A Colorimetric LAMP Assay for Salmonella spp. Detection: Towards a DNA Extraction-Free Approach for Pathogen Screening
Abstract
1. Introduction
2. Materials and Methods
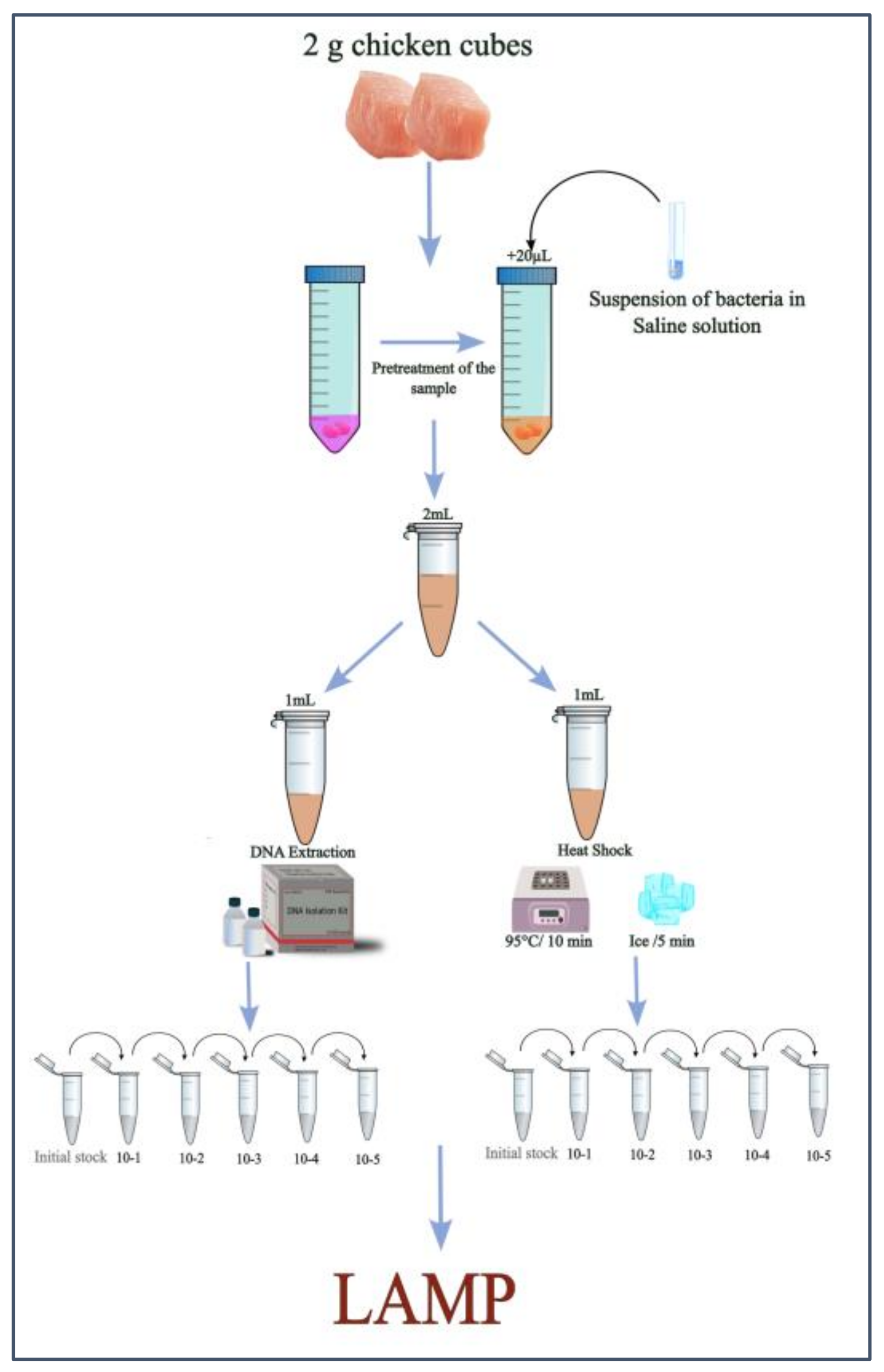
2.1. Experimental Design
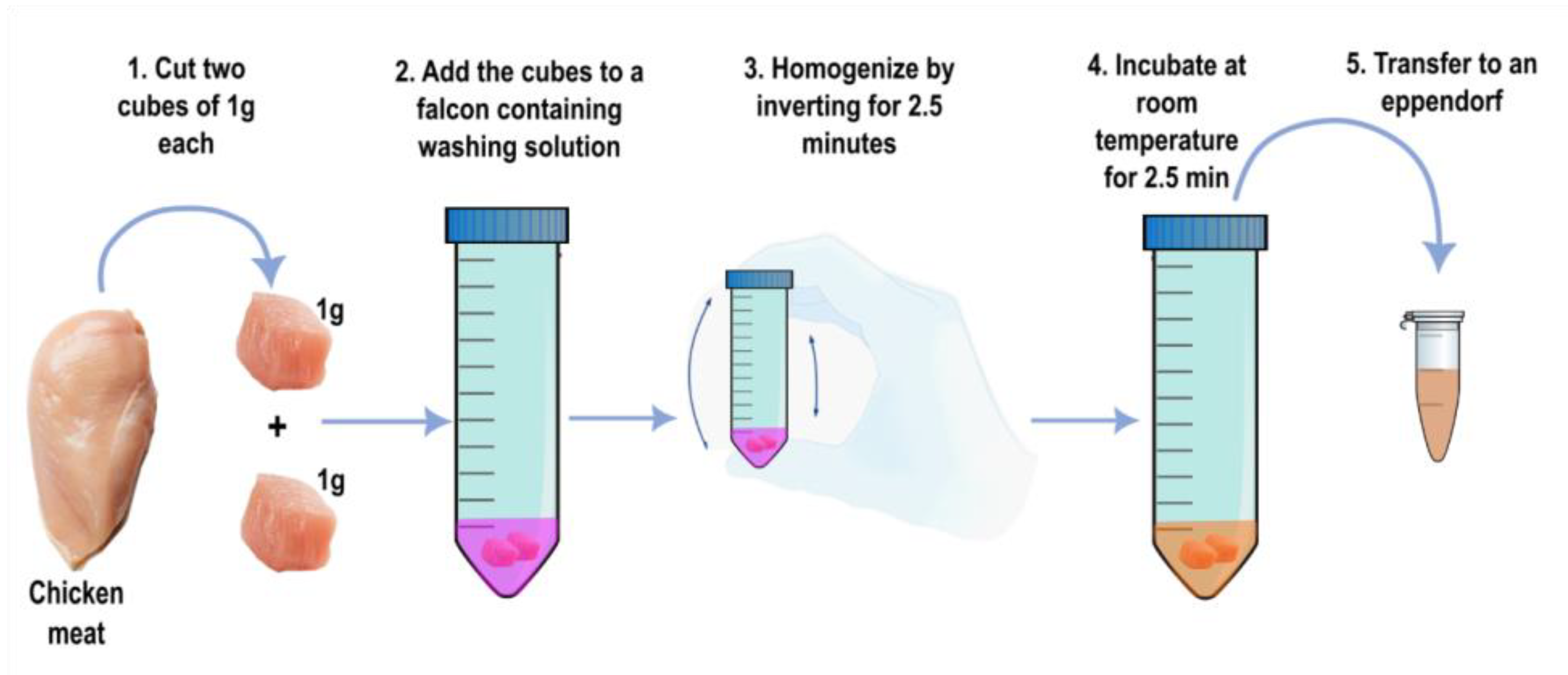
2.2. Pretreatment of Chicken Samples and Artificial Contamination
2.3. DNA Extraction
2.4. Heat-Treatment Protocol
2.5. cLAMP Assay
2.5.1. In-House Colorimetric Buffer Composition
2.5.2. Primer Design and Synthesis
2.5.3. cLAMP Sensitivity
2.5.4. cLAMP Specificity
2.5.5. cLAMP Reaction Mix
2.6. Amplification Confirmation
2.7. Real-Time PCR
3. Results
3.1. Pretreatment of Chicken Samples
3.2. Sensitivity of the Colorimetric LAMP Assay
3.3. Comparison with Real-Time PCR
3.4. cLAMP Specificity Results
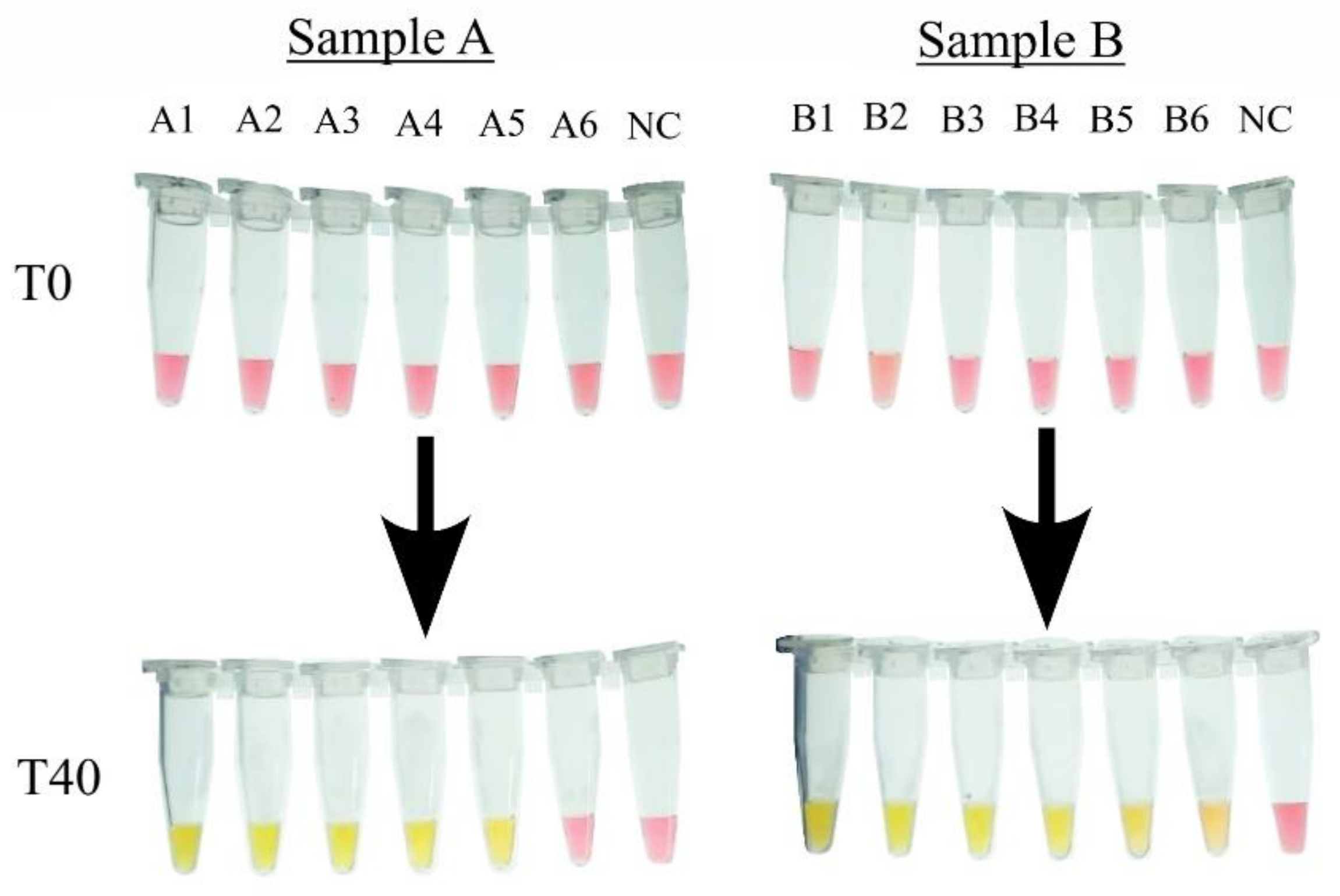
3.5. Application of the cLAMP with Artificially Contaminated Samples
3.5.1. cLAMP with Extracted DNA
3.5.2. cLAMP with Heat-Treatment Products
3.5.3. Confirmation of Results with Gel Electrophoresis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salmonella (Non-Typhoidal). Available online: https://www.who.int/news-room/fact-sheets/detail/Salmonella-(non-typhoidal) (accessed on 5 March 2024).
- Teklemariam, A.D.; Al-Hindi, R.R.; Albiheyri, R.S.; Alharbi, M.G.; Alghamdi, M.A.; Filimban, A.A.R.; Al Mutiri, A.S.; Al-Alyani, A.M.; Alseghayer, M.S.; Almaneea, A.M.; et al. Human Salmonellosis: A Continuous Global Threat in the Farm-to-Fork Food Safety Continuum. Foods 2023, 12, 1756. [Google Scholar] [CrossRef] [PubMed]
- ONSSA. Arrêté Conjoint du Ministre de l’Agriculture, de la Pêche Maritime, du Développement Rural et des Eaux et Forêts et du Ministre de la Santé n°293-19 du 9 Joumada II 1440 (15 Février 2019) Fixant la Liste et les Limites des Critères Microbiologiques Autorisées dans les Produits Primaires et les Produits Alimentaires; BOn°6796 du18/07/2019; ONSSA: Rabat, Morocco, 2019; p. 1686.
- ISO 6579-1:2017(Fr); Microbiologie de La Chaîne Alimentaire—Méthode Horizontale Pour La Recherche, Le Dénombrement et Le Sérotypage Des Salmonella—Partie 1: Recherche Des Salmonella Spp. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/obp/ui/fr/#iso:std:iso:6579:-1:ed-1:v1:fr (accessed on 7 March 2024).
- ISO 20837:2006(En); Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens—Requirements for Sample Preparation for Qualitative Detection. ISO: Geneva, Switzerland, 2006. Available online: https://www.iso.org/obp/ui/#iso:std:iso:20837:ed-1:v1:en (accessed on 7 March 2024).
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-Mediated Isothermal Amplification (LAMP): A Novel Rapid Detection Platform for Pathogens. Microb. Pathog. 2017, 107, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Treglia, I.; Ciccaglioni, G.; Ortoffi, M.F.; Gattuso, A. Application of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Detection of Listeria Monocytogenes in Cooked Ham. Foods 2023, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Yinur, D.; Moges, B.; Hassen, A.; Tessema, T.S. Loop Mediated Isothermal Amplification as a Molecular Diagnostic Assay: Application and Evaluation for Detection of Enterohaemorrhagic Escherichia Coli (O157:H7). Pract. Lab. Med. 2023, 37, e00333. [Google Scholar] [CrossRef] [PubMed]
- Zendrini, A.; Carta, V.; Filipello, V.; Ragni, L.; Cosciani-Cunico, E.; Arnaboldi, S.; Bertasi, B.; Franceschi, N.; Ajmone-Marsan, P.; De Medici, D.; et al. One-Day Molecular Detection of Salmonella and Campylobacter in Chicken Meat: A Pilot Study. Foods 2021, 10, 1132. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.T.; Layne, T.R.; O’Connell, K.C.; Tanner, N.A.; Landers, J.P. Comparative Evaluation and Quantitative Analysis of Loop-Mediated Isothermal Amplification Indicators. Anal. Chem. 2020, 92, 13343–13353. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Sano, S.; Takahashi, K.; Jikihara, T. Method for Colorimetric Detection of Double-Stranded Nucleic Acid Using Leuco Triphenylmethane Dyes. Anal. Biochem. 2015, 473, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Nzelu, C.O.; Gomez, E.A.; Cáceres, A.G.; Sakurai, T.; Martini-Robles, L.; Uezato, H.; Mimori, T.; Katakura, K.; Hashiguchi, Y.; Kato, H. Development of a Loop-Mediated Isothermal Amplification Method for Rapid Mass-Screening of Sand Flies for Leishmania Infection. Acta Trop. 2014, 132, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Visual Detection of Isothermal Nucleic Acid Amplification Using pH-Sensitive Dyes. BioTechniques 2015, 58, 59–68. [Google Scholar] [CrossRef]
- Bajaj, V.; Hwang, C.; Lee, C.A. hilA Is a Novel ompR/toxR Family Member That Activates the Expression of Salmonella Typhimurium Invasion Genes. Mol. Microbiol. 1995, 18, 715–727. [Google Scholar] [CrossRef]
- Chu, J.; Shin, J.; Kang, S.; Shin, S.; Chung, Y.-J. Rapid and Sensitive Detection of Salmonella Species Targeting the hilA Gene Using a Loop-Mediated Isothermal Amplification Assay. Genom. Inform. 2021, 19, e30. [Google Scholar] [CrossRef]
- Notomi, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Yang, Q.; Meyerson, N.R.; Clark, S.K.; Paige, C.L.; Fattor, W.T.; Gilchrist, A.R.; Barbachano-Guerrero, A.; Healy, B.G.; Worden-Sapper, E.R.; Wu, S.S.; et al. Saliva TwoStep for Rapid Detection of Asymptomatic SARS-CoV-2 Carriers. eLife 2021, 10, e65113. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yang, S.; Kim, H. Rapid Detection of Salmonella Enterica Subsp. Enterica Serovar Thompson by Real-Time Fluorescence Loop-Mediated Isothermal Amplification (LAMP) and Colorimetric LAMP Combined without DNA Extraction in the Field. LWT Food Sci. Technol. 2023, 182, 114850. [Google Scholar] [CrossRef]
- Wu, G.P.; Levin, R.E. Rapid and Sensitive Detection of Salmonella Enterica Ser. Enteritis Retrieved from Lettuce Using a Real-Time Loop-Mediated Amplification Isothermal Assay Without Enrichment. Food Biotechnol. 2015, 29, 263–275. [Google Scholar] [CrossRef]







| Primer | Sequence | Length (bp) |
|---|---|---|
| Forward outer (F3) | 5′-GCTACGCTCAGAAAAGAAAGTC-3′ | 22 |
| Backward outer (B3) | 5′-GTAACTTTGTGACATGCCTGTC-3′ | 22 |
| Forward inner (FIP) | 5′-GAATATGCCGTTCTGGTCATCC TTTGTGGAATGACCTGGTCCATACC-3′ | 47 |
| Backward inner (BIP) | 5′-ACAGCCTTCTATTTCTCGTAGCGC GACGCGGAAGTTAACGAAGA-3′ | 44 |
| Loop forward (LF) | 5′-CGGCCGCTCTAACACTCATT-3′ | 22 |
| Loop backward (LB) | 5′-CGCTGTATTTATGCCTTACGACG-3′ | 23 |
| NaoH Concentration | 6 mM | 7.5 mM | 10 mM | 12.5 mM | 13 mM | 15 mM |
|---|---|---|---|---|---|---|
| pH range | 6.4–5.7 | 6.8–6.2 | 7.5–6.9 | 7.7–7.2 | 8–7.5 | 8.3–7.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skenndri, S.; Nassik, S.; Lakhmi, R.; Anneggah, B.E.; Lahkak, F.E.; Moumen, A.; Abdellaoui Maane, I. A Colorimetric LAMP Assay for Salmonella spp. Detection: Towards a DNA Extraction-Free Approach for Pathogen Screening. Foods 2025, 14, 521. https://doi.org/10.3390/foods14030521
Skenndri S, Nassik S, Lakhmi R, Anneggah BE, Lahkak FE, Moumen A, Abdellaoui Maane I. A Colorimetric LAMP Assay for Salmonella spp. Detection: Towards a DNA Extraction-Free Approach for Pathogen Screening. Foods. 2025; 14(3):521. https://doi.org/10.3390/foods14030521
Chicago/Turabian StyleSkenndri, Safae, Saâdia Nassik, Rabab Lakhmi, Badr Eddine Anneggah, Fatima Ezzahra Lahkak, Abdeladim Moumen, and Imane Abdellaoui Maane. 2025. "A Colorimetric LAMP Assay for Salmonella spp. Detection: Towards a DNA Extraction-Free Approach for Pathogen Screening" Foods 14, no. 3: 521. https://doi.org/10.3390/foods14030521
APA StyleSkenndri, S., Nassik, S., Lakhmi, R., Anneggah, B. E., Lahkak, F. E., Moumen, A., & Abdellaoui Maane, I. (2025). A Colorimetric LAMP Assay for Salmonella spp. Detection: Towards a DNA Extraction-Free Approach for Pathogen Screening. Foods, 14(3), 521. https://doi.org/10.3390/foods14030521








