Role of Lysogenic Phages in the Dissemination of Antibiotic Resistance Genes Applied in the Food Chain
Abstract
1. Introduction
2. Horizontal Gene Transfer
2.1. Generalized Transduction
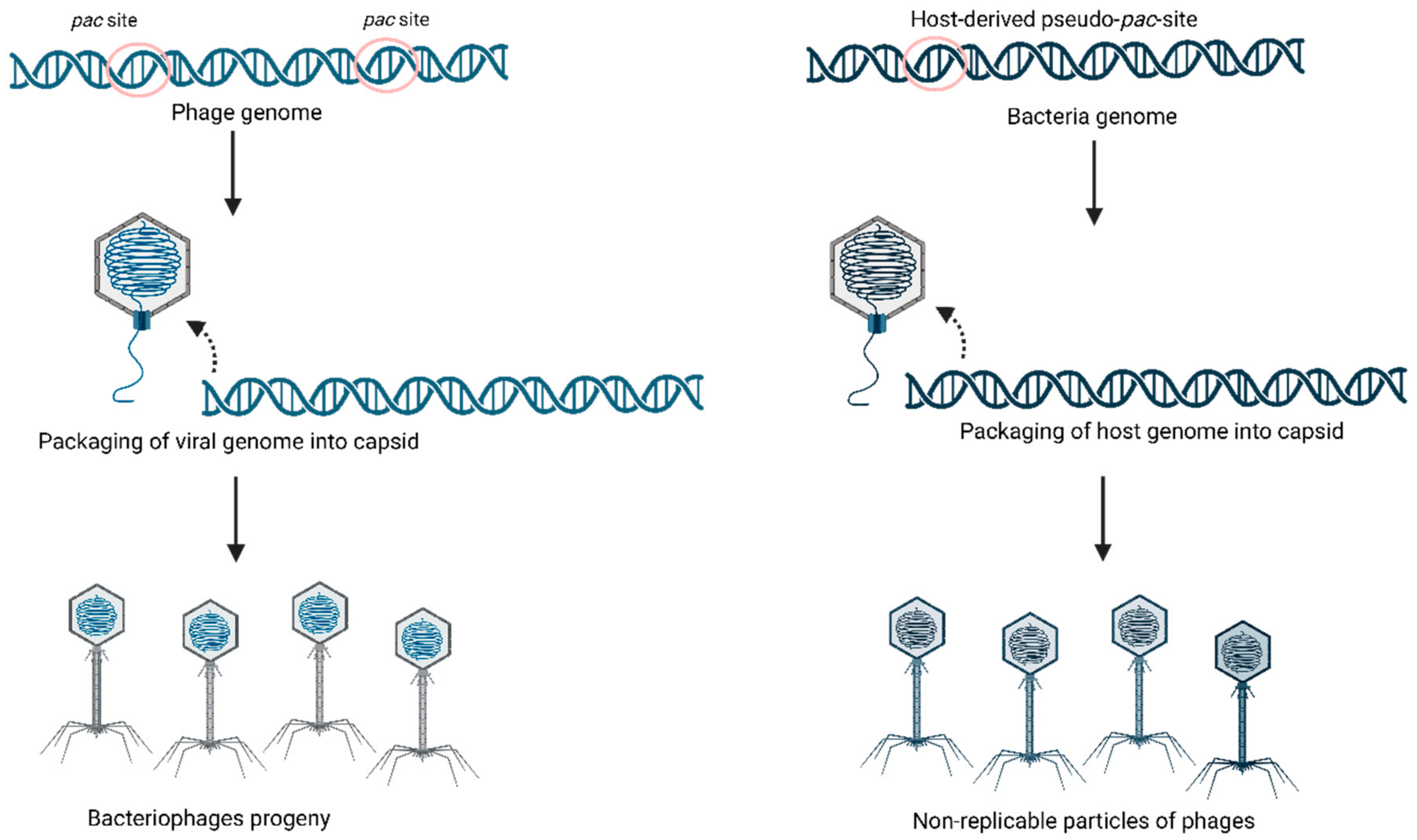
2.2. Specialized Transduction
2.3. Lateral Transduction
3. Natural Reservoirs of ARGs and Influence on Phage Transference
4. Dissemination of ARGs by Lysogenic Phages in the Food Chain
4.1. ARGs in Animal Products and Their Persistence in the Food Chain
4.2. Comparative Analyses in Poultry
4.3. Phages in Dairy Products
5. Farm to Fork
5.1. Agricultural Practices
5.2. Food Processing and Storage Environments
5.3. Retail and Consumer-Level Interactions
5.3.1. Mechanisms by Which Lysogenic Phages Spread ARGs in the Food Chain
5.3.2. Contamination During Food Processing
6. Uncertainties and Challenges in Understanding Lateral Transduction
7. Conclusions
8. Future Perspectives and Research Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARG | Antibiotic genes resistance |
| HGT | Horizontal gene transfer |
| GTA | Gene transfer agent |
| PICI | Phage-inducible chromosomal islands |
| VLP | Virus like particle |
| MCP | Multicopy plasmids |
| ARM | Antimicrobial resistance |
| STEC | Shiga toxin-producing E. coli |
| TCP | Toxin-coregulated pilus |
| TC | Cholera toxin |
| VPI | V. cholerae pathogenicity island |
References
- Moelling, K.; Broecker, F.; Willy, C. A Wake-Up Call: We Need Phage Therapy Now. Viruses 2018, 10, 688. [Google Scholar] [CrossRef]
- Sharma, S.; Chatterjee, S.; Datta, S.; Prasad, R.; Dubey, D.; Prasad, R.K.; Vairale, M.G. Bacteriophages and its applications: An overview. Folia Microbiol. 2017, 62, 17–55. [Google Scholar] [CrossRef]
- Twort, F.W. An Investigation on the Nature of Ultra-Microscopic Viruses. Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef]
- Le Romancer, M.; Gaillard, M.; Geslin, C.; Prieur, D. Viruses in extreme environments. Life Extrem. Environ. 2006, 9781402062858, 99–113. [Google Scholar] [CrossRef]
- Lin, L.; Hong, W.; Ji, X.; Han, J.; Huang, L.; Wei, Y. Isolation and characterization of an extremely long tail Thermus bacteriophage from Tengchong hot springs in China. J. Basic Microbiol. 2010, 50, 452–456. [Google Scholar] [CrossRef]
- Säwström, C.; Lisle, J.; Anesio, A.M.; Priscu, J.C.; Laybourn-Parry, J. Bacteriophage in polar inland waters. Extremophiles 2008, 12, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M. Marine viruses: Truth or dare. Ann. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef]
- Naureen, Z.; Dautaj, A.; Anpilogov, K.; Camilleri, G.; Dhuli, K.; Tanzi, B.; Maltese, P.E.; Cristofoli, F.; De Antoni, L.; Beccari, T.; et al. Bacteriophages presence in nature and their role in the natural selection of bacterial populations. Acta Bio Med. Atenei Parm. 2020, 91 (Suppl. S13), e2020024. [Google Scholar] [CrossRef]
- Elois, M.A.; Silva R da Pilati, G.V.T.; Rodríguez-Lázaro, D.; Fongaro, G. Bacteriophages as Biotechnological Tools. Viruses 2023, 15, 349. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Low, S.J.; Džunková, M.; Chaumeil, P.A.; Parks, D.H.; Hugenholtz, P. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat. Microbiol. 2019, 4, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H.; Canchaya, C.; Hardt, W.D. Phages and the Evolution of Bacterial Pathogens: From Genomic Rearrangements to Lysogenic Conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Fournous, G.; Chibani-Chennoufi, S.; Dillmann, M.L.; Brüssow, H. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 2003, 6, 417–424. [Google Scholar] [CrossRef]
- Meng, M.; Li, Y.; Yao, H. Plasmid-Mediated Transfer of Antibiotic Resistance Genes in Soil. Antibiotics 2022, 11, 525. [Google Scholar] [CrossRef]
- Penadés, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef]
- Kimchi, O.; Meir, Y.; Wingreen, N.S. Lytic and temperate phage naturally coexist in a dynamic population model. ISME J. 2024, 18, 93. [Google Scholar] [CrossRef]
- Weld, R.J.; Butts, C.; Heinemann, J.A. Models of phage growth and their applicability to phage therapy. J. Theor. Biol. 2004, 227, 1–11. [Google Scholar] [CrossRef]
- Roy, K.; Ghosh, D.; DeBruyn, J.M.; Dasgupta, T.; Wommack, K.E.; Liang, X.; Wagner, R.E.; Radosevich, M. Temporal Dynamics of Soil Virus and Bacterial Populations in Agricultural and Early Plant Successional Soils. Front. Microbiol. 2020, 11, 1494. [Google Scholar] [CrossRef]
- Thierauf, A.; Perez, G.; Maloy, A.S. Generalized transduction. Methods Mol. Biol. 2009, 501, 267–286. [Google Scholar] [CrossRef]
- Chiang, Y.N.; Penadés, J.R.; Chen, J. Genetic transduction by phages and chromosomal islands: The new and noncanonical. PLoS Pathog. 2019, 15, e1007878. [Google Scholar] [CrossRef]
- Gatica, J.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome. Environ. Sci. Pollut. Res. 2013, 20, 3529–3538. [Google Scholar] [CrossRef]
- Bezuidt, O.K.I.; Lebre, P.H.; Pierneef, R.; León-Sobrino, C.; Adriaenssens, E.M.; Cowan, D.A.; Van de Peer, Y.; Makhalanyane, T.P. Phages Actively Challenge Niche Communities in Antarctic Soils. mSystems 2020, 5, 10-1128. [Google Scholar] [CrossRef]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of “historical” antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef]
- Walsh, F.; Duffy, B. The Culturable Soil Antibiotic Resistome: A Community of Multi-Drug Resistant Bacteria. PLoS ONE 2013, 8, e65567. [Google Scholar] [CrossRef]
- Naidoo, I.; Mabaso, M.; Moshabela, M.; Sewpaul, R.; Reddy, S.P. South African health professionals’ state of well-being during the emergence of COVID-19. S. Afr. Med. J. 2020, 110, 956. [Google Scholar] [CrossRef]
- Perry, J.A.; Wright, G.D. The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front. Microbiol. 2013, 4, 138. [Google Scholar] [CrossRef]
- Aminov, R.I. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011, 2, 158. [Google Scholar] [CrossRef]
- Humeniuk, C.; Arlet, G.; Gautier, V.; Grimont, P.; Labia, R.; Philippon, A. β-Lactamases of Kluyvera ascorbata, Probable Progenitors of Some Plasmid-Encoded CTX-M Types. Antimicrob. Agents Chemother. 2002, 46, 3045–3049. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.I.; Graham, D.W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010, 44, 580–587. [Google Scholar] [CrossRef]
- Hughes, V.M.; Datta, N. Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature 1983, 302, 725–726. [Google Scholar] [CrossRef]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.-J.; Lee, S.H.; Cho, J.-C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef]
- Naidoo, Y.; Valverde, A.; Cason, E.D.; Pierneef, R.E.; Cowan, D.A. A clinically important, plasmid-borne antibiotic resistance gene (β-lactamase TEM-116) present in desert soils. Sci. Total Environ. 2020, 719, 137497. [Google Scholar] [CrossRef]
- Enault, F.; Briet, A.; Bouteille, L.; Roux, S.; Sullivan, M.B.; Petit, M.A. Phages rarely encode antibiotic resistance genes: A cautionary tale for virome analyses. ISME J. 2017, 11, 237–247. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Muniesa, M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). ECDC/EFSA/EMA first joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. EFSA J. 2015, 13, 4006. [Google Scholar] [CrossRef]
- Schmerold, I.; van Geijlswijk, I.; Gehring, R. European regulations on the use of antibiotics in veterinary medicine. Eur. J. Pharm. Sci. 2023, 189, 106473. [Google Scholar] [CrossRef]
- Wallinga, D.; Smit, L.A.M.; Davis, M.F.; Casey, J.A.; Nachman, K.E. A Review of the Effectiveness of Current US Policies on Antimicrobial Use in Meat and Poultry Production. Curr. Environ. Health Rep. 2022, 9, 339–354. [Google Scholar] [CrossRef]
- Asfaw, T.; Genetu, D.; Shenkute, D.; Shenkutie, T.T.; Eshetie Amare, Y.; Yitayew, B. Foodborne Pathogens and Antimicrobial Resistance in Ethiopia: An Urgent Call for Action on “One Health”. Infect. Drug Resist. 2022, 15, 5265–5274. [Google Scholar] [CrossRef]
- Antunes, P.; Novais, C.; Peixe, L. Food-to-Humans Bacterial Transmission. Microbiol. Spectr. 2020, 8, 1–26. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Imamovic, L.; Jofre, J.; Muniesa, M. Bacteriophages Carrying Antibiotic Resistance Genes in Fecal Waste from Cattle, Pigs, and Poultry. Antimicrob. Agents Chemother. 2011, 55, 4908–4911. [Google Scholar] [CrossRef]
- Al-Mustapha, A.I.; Raufu, I.A.; Ogundijo, O.A.; Odetokun, I.A.; Tiwari, A.; Brouwer, M.S.; Adetunji, V.; Heikinheimo, A. Antibiotic resistance genes, mobile elements, virulence genes, and phages in cultivated ESBL-producing Escherichia coli of poultry origin in Kwara State, North Central Nigeria. Int. J. Food Microbiol. 2023, 389, 110086. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Liang, Y.; Du, X.; Yang, C.; Yang, L.; Xie, J.; Zhao, R.; Tong, Y.; Qiu, S.; et al. Phage-delivered sensitisation with subsequent antibiotic treatment reveals sustained effect against antimicrobial resistant bacteria. Theranostics 2020, 10, 6310. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, X.; Tang, M.; Liu, J.; Tuo, H.; Gu, J.; Tang, Y.; Lei, C.; Wang, H.; Zhang, A. Exploring the profile of antimicrobial resistance genes harboring by bacteriophage in chicken feces. Sci. Total Environ. 2020, 700, 134446. [Google Scholar] [CrossRef]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; et al. A Phage-Like IncY Plasmid Carrying the mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02035-16. [Google Scholar] [CrossRef]
- Shousha, A.; Awaiwanont, N.; Sofka, D.; Smulders, F.J.; Paulsen, P.; Szostak, M.P.; Humphrey, T.; Hilbert, F. Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl. Environ. Microbiol. 2015, 81, 4600–4606. [Google Scholar] [CrossRef]
- Anand, T.; Bera, B.C.; Vaid, R.K.; Barua, S.; Riyesh, T.; Virmani, N.; Hussain, M.; Singh, R.K.; Tripathi, B.N. Abundance of antibiotic resistance genes in environmental bacteriophages. J. Gen. Virol. 2016, 97, 3458–3466. [Google Scholar] [CrossRef]
- Wang, M.; Liu, P.; Zhou, Q.; Tao, W.; Sun, Y.; Zeng, Z. Estimating the contribution of bacteriophage to the dissemination of antibiotic resistance genes in pig feces. Environ. Pollut. 2018, 238, 291–298. [Google Scholar] [CrossRef]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Gummalla, V.S.; Zhang, Y.; Liao, Y.-T.; Wu, V.C.H. The Role of Temperate Phages in Bacterial Pathogenicity. Microorganisms 2023, 11, 541. [Google Scholar] [CrossRef]
- Penadés, J.R.; Christie, G.E. The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites. Annu. Rev. Virol. 2015, 2, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Schroven, K.; Aertsen, A.; Lavigne, R. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation. FEMS Microbiol. Rev. 2021, 45, 1–15. [Google Scholar] [CrossRef]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic Colitis Associated with a Rare Escherichia coli Serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef]
- Kim, J.; Cho, Y.; Lim, S.-K.; Seo, M.-R.; Sohn, J.W.; Kim, B.; Rho, M.; Pai, H. Comparative analyses of the faecal resistome against β-lactam and quinolone antibiotics in humans and livestock using metagenomic sequencing. Sci. Rep. 2023, 13, 20993. [Google Scholar] [CrossRef]
- Schroeder, C.M.; Meng, J.; Zhao, S.; DebRoy, C.; Torcolini, J.; Zhao, C.; McDermott, P.F.; Wagner, D.D.; Walker, R.D.; White, D.G. Antimicrobial resistance of Escherichia coli O26, O103, O111, O128, and O145 from animals and humans. Emerg. Infect. Dis. 2002, 8, 1409–1414. [Google Scholar] [CrossRef]
- Walsh, C.; Duffy, G.; O’Mahony, R.; Fanning, S.; Blair, I.S.; McDowell, D.A. Antimicrobial resistance in Irish isolates of verocytotoxigenic Escherichia coli (E. coli)—VTEC. Int. J. Food Microbiol. 2006, 109, 173–178. [Google Scholar] [CrossRef]
- Marinus, M.G.; Poteete, A.R. High efficiency generalized transduction in Escherichia coli O157:H7. F1000Research 2013, 2, 7. [Google Scholar] [CrossRef]
- Serra-Moreno, R.; Jofre, J.; Muniesa, M. The CI repressors of Shiga toxin-converting prophages are involved in coinfection of Escherichia coli strains, which causes a down regulation in the production of Shiga toxin 2. J. Bacteriol. 2008, 190, 4722–4735. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodriguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019, 9, 13281. [Google Scholar] [CrossRef]
- Blanco-Picazo, P.; Gómez-Gómez, C.; Tormo, M.; Ramos-Barbero, M.D.; Rodríguez-Rubio, L.; Muniesa, M. Prevalence of bacterial genes in the phage fraction of food viromes. Food Res. Int. 2022, 156, 111342. [Google Scholar] [CrossRef] [PubMed]
- Martín-Díaz, J.; Lucena, F.; Blanch, A.R.; Jofre, J. Review: Indicator bacteriophages in sludge, biosolids, sediments and soils. Environ. Res. 2020, 182, 109133. [Google Scholar] [CrossRef]
- Worley-Morse, T.; Mann, M.; Khunjar, W.; Olabode, L.; Gonzalez, R. Evaluating the fate of bacterial indicators, viral indicators, and viruses in water resource recovery facilities. Water Environ. Res. 2019, 91, 830–842. [Google Scholar] [CrossRef]
- Chopin, A.; Bolotin, A.; Sorokin, A.; Ehrlich, S.D.; Chopin, M.C. Analysis of Six Prophages in Lactococcus Lactis IL1403: Different Genetic Structure of Temperate and Virulent Phage Populations. Nucleic Acids Res. 2001, 29, 644–651. [Google Scholar] [CrossRef]
- Madera, C.; Monjardín, C.; Suárez, J.E. Milk contamination and resistance to processing conditions determine the fate of Lactococcus lactis bacteriophages in dairies. Appl. Environ. Microbiol. 2004, 70, 7365–7371. [Google Scholar] [CrossRef]
- Blanco-Picazo, P.; Gómez-Gómez, C.; Morales-Cortes, S.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic resistance in the viral fraction of dairy products and a nut-based milk. Int. J. Food Microbiol. 2022, 367, 109590. [Google Scholar] [CrossRef]
- Kamiński, B.; Paczesny, J. Bacteriophage Challenges in Industrial Processes: A Historical Unveiling and Future Outlook. Pathogens 2024, 13, 152. [Google Scholar] [CrossRef]
- Garneau, J.E.; Moineau, S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 2011, 10 (Suppl. S1), S20. [Google Scholar] [CrossRef]
- Pujato, S.A.; Quiberoni, A.; Mercanti, D.J. Bacteriophages on dairy foods. J. Appl. Microbiol. 2019, 126, 14–30. [Google Scholar] [CrossRef]
- Kerek, Á.; Németh, V.; Szabó, Á.; Papp, M.; Bányai, K.; Kardos, G.; Kaszab, E.; Bali, K.; Nagy, Z.; Süth, M.; et al. Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption. Vet. Sci. 2024, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Moineau, S.; Lévesque, C. Control of Bacteriophages in Industrial Fermentations. In Bacteriophages; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar] [CrossRef]
- Larrañaga, O.; Brown-Jaque, M.; Quirós, P.; Gómez-Gómez, C.; Blanch, A.R.; Rodriguez-Rubio, L.; Muniesa, M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018, 115, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Topp, E. Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microbiol. 2015, 81, 7905–7913. [Google Scholar] [CrossRef]
- Marti, R.; Tien, Y.C.; Murray, R.; Scott, A.; Sabourin, L.; Topp, E. Safely coupling livestock and crop production systems: How rapidly do antibiotic resistance genes dissipate in soil following a commercial application of swine or dairy manure? Appl. Environ. Microbiol. 2014, 80, 3258–3265. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.; Viñas, I.; Usall, J.; Anguera, M.; Abadias, M. Presence and survival of Escherichia coli O157:H7 on lettuce leaves and in soil treated with contaminated compost and irrigation water. Int. J. Food Microbiol. 2012, 156, 133–140. [Google Scholar] [CrossRef]
- Dharmarha, V.; Guron, G.; Boyer, R.R.; Niemira, B.A.; Pruden, A.; Strawn, L.K.; Ponder, M.A. Gamma irradiation influences the survival and regrowth of antibiotic-resistant bacteria and antibiotic-resistance genes on romaine lettuce. Front. Microbiol. 2019, 10, 710. [Google Scholar] [CrossRef]
- Wang, F.H.; Qiao, M.; Chen, Z.; Su, J.Q.; Zhu, Y.G. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 2015, 299, 215–221. [Google Scholar] [CrossRef]
- Alexander, T.W.; Yanke, L.J.; Topp, E.; Olson, M.E.; Read, R.R.; Morck, D.W.; McAllister, T.A. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 2008, 74, 4405–4416. [Google Scholar] [CrossRef]
- Arikan, O.A.; Sikora, L.J.; Mulbry, W.; Khan, S.U.; Foster, G.D. Composting rapidly reduces levels of extractable oxytetracycline in manure from therapeutically treated beef calves. Bioresour. Technol. 2007, 98, 169–176. [Google Scholar] [CrossRef]
- Winckler, C.; Grafe, A. Use of veterinary drugs in intensive animal production evidence for persistence of tetracycline in pig slurry. J. Soils Sediments 2001, 1, 66–70. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and Transport of Antibiotic Residues and Antibiotic Resistance Genes following Land Application of Manure Waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef] [PubMed]
- Mckinney, C.W.; Dungan, R.S.; Moore, A.; Leytem, A.B. Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 2018, 94, 10. [Google Scholar] [CrossRef] [PubMed]
- Nõlvak, H.; Truu, M.; Kanger, K.; Tampere, M.; Espenberg, M.; Loit, E.; Raave, H.; Truu, J. Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total Environ. 2016, 562, 678–689. [Google Scholar] [CrossRef]
- Huang, X.; Liu, C.; Li, K.; Liu, F.; Liao, D.; Liu, L.; Zhu, G.; Liao, J. Occurrence and distribution of veterinary antibiotics and tetracycline resistance genes in farmland soils around swine feedlots in Fujian Province, China. Environ. Sci. Pollut. Res. 2013, 20, 9066–9074. [Google Scholar] [CrossRef]
- Haaber, J.; Leisner, J.J.; Cohn, M.T.; Catalan-Moreno, A.; Nielsen, J.B.; Westh, H.; Penadés, J.R.; Ingmer, H. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 2016, 7, 13333. [Google Scholar] [CrossRef]
- Ruan, C.; Ramoneda, J.; Kan, A.; Rudge, T.J.; Wang, G.; Johnson, D.R. Phage predation accelerates the spread of plasmid-encoded antibiotic resistance. Nat. Commun. 2024, 15, 5397. [Google Scholar] [CrossRef]
- Pfeifer, E.; Bonnin, R.A.; Rocha, E.P.C. Phage-Plasmids Spread Antibiotic Resistance Genes through Infection and Lysogenic Conversion. mBio 2022, 13, e01851-22. [Google Scholar] [CrossRef]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family—A review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Brusa, V.; Costa, M.; Padola, N.L.; Etcheverría, A.; Sampedro, F.; Fernandez, P.S.; Leotta, G.A.; Signorini, M.L. Quantitative risk assessment of haemolytic uremic syndrome associated with beef consumption in Argentina. Sant’Ana A de S, ed. PLoS ONE 2020, 15, e0242317. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.; Nguyen, T.H.; Iwashita, H.; Takemura, T.; Morita, K.; Yamashiro, T. Comparative analyses of CTX prophage region of Vibrio cholerae seventh pandemic wave 1 strains isolated in Asia. Microbiol. Immunol. 2018, 62, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Fidelma Boyd, E.; Moyer, K.E.; Shi, L.; Waldor, M.K. Infectious CTX and the Vibrio Pathogenicity Island Prophage in Vibrio Mimicus: Evidence for Recent Horizontal Transfer between V. Mimicus and V. Cholerae. Infect. Immun. 2000, 68, 1507–1513. [Google Scholar]
- Keen, E.C.; Bliskovsky, V.V.; Malagon, F.; Baker, J.D.; Prince, J.S.; Klaus, J.S.; Adhya, S.L. Novel “Superspreader” Bacteriophages Promote Horizontal Gene Transfer by Transformation. mBio 2017, 8, e02115-16. [Google Scholar] [CrossRef]
- Fillol-Salom, A.; Rostøl, J.T.; Ojiogu, A.D.; Chen, J.; Douce, G.; Humphrey, S.; Penadés, J.R. Bacteriophages benefit from mobilizing pathogenicity islands encoding immune systems against competitors. Cell 2022, 185, 3248–3262.e20. [Google Scholar] [CrossRef]
- Braga, L.P.P.; Spor, A.; Kot, W.; Breuil, M.-C.; Hansen, L.H.; Setubal, J.C.; Philippot, L. Impact of phages on soil bacterial communities and nitrogen availability under different assembly scenarios. Microbiome 2020, 8, 52. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Serna, C.; Ares-Arroyo, M.; Matamoros, B.R.; Delgado-Blas, J.F.; Montero, N.; Bernabe-Balas, C.; Wedel, E.F.; Mendez, I.S.; Muniesa, M.; et al. Extensive antimicrobial resistance mobilization via multicopy plasmid encapsidation mediated by temperate phages. J. Antimicrob. Chemother. 2020, 75, 3173–3180. [Google Scholar] [CrossRef]
- Fillol-Salom, A.; Bacigalupe, R.; Humphrey, S.; Chiang, Y.N.; Chen, J.; Penadés, J.R. Lateral transduction is inherent to the life cycle of the archetypical Salmonella phage P22. Nat. Commun. 2021, 12, 6510. [Google Scholar] [CrossRef]
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penadés, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207–212. [Google Scholar] [CrossRef]
- González-Villalobos, E.; Balcázar, J.L. Does phage-mediated horizontal gene transfer represent an environmental risk? Trends Microbiol. 2022, 30, 1022–1024. [Google Scholar] [CrossRef]
- Kleiner, M.; Bushnell, B.; Sanderson, K.E.; Hooper, L.V.; Duerkop, B.A. Transductomics: Sequencing-based detection and analysis of transduced DNA in pure cultures and microbial communities. Microbiome 2020, 8, 158. [Google Scholar] [CrossRef]
- Lekunberri, I.; Subirats, J.; Borrego, C.M.; Balcázar, J.L. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 2017, 220, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.S.; Zhaxybayeva, O.; Beatty, J.T. Gene transfer agents: Phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012, 10, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Bárdy, P.; Füzik, T.; Hrebík, D.; Pantůček, R.; Thomas Beatty, J.; Plevka, P. Structure and mechanism of DNA delivery of a gene transfer agent. Nat. Commun. 2020, 11, 3034. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadamuro, R.D.; Elois, M.A.; Pilati, G.V.T.; Savi, B.P.; Pessi, L.; Jempierre, Y.F.S.H.; Rodríguez-Lázaro, D.; Fongaro, G. Role of Lysogenic Phages in the Dissemination of Antibiotic Resistance Genes Applied in the Food Chain. Foods 2025, 14, 1082. https://doi.org/10.3390/foods14071082
Cadamuro RD, Elois MA, Pilati GVT, Savi BP, Pessi L, Jempierre YFSH, Rodríguez-Lázaro D, Fongaro G. Role of Lysogenic Phages in the Dissemination of Antibiotic Resistance Genes Applied in the Food Chain. Foods. 2025; 14(7):1082. https://doi.org/10.3390/foods14071082
Chicago/Turabian StyleCadamuro, Rafael Dorighello, Mariana Alves Elois, Giulia Von Tönnemann Pilati, Beatriz Pereira Savi, Leonardo Pessi, Yasmin Ferreira Souza Hoffmann Jempierre, David Rodríguez-Lázaro, and Gislaine Fongaro. 2025. "Role of Lysogenic Phages in the Dissemination of Antibiotic Resistance Genes Applied in the Food Chain" Foods 14, no. 7: 1082. https://doi.org/10.3390/foods14071082
APA StyleCadamuro, R. D., Elois, M. A., Pilati, G. V. T., Savi, B. P., Pessi, L., Jempierre, Y. F. S. H., Rodríguez-Lázaro, D., & Fongaro, G. (2025). Role of Lysogenic Phages in the Dissemination of Antibiotic Resistance Genes Applied in the Food Chain. Foods, 14(7), 1082. https://doi.org/10.3390/foods14071082







