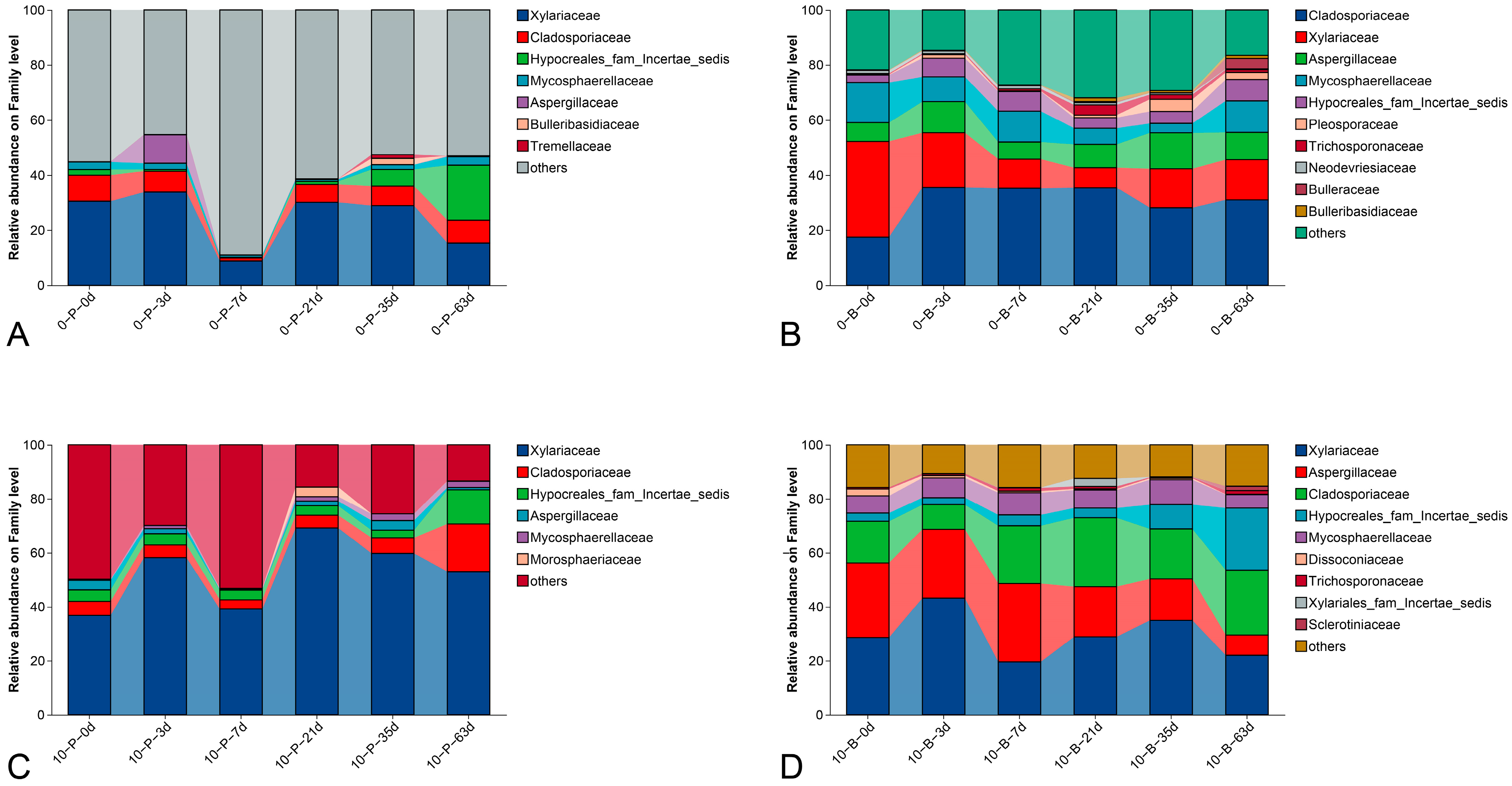
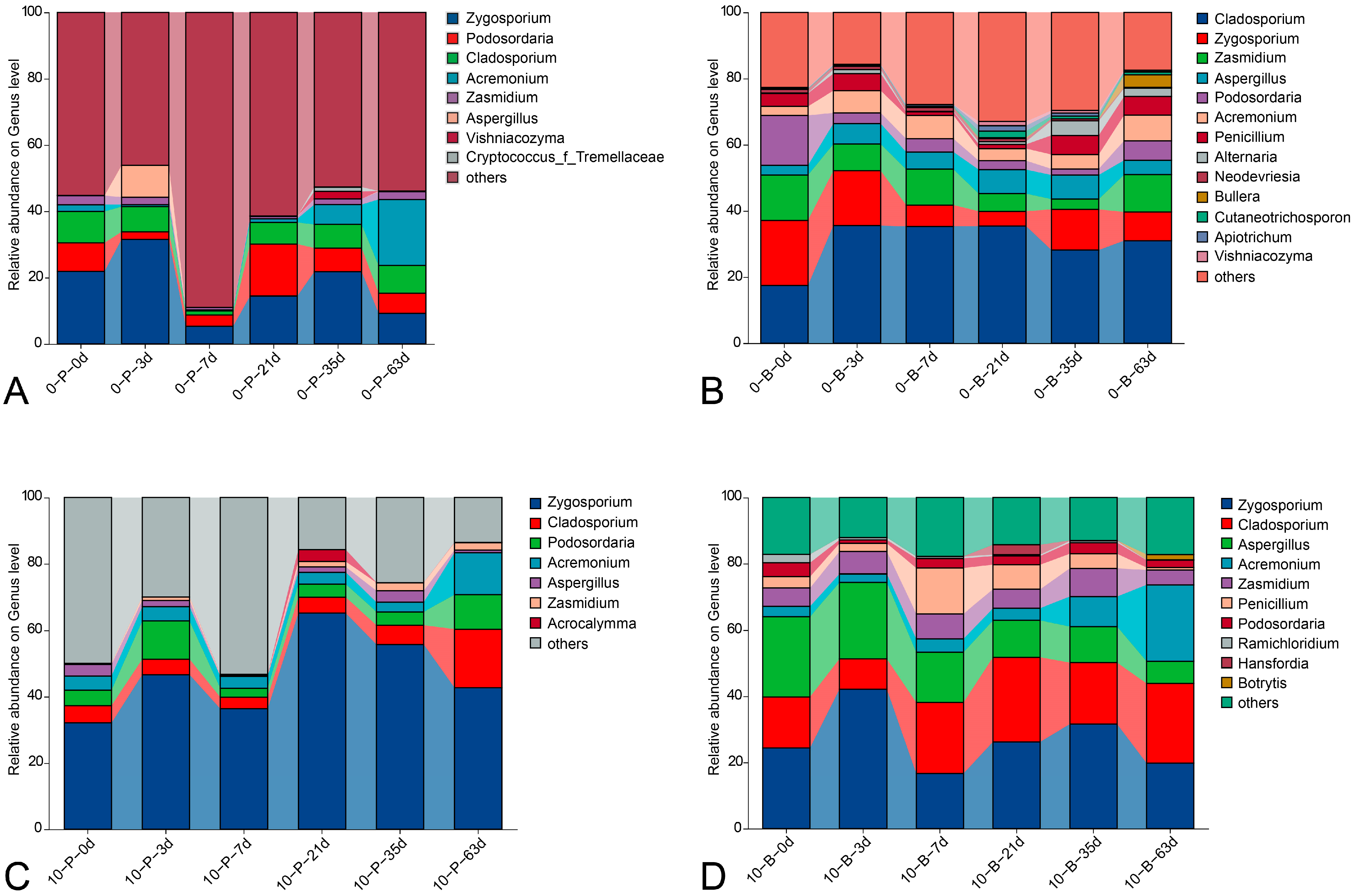
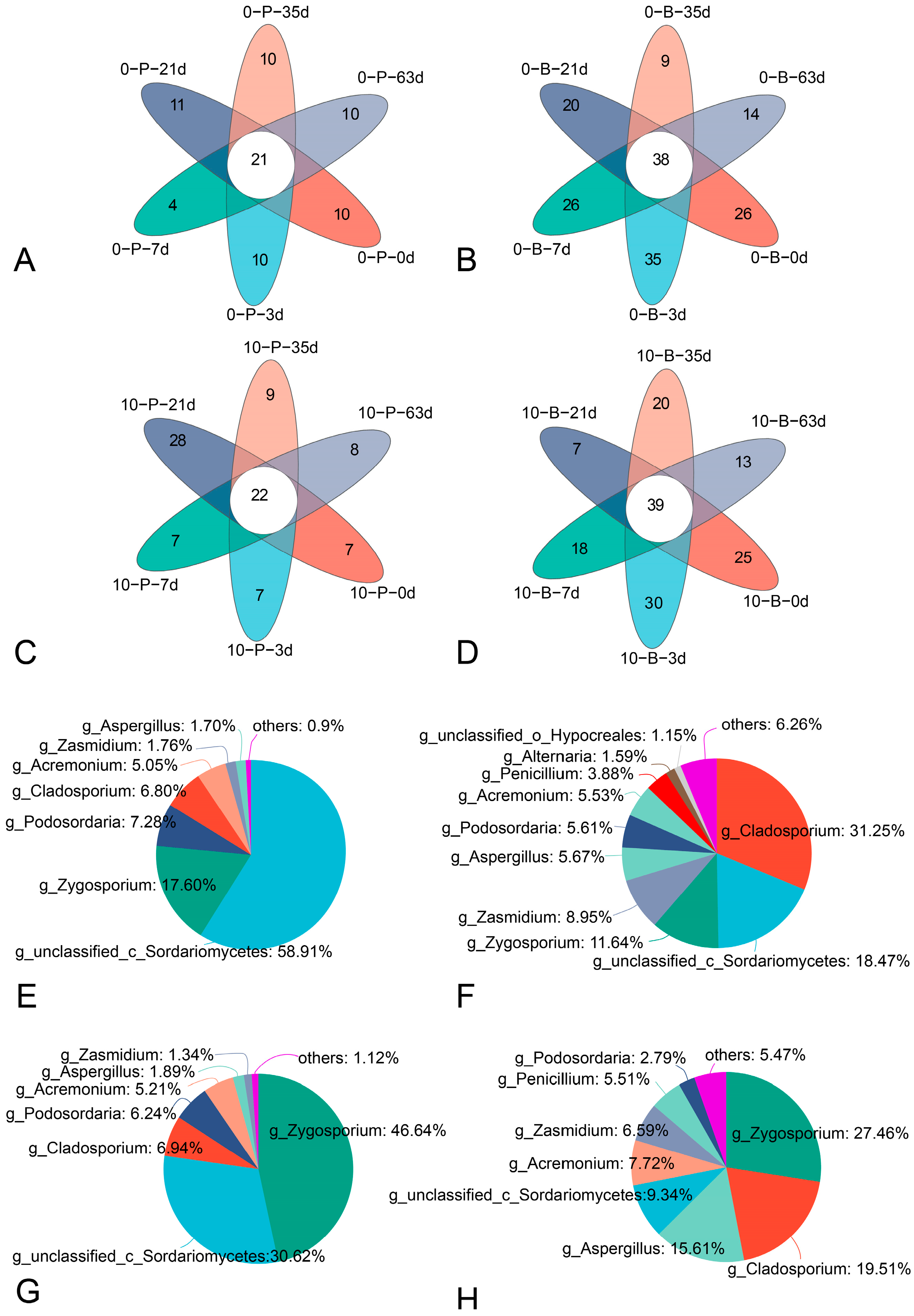
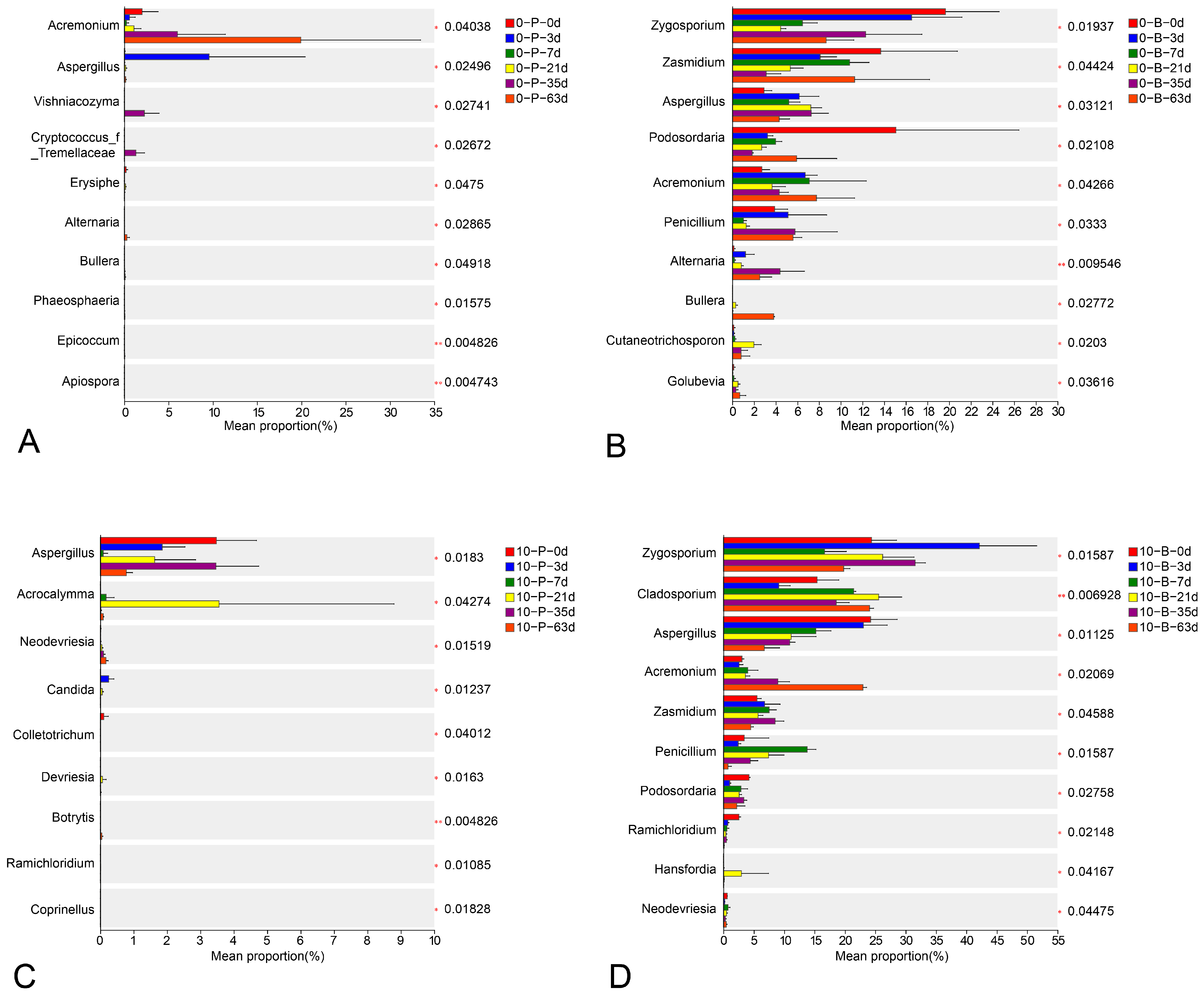
3.4. Taxonomic Composition
3.5. Core Fungal Community of Grape Bunches at Different Storage Temperatures
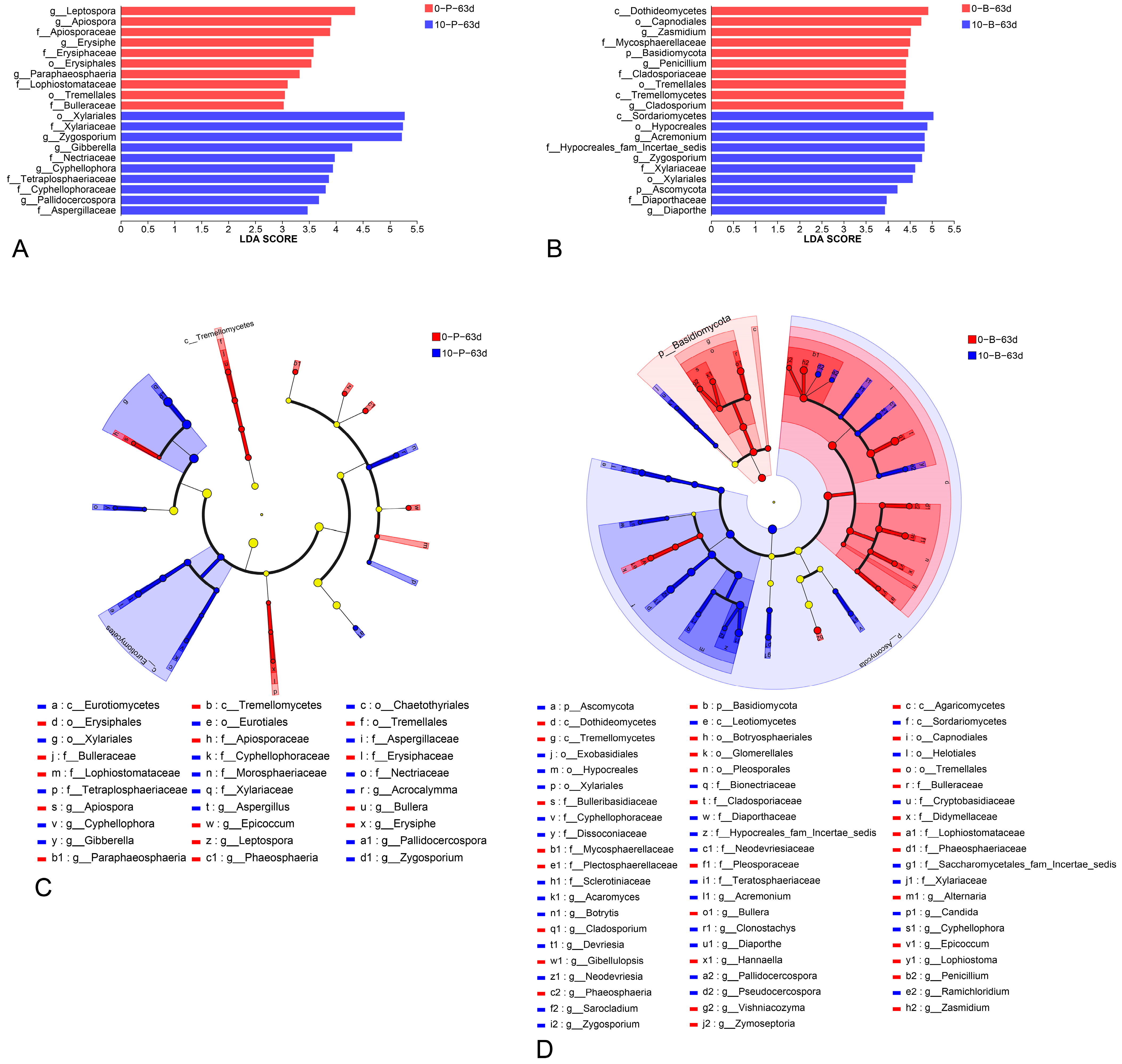
3.6. Interactions Among Ecological Communities at Varying Storage Temperatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahab, M.; Roberto, S.R.; Ahmed, S.; Colombo, R.C.; Silvestre, J.P.; Koyama, R.; de Souza, R.T. Anthocyanin accumulation and color development of ‘Benitaka’ table grape subjected to exogenous abscisic acid application at different timings of ripening. Agronomy 2019, 9, 164. [Google Scholar] [CrossRef]
- Lufu, R.; Ambaw, A.; Opara, U.L. Water loss of fresh fruit: Influencing pre-harvest, harvest and postharvest factors. Sci. Hortic. 2020, 272, 109519. [Google Scholar] [CrossRef]
- de Aguiar, A.C.; Higuchi, M.T.; Yamashita, F.; Roberto, S.R. SO2-Generating Pads and Packaging Materials for Postharvest Conservation of Table Grapes: A Review. Horticulturae 2023, 9, 724. [Google Scholar] [CrossRef]
- Liu, N.; Xie, G.; Yin, W.; Wang, X. Quality characteristics of Niagara grapes and their storage life as affected by 1-MCP combined with sulfur dioxide treatment and modified atmosphere packaging. Int. J. Food Prop. 2022, 25, 159–169. [Google Scholar] [CrossRef]
- Esteves, M.; Lage, P.; Sousa, J.; Centeno, F.; de Fátima Teixeira, M.; Tenreiro, R.; Mendes-Ferreira, A. Biocontrol potential of wine yeasts against four grape phytopathogenic fungi disclosed by time-course monitoring of inhibitory activities. Front. Microbiol. 2023, 14, 1146065. [Google Scholar] [CrossRef]
- Chen, N.; Wei, R.; Cao, X.; Duan, X.; Li, H.; Wang, H. Evaluation of inter-row cover crops effects on the microbial diversity during Cabernet Sauvignon (Vitis vinifera L.) maturation. Food Res. Int. 2022, 162 Pt B, 112113. [Google Scholar] [CrossRef]
- Wei, R.; Ding, Y.; Gao, F.; Zhang, L.; Wang, L.; Li, H.; Wang, H. Community succession of the grape epidermis microbes of cabernet sauvignon (Vitis vinifera L.) from different regions in China during fruit development. Int. J. Food Microbiol. 2022, 362, 109475. [Google Scholar] [CrossRef]
- Knight, S.J.; Karon, O.; Goddard, M.R. Small scale fungal community differentiation in a vineyard system. Food Microbiol. 2020, 87, 103358. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, X.; Zhong, Q.; Zhuang, X.; Bai, Z. Microbial Community Analyses Associated with Nine Varieties of Wine Grape Carposphere Based on High-Throughput Sequencing. Microorganisms 2019, 7, 668. [Google Scholar] [CrossRef]
- Odeyemi, O.A.; Burke, C.M.; Bolch, C.C.J.; Stanley, R. Spoilage microbial community profiling by 16S rRNA amplicon sequencing of modified atmosphere packaged live mussels stored at 4(o)C. Food Res. Int. 2019, 121, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Keshri, J.; Krouptiski, Y.; Abu-Fani, L.; Achmon, Y.; Bauer, T.S.; Zarka, O.; Maler, I.; Pinto, R.; Sela Saldinger, S. Dynamics of bacterial communities in alfalfa and mung bean sprouts during refrigerated conditions. Food Microbiol. 2019, 84, 103261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, Y.; Gao, L.; Wang, Z.; Xu, B. Characterization of a microbial community developing during refrigerated storage of vacuum packed Yao meat, a Chinese traditional food. LWT-Food Sci. Technol. 2018, 90, 562–569. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.M.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, S.J.; Yan, J.; Tang, Y.; Cheng, J.P.; Gao, A.J.; Yao, X.; Ruan, J.J.; Xu, B.L. Research progress on phytopathogenic fungi and their role as biocontrol agents. Front. Microbiol. 2021, 12, 670135. [Google Scholar] [CrossRef]
- Adedayo, A.A.; Babalola, O.O. Fungi that promote plant growth in the rhizosphere boost crop growth. J. Fungi 2023, 9, 239. [Google Scholar] [CrossRef]
- Lutz, S.; Bodenhausen, N.; Hess, J.; Valzano-Held, A.; Waelchli, J.; Deslandes-Hérold, G.; Schlaeppi, K.; van der Heijden, M.G.A. Soil microbiome indicators can predict crop growth response to large-scale inoculation with arbuscular mycorrhizal fungi. Nat. Microbiol. 2023, 8, 2277–2289. [Google Scholar] [CrossRef]
- Allen, T.W.; Burpee, L.L.; Buck, J.W. Variable adhesion and diurnal population patterns of epiphytic yeasts on creeping bentgrass. Can. J. Microbiol. 2006, 52, 404–410. [Google Scholar] [CrossRef]
- Martins, G.; Vallance, J.; Mercier, A.; Albertin, W.; Stamatopoulos, P.; Rey, P.; Lonvaud, A.; Masneuf-Pomarède, I. Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int. J. Food Microbiol. 2014, 177, 21–28. [Google Scholar] [CrossRef]
- Portillo Mdel, C.; Franquès, J.; Araque, I.; Reguant, C.; Bordons, A. Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 2016, 219, 56–63. [Google Scholar] [CrossRef]
- Taylor, M.W.; Tsai, P.; Anfang, N.; Ross, H.A.; Goddard, M.R. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 2014, 16, 2848–2858. [Google Scholar] [CrossRef]
- Setati, M.E.; Jacobson, D.; Bauer, F.F. Sequence-based Analysis of the Vitis vinifera L. cv Cabernet Sauvignon Grape Must Mycobiome in Three South African Vineyards Employing Distinct Agronomic Systems. Front. Microbiol. 2015, 6, 1358. [Google Scholar] [CrossRef] [PubMed]
- De Beeck, M.O.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and Validation of Some ITS Primer Pairs Useful for Fungal Metabarcoding Studies. PLoS ONE 2014, 9, 0097629. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, Z.; Zhao, F.; Zhao, B.; Phillip, F.O.; Feng, J.; Liu, H.; Yu, K. Increased Organic Fertilizer and Reduced Chemical Fertilizer Increased Fungal Diversity and the Abundance of Beneficial Fungi on the Grape Berry Surface in Arid Areas. Front. Microbiol. 2021, 12, 628503. [Google Scholar] [CrossRef]
- Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Toamy, M.A.; Abdelfattaah, A.; Medina, S.; Freilich, S.; Wisniewski, M.; et al. Compositional shifts in the strawberry fruit microbiome in response to near-harvest application of Metschnikowia fructicola, a yeast biocontrol agent. Postharvest Biol. Technol. 2021, 175, 111469. [Google Scholar] [CrossRef]
- Bill, M.; Gokul, J.K.; Viljoen, F.; Korsten, L. Fungal microbiome shifts on avocado fruit associated with a combination of postharvest chemical and physical interventions. J. Appl. Microbiol. 2022, 133, 1905–1918. [Google Scholar] [CrossRef]
- Soldateli, F.J.; Both, V.; Thewes, F.R.; Wendt, L.M.; Ludwig, V.; Rossato, F.P.; Thewes, F.R.; Batista, C.B.; Brackmann, A.; Wagner, R. Overall quality, phenolic compounds, and volatile profile of ‘BRS Isis’ seedless table grapes after long-term storage: Effect of ethanol and high CO2. Sci. Hortic. 2023, 312, 111862. [Google Scholar] [CrossRef]
- Treiber, E.L.; Moreira, L.S.; Clark, M.D. Post-harvest potential of cold-hardy table grapes. Hortscience 2022, 57, 1242. [Google Scholar] [CrossRef]
- Godana, E.A.; Yang, Q.; Liu, J.; Li, J.; Zhang, X.; Zhao, L.; Wang, K.; Zhang, H. Consumer evaluation of sensory properties of table grapes treated with yeast Pichia anomala induced by chitosan. Biol. Control 2022, 170, 104939. [Google Scholar] [CrossRef]
- Su, H.; Zhang, H.; Wang, C.X.; Huang, J.Q.; Shang, J.Y.; Zhang, N.; Wang, D.; Li, K. Grape pruning material improves root development and soil microecology in ‘shine muscat’ grape soils. Hortscience 2020, 55, 2011–2022. [Google Scholar] [CrossRef]
- Liu, S.Y.; Zhang, P.Z.; Wang, X.C.; Hakeem, A.; Niu, M.X.; Song, S.Y.; Fang, J.G.; Shangguan, L.F. Comparative analysis of different bio-organic fertilizers on growth and rhizosphere environment of grapevine seedlings. Sci. Hortic. 2024, 324, 112587. [Google Scholar] [CrossRef]
- Wu, Y.X.; Luo, Z.C.; Qi, L.; Zhang, R.; Wang, Y.X. A study of the relationship between initial grape yield and soil properties based on organic fertilization. Agronomy 2024, 14, 861. [Google Scholar] [CrossRef]
- Gomes, E.P.; Vanz Borges, C.; Monteiro, G.C.; Filiol Belin, M.A.; Minatel, I.O.; Pimentel Junior, A.; Tecchio, M.A.; Lima, G.P.P. Preharvest salicylic acid treatments improve phenolic compounds and biogenic amines in ‘Niagara Rosada’ table grape. Postharvest Biol. Technol. 2021, 176, 111505. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar]
- Mago, T.; Salzberg, S.L. Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Edgar, R.C. Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.; Tiedje, J.; Cole, J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar]
- Matsumoto, H.; Ikoma, Y. Effect of postharvest temperature on the muscat flavor and aroma volatile content in the berries of ‘Shine Muscat’ (Vitis labruscana Baily × V. vinifera L.). Postharvest Biol. Technol. 2016, 112, 256–265. [Google Scholar]
- Jia, H.; Su, Z.; Ren, Y.; Pei, D.; Yu, K.; Dong, T.; Zhang, Y.; Jia, H. Exploration of 5-Azacytidine and Trichostatin A in the modulation of postharvest quality of ‘Shine Muscat’ grape berries. Postharvest Biol. Technol. 2024, 209, 112719. [Google Scholar]
- De Simone, N.; Pace, B.; Grieco, F.; Chimienti, M.; Tyibilika, V.; Santoro, V.; Capozzi, V.; Colelli, G.; Spano, G.; Russo, P. Botrytis cinerea and table grapes: A review of the main physical, chemical, and bio-based control treatments in post-harvest. Foods 2020, 9, 9091138. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Sun, C.; Wang, L.; Zhu, C.; Ren, D.; Wang, T.; Wang, L.; Cai, Y.; Wang, Y.; Zhu, P.; et al. Postharvest disease, latent infection, and preharvest control of ‘Shine-Muscat’ grapes. Postharvest Biol. Technol. 2024, 214, 112989. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Wisniewski, M.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Schena, L. Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PLoS ONE 2016, 11, e0160470. [Google Scholar] [CrossRef]
- Carmichael, P.C.; Siyoum, N.; Chidamba, L.; Korsten, L. Exploring the microbial communities associated with Botrytis cinerea during berry development in table grape with emphasis on potential biocontrol yeasts. Eur. J. Plant Pathol. 2019, 154, 919–930. [Google Scholar] [CrossRef]
- Ahmed, S.; Roberto, S.R.; Domingues, A.R.; Shahab, M.; Junior, O.J.C.; Sumida, C.H.; De Souza, R.T. Effects of Different Sulfur Dioxide Pads on Botrytis Mold in ‘Italia’ Table Grapes under Cold Storage. Horticulturae 2018, 4, 29. [Google Scholar] [CrossRef]
- Wang, F.; Saito, S.; Xiao, C.-L. Postharvest application of natamycin to control gray mold in table grapes. Postharvest Biol. Technol. 2024, 210, 112777. [Google Scholar] [CrossRef]
- Torres-Palazzolo, C.; Ferreyra, S.; Hugalde, I.P.; Kuhn, Y.; Combina, M.; Ponsone, M.L. Recent advances in biocontrol and other alternative strategies for the management of postharvest decay in table grapes. Int. J. Food Microbiol. 2024, 420, 110766. [Google Scholar] [CrossRef]
- Li, W.; Xiao, Y.; Wang, C.; Dang, J.; Chen, C.; Gao, L.; Batzer, J.C.; Sun, G.; Gleason, M.L. A new species of Devriesia causing sooty blotch and flyspeck on Rubber Trees in China. Mycol. Prog. 2013, 12, 733–738. [Google Scholar] [CrossRef]
- Huang, F.; Hou, X.; Dewdney, M.M.; Fu, Y.; Chen, G.; Hyde, K.D.; Li, H. Diaporthe species occurring on citrus in China. Fungal Divers. 2013, 61, 237–250. [Google Scholar] [CrossRef]
- Hilário, S.; Gonçalves MF, M.; Fidalgo, C.; Tacão, M.; Alves, A. Genome Analyses of Two Blueberry Pathogens: Diaporthe amygdali CAA958 and Diaporthe eres CBS 160.32. J. Fungi 2022, 8, 804. [Google Scholar] [CrossRef]
- Wang, J.Y.; Shi, C.; Fang, D.L.; Che, J.L.; Wu, W.L.; Lyu, L.; Li, W.L. The impact of storage temperature on the development of microbial communities on the surface of blueberry fruit. Foods 2023, 12, 1611. [Google Scholar] [CrossRef]
- Sun, D.; Qu, J.; Huang, Y.; Lu, J.; Yin, L. Analysis of microbial community diversity of muscadine grape skins. Food Res. Int. 2021, 145, 110417. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wei, R.; Wang, L.; Wang, W.; Wang, H.; Li, H. Exploring the ecological characteristics of natural microbial communities along the continuum from grape berries to winemaking. Food Res. Int. 2023, 167, 112718. [Google Scholar] [CrossRef]
- Martiniuk, J.T.; Hamilton, J.; Dodsworth, T.; Measday, V. Grape-associated fungal community patterns persist from berry to wine on a fine geographical scale. FEMS Yeast Res. 2023, 23, foac067. [Google Scholar] [CrossRef] [PubMed]
- Lane, C.; Al Shoffe, Y.; Schafran, P.; Li, F.W.; Kao-Kniffin, J.; Watkins, C.B. Amplicon sequencing and shotgun metagenomics reveal functional impacts of amino ethoxyvinylglycine-mediated ripening and cold storage on the microbiome of ‘NY1’ apples. Postharvest Biol. Technol. 2024, 213, 112969. [Google Scholar] [CrossRef]
- Huang, P.W.; Li, J.; Gong, Q.H.; Zhang, Z.H.; Wang, B.; Yang, Z.Q.; Zheng, X.F. Characterization and analysis of dynamic changes of microbial community associated with grape decay during storage. Food Microbiol. 2024, 123, 104581. [Google Scholar] [CrossRef]
- Gao, Q.; Zhang, Y.; Gao, C.C.; Li, H.M.; Cheng, Y.D.; Qian, X.; Zhang, L.S.; Liu, J.Y.; Ogunyemi, S.O.; Guan, J.F. The Microbial Diversity in Relation to Postharvest Quality and Decay: Organic vs. Conventional Pear Fruit. Foods 2023, 12, 1980. [Google Scholar] [CrossRef]
- Kolaríková, Z.; Slavíková, R.; Krüger, C.; Krüger, M.; Kohout, P. PacBio sequencing of Glomeromycota rDNA: A novel amplicon covering all widely used ribosomal barcoding regions and its applicability in taxonomy and ecology of arbuscular mycorrhizal fungi. New Phytol. 2021, 231, 490–499. [Google Scholar] [CrossRef]
- Du, H.; Sun, J.; Zhou, T.C.; Xu, Y. A High-Throughput Absolute Abundance Quantification Method for the Characterisation of Daqu Core Fungal Communities. Fermentation 2022, 8, 345. [Google Scholar] [CrossRef]

| ID | Ascomycota (%) | Basidiomycota (%) | Others (%) | ID | Ascomycota (%) | Basidiomycota (%) | Others (%) |
|---|---|---|---|---|---|---|---|
| 0-B-0d | 98.89 ± 0.40 | 1.08 ± 0.39 | 0.04 ± 0.04 | 0-P-0d | 99.72 ± 0.15 | 0.28 ± 0.15 | 0.00 ± 0.00 |
| 0-B-3d | 98.70 ± 0.28 | 1.24 ± 0.28 | 0.06 ± 0.01 | 0-P-3d | 99.70 ± 0.23 | 0.30 ± 0.23 | 0.00 ± 0.00 |
| 0-B-7d | 98.03 ± 0.49 | 1.96 ± 0.49 | 0.02± 0.01 | 0-P-7d | 99.95 ±0.02 | 0.05 ±0.02 | 0.00 ± 0.00 |
| 0-B-21d | 91.97 ± 0.13 | 7.83 ± 1.22 | 0.20 ± 0.16 | 0-P-21d | 99.69 ± 0.12 | 0.31 ± 0.12 | 0.00 ± 0.00 |
| 0-B-35d | 95.61 ± 1.02 | 4.22 ± 1.14 | 0.16 ± 0.13 | 0-P-35d | 96.08 ± 1.80 | 3.92 ± 1.80 | 0.00 ± 0.00 |
| 0-B-63d | 91.88 ± 1.13 | 8.12 ± 1.13 | 0.00 ± 0.00 | 0-P-63d | 99.32 ± 0.37 | 0.68 ± 0.37 | 0.00 ± 0.00 |
| 10-B-0d | 98.69 ± 0.12 | 1.31 ± 0.12 | 0.00 ± 0.00 | 10-P-0d | 99.93 ± 0.03 | 0.07 ± 0.03 | 0.00 ± 0.00 |
| 10-B-3d | 98.75 ± 0.73 | 1.24 ± 0.71 | 0.01 ± 0.02 | 10-P-3d | 99.94 ± 0.02 | 0.06 ± 0.02 | 0.00 ± 0.00 |
| 10-B-7d | 96.95 ± 0.79 | 2.87 ± 0.55 | 0.18 ± 0.24 | 10-P-7d | 99.74 ± 0.21 | 0.23 ± 0.18 | 0.03 ±0.04 |
| 10-B-21d | 98.05 ± 0.70 | 1.69 ± 0.59 | 0.26 ± 0.11 | 10-P-21d | 99.70 ± 0.20 | 0.29 ± 0.20 | 0.00 ± 0.00 |
| 10-B-35d | 98.90 ± 0.10 | 1.08 ± 0.09 | 0.03 ± 0.02 | 10-P-35d | 99.92 ± 0.04 | 0.08 ± 0.04 | 0.00 ± 0.00 |
| 10-B-63d | 97.54 ± 1.17 | 2.44 ± 1.19 | 0.02 ± 0.01 | 10-P-63d | 99.80 ± 0.06 | 0.20 ± 0.06 | 0.00 ± 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Xie, L.; Zhou, Y.; Ji, X.; Wang, H.; Pang, L.; Liang, F.; Cheng, G.; Zhang, J. Compositional Shifts in the Mycobiota of ‘Shine Muscat’ Grape (Vitis labruscana Baily × V. vinifera L.) Bunches During Cold Storage at Different Temperatures. Foods 2025, 14, 1169. https://doi.org/10.3390/foods14071169
Li W, Xie L, Zhou Y, Ji X, Wang H, Pang L, Liang F, Cheng G, Zhang J. Compositional Shifts in the Mycobiota of ‘Shine Muscat’ Grape (Vitis labruscana Baily × V. vinifera L.) Bunches During Cold Storage at Different Temperatures. Foods. 2025; 14(7):1169. https://doi.org/10.3390/foods14071169
Chicago/Turabian StyleLi, Wei, Linjun Xie, Yongmei Zhou, Xian Ji, Haijun Wang, Liting Pang, Feicui Liang, Guo Cheng, and Jin Zhang. 2025. "Compositional Shifts in the Mycobiota of ‘Shine Muscat’ Grape (Vitis labruscana Baily × V. vinifera L.) Bunches During Cold Storage at Different Temperatures" Foods 14, no. 7: 1169. https://doi.org/10.3390/foods14071169
APA StyleLi, W., Xie, L., Zhou, Y., Ji, X., Wang, H., Pang, L., Liang, F., Cheng, G., & Zhang, J. (2025). Compositional Shifts in the Mycobiota of ‘Shine Muscat’ Grape (Vitis labruscana Baily × V. vinifera L.) Bunches During Cold Storage at Different Temperatures. Foods, 14(7), 1169. https://doi.org/10.3390/foods14071169





