Abstract
This study investigates the occurrence, antimicrobial resistance (AMR) profiles, virulence factors, and plasmid composition of Enterococcus species isolated from salad ingredients in the United Arab Emirates (UAE). Four hundred salad vegetable items collected from local markets, over ten months through 2023, were screened, yielding an Enterococcus detection rate of 85.5% (342/400). E. casseliflavus was the most commonly identified species (50%), followed by E. faecium (20%) and E. faecalis (16%). Among 85 Enterococcus isolates tested for antimicrobial susceptibility, 55.3% displayed resistance to at least one agent, with 18.8% classified as multidrug-resistant (MDR). All isolates were not resistant to ampicillin, linezolid, teicoplanin, tigecycline, and high-level gentamicin. Intrinsic phenotypic resistance to vancomycin was found in E. gallinarum and E. casseliflavus, while low-level (<5%) ciprofloxacin and erythromycin resistance was sporadically detected in E. faecium and E. faecalis. Whole-genome sequencing (WGS) of 14 isolates (nine E. faecium, four E. faecalis, and one E. casseliflavus) unveiled a complex resistome. We report the first detection in salad vegetables of vancomycin resistance genes (vanC, vanXY-C2) in a vancomycin-susceptible E. faecalis isolate. Identifying tetM, ermB, and optrA genes in the studied isolates further underscored emerging resistance to tetracyclines, macrolides, and oxazolidinones. Concurrently, virulence gene analysis revealed 74 putative virulence factors, with E. faecalis harboring a higher diversity of biofilm-related and exoenzyme-encoding genes. One E. faecalis strain carried the cytolysin cluster (cylI, cylS, cylM), highlighting its pathogenic potential. Plasmid profiling identified 19 distinct plasmids, ranging from 3845 bp to 133,159 bp. Among the genome-sequenced isolates, mobilizable plasmids (47.3%) commonly carried AMR genes, especially tet(L) and tet(M), whereas conjugative plasmids (10.5%) did not harbor resistance determinants. These findings highlight that salad vegetables can still harbor and potentially transmit Enterococcus strains with clinically relevant resistance determinants and virulence traits. Enhancing foodborne AMR surveillance with WGS and targeted interventions is key to controlling its spread in the food.
1. Introduction
Salad vegetables are a vital component of a balanced meal and are highly valued for their numerous nutritional advantages. However, they are susceptible to microbial contamination from contact with soil, animal manure, contaminated irrigated water, and other environmental sources [1]. Thus, monitoring the occurrence and diversity of foodborne pathogens and hygienic bacterial indicator bacteria, including Enterococcus species, is critical to safeguarding consumers [2]. While Enterococcus species typically cause infections only under certain conditions, they have emerged as among the leading contributors to nosocomial infections in humans [2,3]. Among the various Enterococcus species identified in food and clinical-related samples, Enterococcus faecalis and Enterococcus faecium are the most prevalent [3]. The threat posed by species other than faecium and faecalis (e.g., Enterococcus gallinarum (particularly in immunocompromised individuals) and Enterococcus casseliflavus (linked to opportunistic infections, including bacteremia and urinary tract infections)) should not be overlooked, as these bacteria are also linked to human health [4,5].
Enterococcus species exhibit inherent resistance to a range of antibiotics, creating significant challenges for treatment on a global scale [3]. The degree of resistance varies by species, but they are inherently resistant to low-dose aminoglycosides, cephalosporins, clindamycin, carbapenems, polymyxins, and lincomycin [4,5]. Research investigating the resistance profiles of Enterococcus species has confirmed a global rise in multidrug-resistant strains, with a significant prevalence of resistance to vancomycin and tetracyclines [4]. It is worth mentioning that vancomycin-resistant E. faecium is included among the World Health Organization Bacterial Priority Pathogens List, 2024, particularly due to its ability to transmit resistance elements across the One Health spectrum [5]. A key factor in Enterococcus species adaptability is their highly flexible genome, enabling them to integrate foreign mobile genetic elements (e.g., plasmids) that harbor resistance determinants through horizontal gene transfer [6]. This highlights the need to monitor antimicrobial resistance in Enterococcus species, especially in the food chain, including salad vegetables typically consumed with minimal heat treatment or even raw, representing a potential threat of transmitting resistant bacteria to humans.
Understanding the genomic landscape of Enterococcus in raw vegetables is crucial for assessing the risks associated with consuming minimally processed foods and tracking the potential role of the global fresh produce trade in disseminating resistant bacteria [7,8]. Hence, this study investigates the diversity, antibiotic resistance profiles, putative virulence, and plasmid composition of Enterococcus species in everyday salad green items from the United Arab Emirates (UAE). Considering that a substantial amount of the UAE’s fresh produce and salad vegetables are sourced from imports, this research carries significant implications beyond the local context. By employing whole-genome sequencing (WGS) and bioinformatic mining of genomic data, this research hopes to generate a comprehensive picture of the resistome, virulome, and plasmid architecture of Enterococcus strains in fresh produce. The novelty of this work lies in providing the first comprehensive genomic characterization of Enterococcus species isolated from salad vegetables consumed raw or minimally processed in the UAE context. The insights gained from this study can strengthen food safety policies, enhance surveillance frameworks, and inform global efforts to curb the propagation of antimicrobial resistance across the food supply.
2. Materials and Methods
2.1. Sources and Sample Selection
A structured sampling approach was implemented by purchasing fresh salad vegetables from retail outlets. The sample size was determined using a presumed detection rate of 50%, a 95% certainty level, and a 5% allowable error [9]. Four hundred samples were gathered and analyzed year-round throughout 2023, excluding July and August due to the summer break. The sampling covered major communal markets (n = 6) and major supermarkets (n = 46) in Dubai, Abu Dhabi, and Al-Ain cities, ensuring a representative selection of locally produced (72.5% of the samples) and imported (27.5% of the samples) items. To maintain sample integrity, only vegetables free from visible dirt and spoilage were selected. Samples were stored refrigerated at 4 °C until testing commenced, wrapped in sterile material, and transported for laboratory analysis on the same collection day.
2.2. Enterococcus Isolation and Characterization
In the course of microbiological testing, 10 g of each sample was blended with 90 mL of Buffered Peptone Water (Oxoid, Basingstoke, UK) for two minutes (BagMixer® 400 P, Interscience, Saint Nom la Brétèche, France). This initial mixture was labeled 10−1, and a 10−2 dilution was subsequently prepared using 0.1% peptone water. From both dilutions (10−1 and 10−2), 100 µL was plated onto Slanetz–Bartley agar (Oxoid, UK) and incubated for 36–48 h at 41.5 °C [10]. Following incubation, as many as five colonies per plate that showed typical Enterococcus characteristics were chosen for species confirmation using the Autobio ms1000 MALDI-TOF system (Autobio Diagnostics, Zhengzhou, China). According to the manufacturer’s protocols, identification scores falling above 9.000 were regarded as reliable for species-level classification.
2.3. Evaluation of Phenotypic AMR
Eighty-five strains representing four Enterococcus species were assessed for their antimicrobial susceptibility profiles using the Vitek-2 automated platform (bioMérieux, Craponne, France). The AST-P592 panel (bioMérieux) was used to test resistance against ten antibiotics: ampicillin, erythromycin, ciprofloxacin, high-level gentamicin, high-level streptomycin, tigecycline, teicoplanin, tetracycline, vancomycin, and linezolid [10]. Classification of isolates as susceptible, intermediate, or resistant followed the manufacturer’s built-in classification, utilizing CLSI standards [11]. Isolates showing resistance to greater than one agent in ≥3 antimicrobial groups were identified as multidrug-resistant (MDR) [12].
2.4. Whole-Genome Sequencing
WGS was conducted on nine E. faecium, four E. faecalis, and one E. casseliflavus, chosen for their notable phenotypic resistance characteristics. The library preparation and sequencing were executed through the service provider Novogene (Cambridge, UK) using the HiSeq platform (Illumina, San Diego, CA, USA). Subsequent analyses were performed through the Solu platform (Solu Healthcare Inc., Helsinki, Finland), enabling batch processing for species determination, multilocus sequence typing (MLST), resistance genes and mutations, and putative virulence determinants [13]. Gene identification employed a ≥95% sequence identity threshold and a minimum 60% sequence coverage cutoff [14]. The raw read data have been deposited in the National Library of Medicine (NCBI) under Bio-Project: PRJNA1231383.
2.5. Plasmid Mobility Evaluation
Plasmid transfer capabilities were investigated using MOB-suite (v3.1.0), which analyzes assembled contigs to detect key plasmid-related features such as relaxase and replicase genes, as well as repetitive regions [15]. The identified plasmid sequences are then matched against a reference database of mobility clusters (MOB-clusters) by calculating the minimal Mash distance, allowing putative plasmids to be grouped into specific MOB-clusters. These plasmids are subsequently labeled as “Conjugative”, “Mobilizable”, or “Non-mobilizable” based on their relaxase gene profiles [16]. The relaxase genes recognize and cleave the origin of transfer (oriT), thereby facilitating horizontal gene transfer and, consequently, the spread of genes conferring resistance and virulence attributes among bacteria [15,16]. After running the MOB-suite on all assemblies, the resulting data were consolidated and examined using the Solu cloud platform, which provides real-time genomic pathogen monitoring [13,15].
3. Results
3.1. Diversity of Enterococcus Species
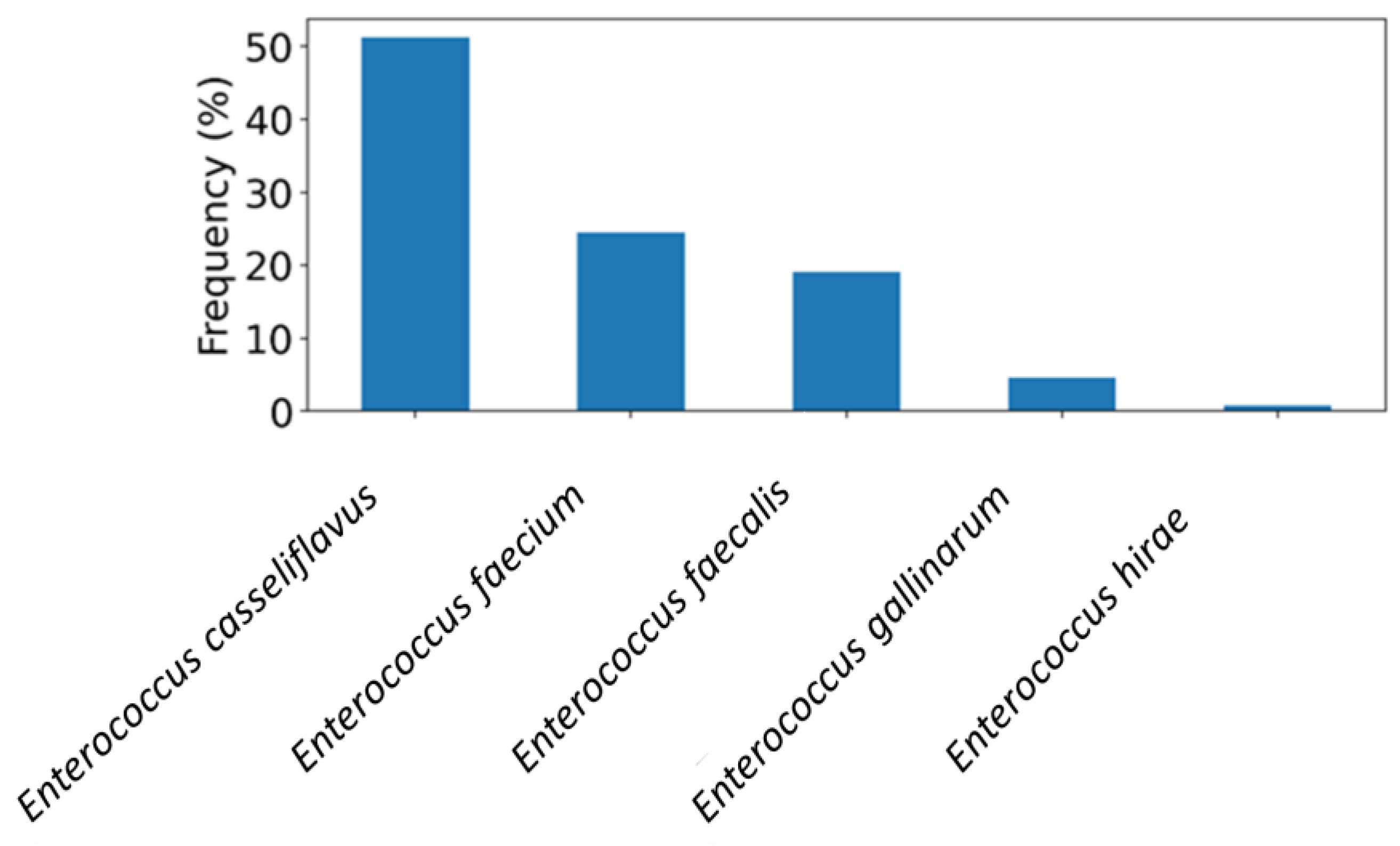
Four hundred salad vegetable samples obtained from retail markets in the UAE were tested to assess the occurrence of Enterococcus species. The rate of detection was 85.5% (342/400), with E. casseliflavus emerging as the most abundant species (50%, n = 171) (Figure 1). E. faecium and E. faecalis were identified in 20% and 16% of the Enterococcus-positive samples, respectively (Figure 1). Additional species were E. gallinarum and E. hirae, detected at lower frequencies as illustrated in Figure 1.
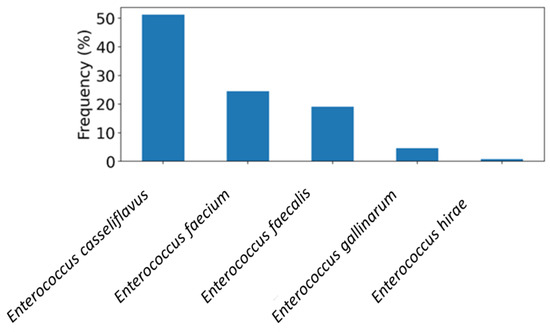
Figure 1.
The frequency distribution of Entercoccus species recovered from salad vegetable samples (n = 400) in the UAE.
3.2. Antimicrobial Susceptibility Profiles Across Enterococcus Species
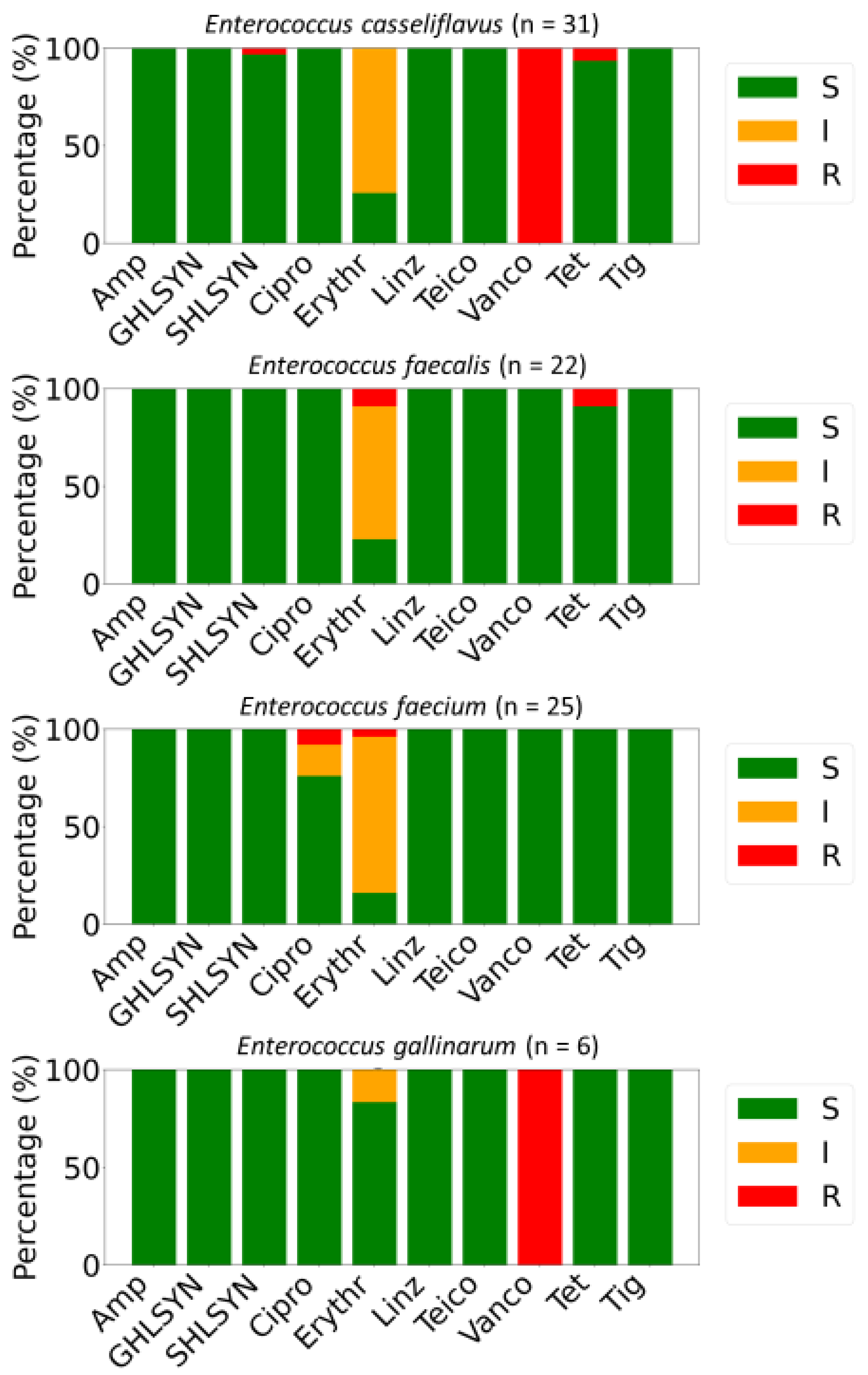
Antimicrobial susceptibility testing of 85 Enterococcus isolates demonstrated variability in resistance patterns among species. Non-susceptibility to a minimum of one antibiotic was observed in 55.3% (n = 47) of the isolates, while 18.8% (n = 16) were classified as MDR (Figure 2). All isolates exhibited full susceptibility to ampicillin, high-level gentamicin (GHLSYN), linezolid (Linz), teicoplanin (Teico), and tigecycline (Tig). Intrinsic resistance toward vancomycin was detected in all E. gallinarum and E. casseliflavus isolates. High-level streptomycin (SHLSYN) resistance was identified in 1.18% of strains, exclusively among E. casseliflavus, while 98.82% remained susceptible. Resistance to ciprofloxacin was revealed in 2.35% of the characterized E. faecium strains, with 4.71% displaying intermediate resistance. Additionally, 3.53% of strains were resistant to erythromycin, whereas a substantial proportion (69.41%) exhibited intermediate susceptibility to this antibiotic (Figure 2).
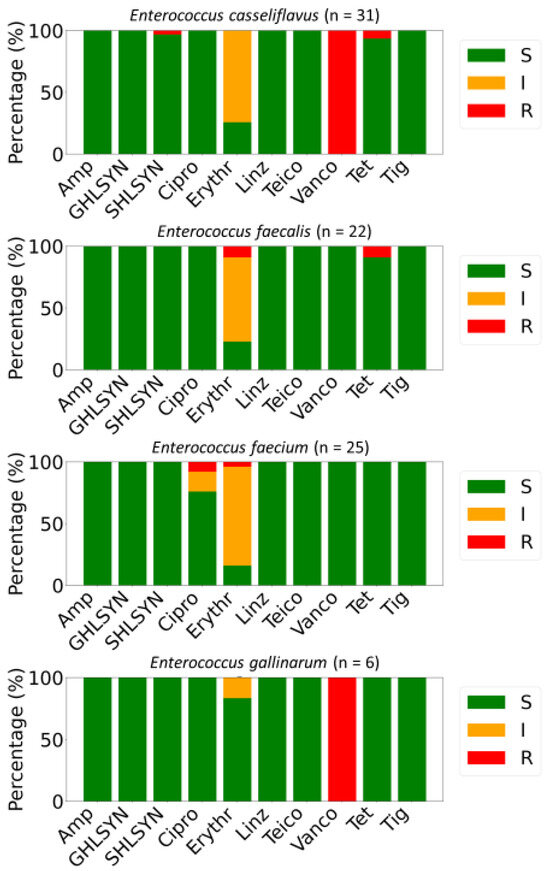
Figure 2.
Distributions of antimicrobially resistant (R, red), susceptible (S, green), and intermediate (I, orange) isolates among Enterococcus spp. isolated from salad greens in the UAE. The panel of antimicrobials included ciprofloxacin (Cipro), tetracycline (Tet), ampicillin (Amp), erythromycin (Erythr), high-level gentamicin (GHLSYN), streptomycin (SHLSYN), vancomycin (Vanco), tigecycline (Tig), teicoplanin (Teico), and linezolid (Linz).
3.3. Whole-Genome Analysis: Resistome Composition
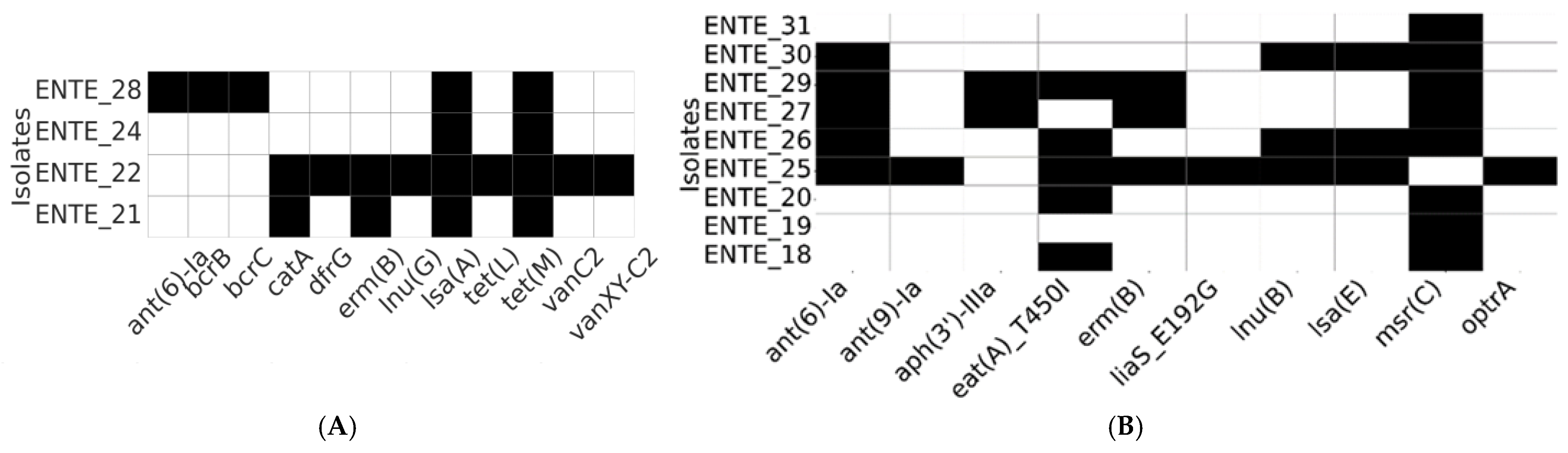
WGS was performed on nine E. faecium and four E. faecalis isolates, selected based on their phenotypic resistance profiles. The resistome analysis identified diverse antimicrobial resistance genes (Figure 3). We also sequenced one E. casseliflavus, where the only AMR genes found in it were those associated with inherent vancomycin resistance (vanC, vanR-C, vanS-C, vanTc, and vanXY-C), and all were predicted to be chromosomal.
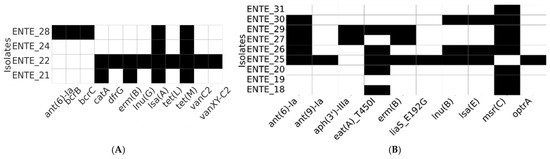
Figure 3.
Resistome profiles of the whole-genome-sequenced E. faecalis (A) and E. faecium (B). The heatmap depicts the presence (black) or absence (white) of antimicrobial resistance determinants.
Vancomycin resistance genes (vanC, vanXY-C2; chromosome located) were detected in a vancomycin-susceptible E. faecalis strain (ENTE-22) recovered from a romaine lettuce sample imported from Jordan. Similarly, the optrA gene (predicted to be chromosome-located), associated with oxazolidinone resistance, was detected in a linezolid-susceptible E. faecium isolate originating from a UAE-grown romaine lettuce sample (Figure 3).
Among the identified resistance determinants, the tetracycline resistance gene (tetM) was exclusively present in all E. faecalis isolates. Additionally, lsa(A), a gene responsible for resistance to lincosamides and streptogramins, was found in every E. faecalis isolate. The msr(C) gene, which mediates macrolide resistance, was widely distributed except for a single E. faecium isolate. Genes conferring aminoglycoside resistance (ant(6)-Ia) and macrolide resistance (ermB) were shared among both E. faecalis and E. faecium (Figure 3).
3.4. Whole-Genome Analysis: Virulome Composition
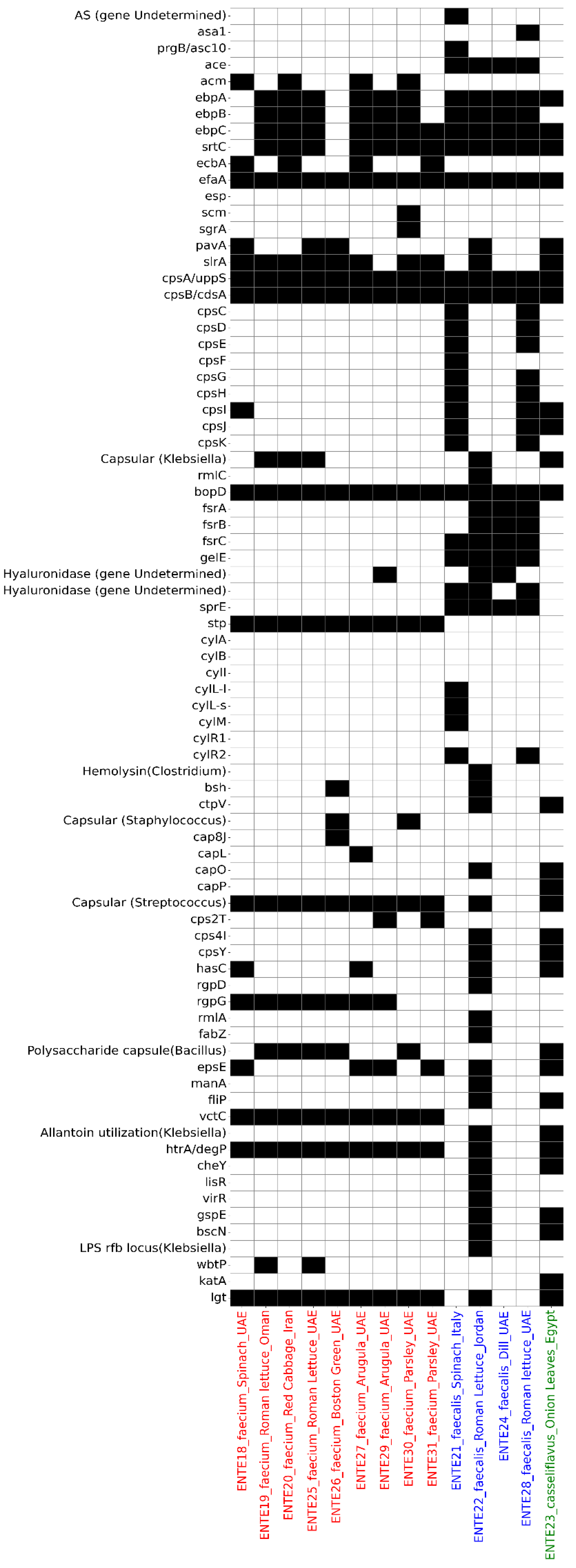
Virulence gene analysis identified 74 putative virulence genes across the 14 sequenced isolates, with the sum of putative virulence genes per isolate ranging from 17 (ENTE-18, E. faecium) to 44 (ENTE-22, E. faecalis) (Figure 4). The sequenced E. casseliflavus also harbored 34 putative virulence genes (Figure 4).

Figure 4.
The putative virulome profile of the whole-genome-sequenced E. faecium (labeled red on the x-axis), E. faecalis (labeled blue on the x-axis), and E. casseliflavus (labeled green on the x-axis). The heatmap depicts the presence (black) or absence (white) of antimicrobial resistance determinants.
The most frequently detected virulence-associated genes were those encoding adherence factors, with E. faecalis carrying the ace gene in 100% of isolates. The efaA gene was universally present across all 14 isolates. Genes associated with biofilm formation, such as those from the fsr operon, were exclusively found in E. faecalis isolates, while they were absent in E. faecium and E. casseliflavus (Figure 4).
Several exoenzyme genes were detected, including gelatinase (gelE), presented in all E. faecalis isolates, and hyaluronidase, identified in three isolates (two E. faecalis and one E. faecium). Toxin-associated genes varied between isolates. The stp gene, encoding serine/threonine phosphatase, was present in all E. faecium isolates, while sprE, encoding a metalloprotease enzyme, was identified in all E. faecalis isolates. The cyl cluster of genes (cylI, cylS, cylM), responsible for cytolysin production, was evident in only one E. faecalis isolate, originating from spinach imported from Italy (Figure 4).
The htrA/degP gene, which plays a critical role in bacterial stress response and survival, was found in all isolates except three E. faecalis isolates. Genes encoding the putative type-II secretion system (gspE) and type-III secretion system (bscN) were also identified in one E. faecalis and one E. casseliflavus isolate. These isolates also carried filP, a gene associated with bacterial invasion and cytoskeletal-like protein synthesis (Figure 4).
3.5. Plasmid Architecture of Enterococcus Isolates
Using MOB-suite bioinformatics tools, plasmid profiling identified 19 distinct plasmids among the sequenced Enterococcus isolates. Plasmid sizes ranged from 3845 bp to 133,159 bp, with the number of plasmids varying from one to four per isolate (Table 1).

Table 1.
Plasmid diversity, mobility, and replicon type analysis of 19 predicted plasmids from the whole-genome analysis of Enterococcus isolated from salad vegetable samples in the UAE.
E. faecium isolates exhibited a greater plasmid diversity than E. faecalis, with replicon typing revealing predominant clusters such as rep_cluster_185, rep_cluster_893, and rep_cluster_1018 (Table 1). Mobility classification categorized plasmids into mobilizable (47.3%), non-mobilizable (42.2%), and conjugative (10.5%) groups. The results in Table 1 indicate that mobilizable plasmids frequently harbored ARGs, particularly tet(L) and (M), which confer resistance to tetracycline. In contrast, conjugative plasmids, predicted in two isolates, did not carry any antimicrobial resistance genes (Table 1).
4. Discussion
In recent years, enterococci have continued to emerge as opportunistic pathogens and a significant cause of nosocomial infections due to their efficient host adaptation and ability to acquire virulence and antibiotic-resistance genes [2,17]. The high prevalence of Enterococcus spp. in salad vegetables observed in this study underscores fresh produce’s potential role in transmitting antimicrobial-resistant bacteria to consumers. The detection rate of 85.5% revealed in this study aligns with previous reports from Oman [18], Korea [19], and Poland [20] that have documented the widespread occurrence of Enterococcus species in fresh produce. Researchers studying plant-derived foods suggest that these products create an environment where enterococci are highly prevalent, mainly due to contact with soil and fertilizers [19]. The predominance of E. casseliflavus over clinically significant species such as E. faecalis and E. faecium warrants further investigation, as this species, though typically considered of lower pathogenic potential, has been associated with intrinsic resistance to vancomycin [21].
The antimicrobial susceptibility profiles for isolates in this study revealed variable resistance among Enterococcus species, with intrinsic vancomycin resistance detected widely in E. casseliflavus and E. gallinarum isolates. This finding was consistent with reports from Oman [18], Poland [20], and Tunisia [22], where these species have well-known chromosomally encoded intrinsic vanC-mediated resistance [23]. Our study reports the first detection in E. faecalis from salad vegetables (Roman lettuce, imported from Jordan) of vancomycin resistance genes (vanC, vanXY-C2) in a vancomycin-susceptible isolate. Even if resistance is not expressed in this isolate, our finding highlights that E. faecalis can acquire resistance genes, potentially from co-inhabiting species within the same food/agriculture ecosystem, that carry them intrinsically (e.g., from the widespread E. casseliflavus in fresh produce) [24,25]. This emphasizes the need for ongoing surveillance of this species. Similarly, identifying optrA in linezolid-susceptible E. faecalis isolates suggests the potential for silent reservoirs of resistance, where genes remain functionally inactive or require specific environmental triggers for expression [10]. Given the clinical significance of vancomycin and linezolid in treating enterococcal infections, surveillance for such latent resistance elements is imperative to mitigate future resistance emergence.
The absence of resistance to high-level gentamicin, linezolid, ampicillin, and tigecycline is encouraging, indicating that these antibiotics remain well preserved through effective stewardship and responsible agricultural practices [26]. However, the presence of MDR strains in 18.8% of isolates is a concern, as it reflects the potential for fresh salad greens to serve as reservoirs for antibiotic-resistant bacteria. Similar resistance patterns have been noted in studies in Oman and South Korea [18,19], highlighting this issue’s global nature. The detection of ciprofloxacin resistance (2.35%) among E. faecium isolates is particularly noteworthy, given the critical role of fluoroquinolones in treating enterococcal infections. In the USA, ciprofloxacin resistance was detected in 5% of E. faecalis and 28% of E. faecium isolates from fresh produce [27]. Notably, these rates of resistance are higher compared to our present study in the UAE, highlighting potential regional differences in resistance patterns to clinically relevant antimicrobials and the influence of local agricultural and environmental factors.
For the first time in the UAE and the Middle East, this study conducted comprehensive whole-genome bioinformatics analyses to examine antimicrobial resistance, plasmid content, and putative virulence genes in Enterococcus spp. from plant-derived foods. Except for one isolate, all E. faecium isolates studied have shown the presence of the chromosomal msrC gene, contributing to the intrinsic resistance to macrolide–streptogramin B [28]. Also, in E. faecium, the gene aac(6′)-Ie-aph(2″)-Ia, hypothesized to be associated with gentamicin resistance (HLGR), was widely presented; however, this was not translated to phenotypic resistance in any of the isolates. Previous research noted that some E. faecium strains with this gene do not exhibit HLGR, due to insertions like IS1216V, which disrupt the gene’s function and lead to a loss of the HLGR phenotype [29]. On the other hand, the tetM gene was found in all E. faecalis isolates characterized in this study and was associated with phenotypical resistance levels. Also, based on MOB-suite analysis [15], we pointed out that such a gene was linked in several isolates to mobile genetic elements (e.g., plasmids), a feature that increases the risk of tetM transfer to other bacterial species [30].
Plasmid-mediated AMR plays a crucial role in the persistence and dissemination of resistance genes in foodborne Enterococcus spp. [30]. This study identified a diverse array of plasmid replicons among isolates, highlighting the capacity of plant-derived isolates to maintain horizontal gene transfer, a critical mechanism in disseminating AMR determinants. The greater plasmid diversity observed in E. faecium isolates compared to E. faecalis is likely attributable to differences in their genomic plasticity and ecological adaptability. E. faecium is widely recognized for its highly flexible genome, enhancing its ability to acquire and maintain diverse mobile genetic elements, including plasmids. This adaptability may reflect E. faecium’s broader environmental niches, stronger selective pressure in various habitats, and higher frequency of horizontal gene transfer events. In contrast, E. faecalis generally exhibits a relatively more stable genome, reducing its capacity for extensive plasmid acquisition and diversity [27,30].
This study also revealed a widespread distribution of putative virulence-associated genes across Enterococcus isolates, including those linked to biofilm formation. These virulence factors enhance the ability of enterococcal isolates to acquire adaptive elements, providing evolutionary advantages that improve their survival and persistence in various environments [30]. While some virulence determinants, such as gelE (gelatinase) and hyl (hyaluronidase), have established roles in pathogenicity, many putative virulence genes remain poorly understood [31]. It should be noted that the detection of multiple virulence genes in non-clinical isolates suggests that their presence alone may not indicate pathogenic potential, reinforcing the need for functional validation studies [32]. Future work should investigate the expression and regulation of these genes in foodborne Enterococcus to better understand their role in bacterial fitness, persistence, and potential virulence within the food supply chain.
This study generates novel insights into the prevalence, antimicrobial resistance, and virulence profiles of Enterococcus spp. in fresh produce, offering the first comprehensive whole-genome bioinformatics analysis of these bacteria in plant-derived foods in the UAE and the Middle East. However, some limitations should be acknowledged. This study was conducted on a specific set of fresh produce samples, which may not fully capture the broader diversity of Enterococcus contamination across different food types and seasons. Additionally, while genetic analyses revealed key antimicrobial resistance and virulence determinants, functional validation of these genes was beyond the scope of this study. Despite these constraints, the findings lay a strong foundation for future research. Future investigations should also explore the impact of agricultural practices and environment on the dissemination of Enterococcus in fresh produce, strengthening food safety strategies from a One Health perspective.
5. Conclusions
To our knowledge, this study presents the first reported Enterococcus spp. genome sequences isolated from fresh salad greens in the UAE and Middle East. This work highlights the need for enhanced surveillance of Enterococcus spp. in fresh produce, particularly in settings dependent on imported food, such as in the UAE. The detection of MDR strains, including those with intrinsic vancomycin resistance, underscores the importance of stricter regulations on antimicrobial use in agriculture. A multidisciplinary approach integrating microbiological surveillance, genomic epidemiology, and One Health policies is crucial to mitigating AMR risks. WGS should be utilized to assess the interplay between resistance and virulence, informing food safety policies and risk assessments. Stricter monitoring is essential to prevent foodborne Enterococcus from becoming reservoirs of clinically relevant resistance genes. To effectively address these findings, regulatory and monitoring agencies should implement targeted screening protocols for Enterococcus spp. in imported produce, integrating WGS into routine surveillance programs to rapidly identify and trace AMR strains. Enhancing international collaboration and data-sharing frameworks can further improve tracking and managing risks associated with antimicrobial-resistant pathogens in global food trade. Additionally, educational initiatives and training for agricultural producers regarding antimicrobial stewardship and hygiene practices can significantly contribute to reducing contamination risks at the source.
Author Contributions
Conceptualization, I.H.; data curation, R.H.A.-R. and A.G.; formal analysis, I.H., M.-Y.I.M. and G.B.L.; investigation, I.H., M.K. and R.H.A.-R.; methodology, I.H., M.K. and A.G.; project administration, I.H., M.-Y.I.M. and G.B.L.; supervision, I.H., M.K. and R.H.A.-R.; writing—original draft preparation, I.H.; writing—review and editing, I.H., G.B.L., M.-Y.I.M., A.G., M.K. and R.H.A.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding from ASPIRE, the technology program management pillar of Abu Dhabi’s Advanced Technology Research Council (ATRC), via the ASPIRE Research Institute for Food Security in the Drylands (ARIFSID) project (Subtheme 4.1—One Health and Antimicrobial Resistance).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author. The raw read data have been deposited in the National Library of Medicine (NCBI) under Bio-Project: PRJNA1231383.
Acknowledgments
The Emirate of Abu Dhabi ASPIRE Award for Research Excellence (AARE-2020), award number AARE20-166, endorsed this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Habib, I.; Khan, M.; Mohamed, M.I.; Ghazawi, A.; Abdalla, A.; Lakshmi, G.; Elbediwi, M.; Al Marzooqi, H.M.; Afifi, H.S.; Shehata, M.G.; et al. Assessing the Prevalence and Potential Risks of Salmonella Infection Associated with Fresh Salad Vegetable Consumption in the United Arab Emirates. Foods 2023, 12, 3060. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Guo, S.; Xu, S.; Wang, J.; Zhang, S.; Kuang, Y.; Jin, P. Enterococci for Human Health: A Friend or Foe? Microb. Pathog. 2025, 201, 107381. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Vancomycin-Resistant Enterococcus faecium: A Current Perspective on Resilience, Adaptation, and the Urgent Need for Novel Strategies. J. Glob. Antimicrob. Resist. 2025, 41, 233–252. [Google Scholar] [CrossRef]
- Abd El-Hamid, M.I.; Abd El-Aziz, N.K.; Ammar, A.M.; Gharib, A.A.; Ibrahim, G.A.; Moawed, B.F.M.; Alshamy, H.; El-Malt, R.M.S. Emergence of Multi-Drug-Resistant, Vancomycin-Resistant, and Multi-Virulent Enterococcus Species from Chicken, Dairy, and Human Samples in Egypt. J. Appl. Microbiol. 2025, 136, lxaf001. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of Animal Origin. Microbiol. Spectr. 2018, 6, 41. [Google Scholar] [CrossRef]
- Kuroda, M.; Sekizuka, T.; Matsui, H.; Suzuki, K.; Seki, H.; Saito, M.; Hanaki, H. Complete Genome Sequence and Characterization of Linezolid-Resistant Enterococcus faecalis Clinical Isolate KUB3006 Carrying a cfr(B)-Transposon on its Chromosome and optrA-Plasmid. Front. Microbiol. 2018, 9, 2576. [Google Scholar] [CrossRef]
- da Costa, P.M.; Loureiro, L.; Matos, A.J. Transfer of Multidrug-Resistant Bacteria between Intermingled Ecological Niches: The Interface between Humans, Animals and the Environment. Int. J. Environ. Res. Public Health 2013, 10, 278–294. [Google Scholar] [CrossRef]
- Kim, M.C.; Cha, M.H.; Ryu, J.G.; Woo, G.J. Characterization of Vancomycin-Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Fresh Produces and Human Fecal Samples. Foodborne Pathog. Dis. 2017, 14, 195–201. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, Q.; Liu, Q.; Dufe, D.; Wouters, P.; Deng, L.; Pong, A. How Many Subjects Are Enough in a Veterinary Trial?—Literature Review and Insights from Industrial Statisticians. Res. Vet. Sci. 2025, 186, 105569. [Google Scholar] [CrossRef]
- Habib, I.; Ghazawi, A.; Lakshmi, G.B.; Mohamed, M.I.; Li, D.; Khan, M.; Sahibzada, S. Emergence and Genomic Characterization of the First Reported optrA-Carrying Linezolid-Resistant Enterococci Isolated from Retail Broiler Meat in the United Arab Emirates. Foods 2022, 11, 3190. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Saratto, T.; Visuri, K.; Lehtinen, J.; Ortega-Sanz, I.; Steenwyk, J.L.; Sihvonen, S. Solu: A Cloud Platform for Real-Time Genomic Pathogen Surveillance. BMC Bioinform. 2025, 26, 12. [Google Scholar] [CrossRef]
- Roberts, L.W.; Forde, B.M.; Hurst, T.; Ling, W.; Nimmo, G.R.; Bergh, H.; George, N.; Hajkowicz, K.; McNamara, J.F.; Lipman, J.; et al. Genomic Surveillance, Characterization and Intervention of a Polymicrobial Multidrug-Resistant Outbreak in Critical Care. Microb. Genom. 2021, 7, mgen000530. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The Diversity of Conjugative Relaxases and Its Application in Plasmid Classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef]
- Habib, I.; Lakshmi, G.B.; Mohamed, M.I.; Ghazawi, A.; Khan, M.; Li, D. Enumeration, Antimicrobial Resistance, and Virulence Genes Screening of Enterococcus spp. Isolated from Retail Chicken Carcasses in the United Arab Emirates. Foodborne Pathog. Dis. 2022, 19, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M. Antimicrobial Susceptibility of Fresh Produce-Associated Enterobacteriaceae and Enterococci in Oman. Foods 2021, 11, 3085. [Google Scholar] [CrossRef]
- Tango, C.N.; Wei, S.; Khan, I.; Hussain, M.S.; Kounkeu, P.N.; Park, J.H.; Kim, S.H.; Oh, D.H. Microbiological Quality and Safety of Fresh Fruits and Vegetables at Retail Levels in Korea. J. Food Sci. 2018, 83, 386–392. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zarzecka, U.; Zadernowska, A. Enterococci Isolated from Plant-Derived Food—Analysis of Antibiotic Resistance and the Occurrence of Resistance Genes. LWT 2021, 139, 110549. [Google Scholar] [CrossRef]
- Mullally, C.A.; Fahriani, M.; Mowlaboccus, S.; Coombs, G.W. Non-faecium Non-faecalis Enterococci: A Review of Clinical Manifestations, Virulence Factors, and Antimicrobial Resistance. Clin. Microbiol. Rev. 2024, 37, e0012123. [Google Scholar] [CrossRef]
- Ben Said, L.; Klibi, N.; Dziri, R.; Borgo, F.; Boudabous, A.; Ben Slama, K.; Torres, C. Prevalence, Antimicrobial Resistance and Genetic Lineages of Enterococcus spp. from Vegetable Food, Soil and Irrigation Water in Farm Environments in Tunisia. J. Sci. Food Agric. 2016, 96, 1627–1633. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Courvalin, P.; Reynolds, P.E. vanC Cluster of Vancomycin-Resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 2000, 44, 1660–1666. [Google Scholar] [CrossRef]
- Schwaiger, K.; Bauer, J.; Hörmansdorfer, S.; Mölle, G.; Preikschat, P.; Kämpf, P.; Bauer-Unkauf, I.; Bischoff, M.; Hölzel, C. Presence of the Resistance Genes vanC1 and pbp5 in Phenotypically Vancomycin- and Ampicillin-Susceptible Enterococcus faecalis. Microb. Drug Resist. 2012, 18, 434–439. [Google Scholar] [CrossRef]
- Hölzel, C.; Bauer, J.; Stegherr, E.M.; Schwaiger, K. Presence of the Vancomycin Resistance Gene Cluster vanC1, vanXYc, and vanT in Enterococcus casseliflavus. Microb. Drug Resist. 2014, 20, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, G.; Calonico, C.; Ducci, B.; Magnanini, A.; Lo Nostro, A. Prevalence and Antibiotic Resistance of Enterococcus spp. Isolated from Retail Cheese, Ready-to-Eat Salads, Ham, and Raw Meat. Food Microbiol. 2014, 41, 1–7. [Google Scholar] [CrossRef]
- Johnston, L.M.; Jaykus, L.A. Antimicrobial Resistance of Enterococcus Species Isolated from Produce. Appl. Environ. Microbiol. 2004, 70, 3133–3137. [Google Scholar] [CrossRef]
- Werner, G.; Hildebrandt, B.; Witte, W. The Newly Described msrC Gene Is Not Equally Distributed among All Isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 3672–3673. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Y.H.; Lin, S.Y.; Lin, Y.T.; Tseng, S.P.; Chang, C.C.; Yu, S.Y.; Hung, W.W.; Jao, Y.T.; Lin, C.Y.; Chen, Y.H.; et al. Emergence of aac(6′)-Ie-aph(2″)-Ia-Positive Enterococci with Non-High-Level Gentamicin Resistance Mediated by IS1216V: Adaptation to Decreased Aminoglycoside Usage in Taiwan. J. Antimicrob. Chemother. 2021, 76, 1689–1697. [Google Scholar] [CrossRef]
- Messele, Y.E.; Trott, D.J.; Hasoon, M.F.; Veltman, T.; McMeniman, J.P.; Kidd, S.P.; Petrovski, K.R.; Low, W.Y. Phylogeny, Virulence, and Antimicrobial Resistance Gene Profiles of Enterococcus faecium Isolated from Australian Feedlot Cattle and Their Significance to Public and Environmental Health. Antibiotics 2023, 12, 1122. [Google Scholar] [CrossRef]
- Shridhar, P.B.; Amachawadi, R.G.; Tokach, M.; Patel, I.; Gangiredla, J.; Mammel, M.; Nagaraja, T.G. Whole Genome Sequence Analyses-Based Assessment of Virulence Potential and Antimicrobial Susceptibilities and Resistance of Enterococcus faecium Strains Isolated from Commercial Swine and Cattle Probiotic Products. J. Anim. Sci. 2022, 100, skac030. [Google Scholar] [CrossRef]
- El Zowalaty, M.E.; Lamichhane, B.; Falgenhauer, L.; Mowlaboccus, S.; Zishiri, O.T.; Forsythe, S.; Helmy, Y.A. Antimicrobial Resistance and Whole Genome Sequencing of Novel Sequence Types of Enterococcus faecalis, Enterococcus faecium, and Enterococcus durans Isolated from Livestock. Sci. Rep. 2023, 13, 18609. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).