Abstract
Bacteria in the genus Vibrio, including at least 152 species, thrive in marine and estuarine environments and are frequently detected in aquatic products worldwide. Of these, 12 species have been implicated in human infectious diseases, such as the life-threatening pandemic cholera, acute gastroenteritis, and severe sepsis. Nevertheless, molecular mechanisms of their pathogenesis are not fully uncovered yet. Prophages are found prevalent in Vibrio spp. genomes, carrying a number of genes with various functions. In this review, we deciphered the evolutionary relationship between prophages and Vibrio species and highlighted the impact of prophages on the bacterial pathogenicity, environmental fitness, and genome evolution, based on 149 newly discovered intact prophages located in the genomes of 82 Vibrio spp., which we searched and collected from Web of Science Core Collection in the most recent 5 years. The effects of prophages on resistance to superinfection, strain competition, and their regulation were also discussed. This review underscored crucial roles of prophages in shaping Vibrio spp. genomes and their implications for food safety and public health.
1. Introduction
The genus Vibrio contains a large subgroup of bacteria in the family Vibrionaceae, the order Vibrionales, and the class Gammaproteobacteria. They thrive in marine and estuarine environments and are found in fish, corals, shrimps, plankton, and mammals [1,2]. For instance, most recently, Zeidler et al. [3] surveyed the Vibrio spp. prevalence in seafood samples (n = 306) including shrimp and mussel collected in Berlin, Germany from March 2023 to January 2024. Their results showed that Vibrio spp. were detected in 56% of the samples, among which Vibrio parahaemolyticus (58%) was the most prevalent species, followed by Vibrio alginolyticus (42%), Vibrio cholerae non-O1/non-O139 (25%), and Vibrio vulnificus (4%) [3]. These bacteria are generally able to grow on marine agar and selective thiosulfate-citrate-bile salt-sucrose (TCBS) agar [4]. The genus Vibrio consists of at least 152 species (https://lpsn.dsmz.de/genus/vibrio, accessed on 10 August 2024), of which 12 species have been linked to human infectious diseases, including V. alginolyticus, Vibrio cincinnatiensis, V. cholerae, Vibrio damsela, Vibrio fluvialis, Vibrio furnissii, Vibrio harveyi, Vibrio hollisae, Vibrio metschnikovii, Vibrio mimicus, V. parahaemolyticus, and V. vulnificus [5].
V. cholerae, V. parahaemolyticus, and V. vulnificus are the most prominent species causing infectious diseases in humans, followed by V. alginolyticus [6]. Of these, V. cholerae can cause cholera, being the most terrifying pandemic diarrhea disease worldwide, usually transmitted via contaminated food or water [7]. Since 1817, seven cholera pandemics have been recorded, among which the first six were caused by V. cholerae of the classical biotype, and the seventh, caused by the El Tor biotype, has been ongoing since 1961 [8]. Approximately 1.3 to 1.4 million people in the world are infected with cholera each year, causing 21,000 to 143,000 people to die (www.cdc.gov/cholera/index.html, accessed on 21 September 2024). Most recently, the 2022–2023 cholera outbreaks led to 59,156 cases and 1771 deaths in Malawi [9]. Recently, Chen et al. [10] were the first to report experimental evidence for potentially toxic V. cholerae in snails. They isolated V. cholerae strains (n = 203) from 36 species of aquatic food animals, nearly two-thirds of which had not been tested before. Although no isolate was found to carry cholera toxin genes ctxAB (0.0%), the isolates were found to be positive for virulence genes tcpA (0.98%), ace (0.5%), and zot (0.5%), which derived from Cipangopaludina chinensis. High percentages of virulence-associated genes were also noted, including hapA (73.4%), rtxCABD (68.0–41.9%), tlh (54.2%), and hlyA (37.9%) [10]. Most recently, Yan et al. [11] demonstrated that different secretomes and proteomes of V. cholerae isolates were triggered by the matrices of eight species of edible aquatic animals, which facilitated the bacterial resistance in the aquatic animals and increased its pathogenicity to the host [11].
Vibriosis is a group of clinical syndromes associated with non-cholera Vibrio spp., such as gastroenteritis, septicemia, and invasive skin and soft tissue infections [5]. For instance, V. parahaemolyticus is a top seafood-borne pathogen in the world and can cause acute gastroenteritis, septicemia, and even death [12]. Increased V. parahaemolyticus contamination has been reported in processed, ready-to-eat foods, implying that this bacterium can survive in the harsh circumstance of food processing [13]. For example, most recently, Wu et al. [14] collected 154 crayfish (Procambarus clarkii) and environmental samples from crayfish farms and wholesale and retail markets from May to September 2024 in Jiangxi Province, China. They found that V. parahaemolyticus was detected in 66% of the crayfish production–sale chain, and in 92% of the market samples. Crayfish tanks contributed to the highest contamination percentage. The crayfish surface, which could be adsorbed by 90% of the V. parahaemolyticus cells in 6 h, was more susceptible to bacterial contamination than the crayfish intestine [14]. V. vulnificus can infect open wounds through contact with saltwater, brackish water, or raw seafood, causing sepsis and cellulitis [15,16]. It occurs in high numbers in seafood such as marine fish, shrimps, oysters, and crabs around the world, particularly in warmer months [17]. V. alginolyticus is the etiologic agent of severe soft tissue infections, sepsis, and other extraintestinal infections [18]. For instance, Abdelsalam et al. [19] reported a disease outbreak featured by caligid copepod infestations and subsequent V. alginolyticus infections in European seabass (Dicentrarchus labrax) and flathead grey mullet (Mugil cephalus) grown at a private facility in Deeba Triangle, Egypt. Moreover, the V. alginolyticus isolates showed resistance to β-lactams, aminoglycosides, and trimethoprim-sulfamethoxazole [19]. Recently, Sun et al. [20] collected 128 V. alginolyticus isolates across China in 2020, which were isolated from seafood (n = 75), freshwater products (n = 51), and other samples (n = 2). They found that 95.31% of the V. alginolyticus isolates were resistant to at least one antibiotic category. Fifteen virulence genes were present in V. alginolyticus isolates (n = 57), such as the type III secretion system (T3SS) translocated factors-related genes vopD, vopB, and vcrH (54.4%, 31/57), T3SS-regulated gene tyeA (54.39%), and T3SS-inner rod protein and needle protein genes vscI and vscF (50.88%) [20]. The results highlighted the prevalence of V. alginolyticus in both freshwater and seafood products. It is estimated that 80,000 cases of human vibriosis occur in the United States each year (www.cdc.gov/vibrio/index.html, accessed on 21 September 2024). The rising incidence of vibriosis is being reported globally. Most recently, Morgado et al. [21] analyzed vibriosis case data for Maryland (2006–2019; n = 611), USA, obtained from the Cholera and Other Vibrio Illness Surveillance (COVIS) system managed by the Centers for Disease Control and Prevention (CDC). They found a 39% (p = 0.01) increase in the average annual incidence rate (per 100,000 population) of vibriosis, comparing the 2006–2012 and 2013–2019 periods. V. vulnificus infections showed the highest percentage increase (53%, p = 0.01), followed by V. parahaemolyticus (47%, p = 0.05). The long-term increases in Vibrio infections are happening in Maryland, together with enhanced rates of hospitalization and average hospital durations [21]. Thus, a better understanding of the pathogenetic underpinnings of these Vibrio spp. should have profound implications for the effective control and prevention of cholera and vibriosis.
Phages, being the most abundant biological entities in the biosphere, are viruses that infect bacteria [22]. After infecting bacterial hosts, virulent phages proliferate in large numbers, lyse host cells, and release progeny phage particles. The life cycle of temperate phages includes a lysogenic conversion in which phages insert into the host chromosomes and become prophages [23]. Studies have indicated that prophages vary in bacterial genomes and can endow their hosts with new biological functions through their carried genes [24,25,26]. Most recently, Steensen et al. [27] reported that tailless and filamentous prophages are very prevalent in marine Vibrio, and proposed that these prophages are active and highly rich drivers of host ecology and evolution, like the well-known tailed ones [27]. Virulence genes in prophages can be transmitted at high frequencies by horizontal gene transfer (HGT), resulting in the emergence of pandemic, epidemic, or pathogenic clones of Vibrio spp., thus posing a high risk to food safety and public health [28,29,30].
To the best of our knowledge, complete genome sequences of 557 phages hosted by Vibrio spp. have been deposited in the Viral Genome Resource at National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/labs/virus/vssi, accessed on 27 August 2024). By searching the Web of Science Core Collection database, we collected 149 intact prophage gene clusters identified in the genomes of 82 Vibrio isolates in the most recent 5 years (Table S1). In this review, we focused on these newly discovered prophages in Vibrio spp., specifically, their impact on the pathogenicity, environmental fitness, and genome evolution of Vibrio spp.
2. Materials and Methods
2.1. Data Source and Searches
In this review, the Web of Science Core Collection database was used to collect the literature. The search terms used were as follows: “Vibrio” and “prophage”. The publications were retrieved from 1 January 2020 to 19 September 2024. The inclusion criteria of the review were met if articles addressed the Vibrio spp., prophages, genome features, resistance, virulence, transmission, evolution, or regulation. Moreover, the studies’ origin, sample size, strain origin, strain identification, sequencing technique, and number of prophages were also assessed. Furthermore, the information in the titles and abstracts was assessed, and the full texts were examined. The exclusion criteria were as follows: studies conducted on non-named strains; research objectives that focused on Vibrio genomes without prophages; studies aimed at the application of phages but that did not have any of the above results; and publications that were not in English. The searches yielded a total of 50 publications, of which 42 articles (Table S2) were chosen and used in this review.
2.2. The Collected Prophage Gene Clusters Identified in the Vibrio spp. Genomes
Based on the retrieved articles, we collected 149 intact prophage gene clusters identified in the genomes of 82 Vibrio isolates, which belonged to seven Vibrio species, including V. alginolyticus, Vibrio campbellii, V. cholerae, Vibrio gazogenes, Vibrio nigripulchritudo, V. parahaemolyticus, and Vibrio penaeicida (Table S1). The relationships of these prophages were plotted and visualized using the SRplot software [31].
3. Results and Discussion
3.1. Prophages Contribute to the Virulence of Vibrio spp.
A well-known mechanism of virulence acquisition in Vibrio is the integration of CTXΦ phage (10,638 bp, GenBank accession no. NC_015209) into V. cholerae [32]. CTXΦ, which commonly exists in the 7th pandemic serogroup O1 and toxigenic serogroup O139 strains, encodes cholera toxin (CT) and toxin-coregulated pilus (TCP) [33]. The CT can lead to severe disruption of intestinal cell function, the watery diarrhea of cholera [34], while TCP is the receptor for entry of CTXΦ into the cell [35]. Another toxin encoded by CTXΦ and found in Vibrio prophages is zonula occludens toxin (Zot), the C-terminal domain of which displays enterotoxic activity [36]. This toxin increases the permeability of the surface of marine plankton or the host intestinal cell [37]. Recently, Castillo et al. reported that 28 of 64 Vibrio species harbored prophages that encoded the Zot toxin [38]. Accessory cholera enterotoxin (Ace) is another toxin found in V. cholerae lysogen that is also responsible for diarrhea in cholera [34]. Most recently, a number of zot-like and ace-like genes were identified in Vibrio phages, such as the VSK (6882 bp, GenBank accession no. NC_003327) in V. cholerae, f237 (8784 bp, GenBank accession no. AP000581) and VF33 (7965 bp, GenBank accession no. NC_005948) in V. parahaemolyticus, and VALGΦ6 (8529 bp, GenBank accession no. MN719123) in V. alginolyticus [39,40,41], which could be biological reservoirs for the emergence of pathogenic clones of Vibrio spp.
Phages can escape degradation in the host cell by methylating their own DNA using DNA adenine methyltransferase (DAM) [42]. Therefore, DAM is considered to be one of the important enzymes related to Vibrio virulence. Previous research has indicated that DAM likely contributed to the switching between lytic and lysogenic life cycles of the V. harveyi Myoviridae-like (VHML) phage (43,198 bp, GenBank accession no. NC_004456) by the methylation and subsequent inhibition of the lytic repressor gene CI, similarly to the rha antirepressor gene of Escherichia coli phage Φ80 (46,150 bp, GenBank accession no. NC_021190) [42]. Some phages are involved in Vibrio virulence by up-regulating key virulence-associated genes. For example, the VHML phage could up-regulate hemolysin excretion in V. harveyi 642 [43]. Interestingly, V. harveyi strains infected with the VHML phage needed fewer nutrients than the uninfected ones, which may help the lysogenized bacteria to be more competitive in nutrient-limited circumstance [43,44]. Recently, Santoriello et al. reported that a type VI secretion system (T6SS) gene cluster Aux3 existed as a mobile and prophage-like element in some environmental V. cholerae, excised from and inserted into the host chromosomes via site-specific recombination, but a stable truncated form existed in pandemic strains in that the recombination is reduced, indicating that pandemic V. cholerae is inactive in site-specific recombination to retain an interbacterial defense mechanism [45,46]. Most recently, Xu et al. [47] reported a prophage gene cluster in V. parahaemolyticus N8-42 genome, which showed sequence similarity to the Vibrio_phage_fs2 (8651 bp, NCBI accession number: NC_001956) and encoded type II secretion system (T2SS) and T3SS family protein (GspD) [47]. GspD constructs the outer membrane channel of T2SS, which secretes diverse toxins causing severe diarrhea and cholera [48].
Some prophages are pathogenic to aquatic animal hosts. For instance, the prophage VfO3K6 (8784 bp, GenBank accession no. NC_002362) in V. parahaemolyticus SAB6 can cause acute hepatopancreatic necrosis disease (AHPND) in shrimps [49]. Khemayan et al. [50] found that the open read frame (ORF) 058 of a virulence-enhancing siphophage VHS1 (81,509 bp, GenBank accession no. JF713456) in V. harveyi likely encoded a toxin. The virulence of V. harveyi for Penaeus monodon was enhanced by more than 100 fold when it was lysogenized with VHS1 [50]. Recently, Wang et al. [51] isolated 33 pathogenic Vibrio spp. strains from aquaculture-grown Penaeus vannamei in Guangdong and Jiangsu Provinces, China from 2019 to 2022. They identified 108 prophages in the genomes of these Vibrio isolates, including intact prophages (n = 40), questionable prophages (n = 11), and incomplete prophages (n = 57), via the online software PHASTER (https://phaster.ca/) to score an identified DNA sequence region on the basis of the coding sequence count and the presence or absence of phage-associated genes. If its score was above 90, between 70 and 90, or below 70, then the predicted prophage was defined as intact, questionable, or incomplete, respectively. Two of these prophages carried virulence genes, e.g., the T6SS virulence genes in the questionable prophage of V. parahaemolyticus A1, and virulence genes wbfV/wcvB and wecA in the incomplete prophage of V. alginolyticus B14 [51]. Most recently, Mesa et al. [52] reported that V. harveyi PH1009, isolated from Masbate Island, Philippines, was co-infected with the white spot syndrome virus in P. monodon. Two prophage regions were found in the V. harveyi PH1009 genome, one of which carried the zot and ace genes.
3.2. Prophages Amplify the Ecological Persistence of Vibrio spp.
Recently, Chen’s research group reported a prophage-like gene cluster in V. parahaemolyticus CHN25, which showed high sequence similarity to the Vibrio phage martha 12B12 (33,277 bp, GenBank accession no. NC_021070). Twenty-four genes were predicted, encoding phage proteins (n = 7, e.g., phage head, tail, and baseplate), regulators (n = 8), and hypothetical proteins (n = 9). They demonstrated that the unknown genes VpaChn25_0724, VpaChn25_0734, VpaChn25_0713, VpaChn25_0714, and VpaChn25_RS25055 play essential roles in the environmental adaptability of the host [53,54,55]. For example, biofilm formation and cytotoxicity were significantly reduced in the ΔVpaChn25_0724, ΔVpaChn25_0734, ΔVpaChn25_0713, ΔVpaChn25_0714, and ΔVpaChn25_RS25055 mutants, as compared to the wild type V. parahaemolyticus CHN25 (p < 0.05). Remarkably, knock out of the VpaChn25_0724 gene caused cell membrane damage and increased cell surface hydrophobicity of V. parahaemolyticus CHN25 [53]. The deletion of VpaChn25_0734 and VpaChn25_RS25055 genes decreased cell surface hydrophobicity of V. parahaemolyticus CHN25; the deletion of the VpaChn25_0713 gene led to a reduction in internal membrane permeability of V. parahaemolyticus CHN25; and the deletion of VpaChn25_RS25055, VpaChn25_0713, and VpaChn25_0714 genes resulted in higher cell membrane fluidity of V. parahaemolyticus CHN25 [54,55]. Comparative secretomic analysis uncovered a significant increase in extracellular proteins, e.g., OmpW, FlaB/D, and FlaA flagellins, aldehyde-alcohol dehydrogenase (AdhE), phage head morphogenesis protein, and D-lactate dehydrogenase (D-LDH), in the ΔVpaChn25_0724 mutant, as compared to the wild-type strain (p < 0.05). Comparative transcriptomic analysis revealed 12 significantly changed metabolic pathways in the ΔVpaChn25_0724 mutant, indicating the shunt transport and carbon source usage, and inhibited energy production and membrane biosynthesis (p < 0.05). Several notably repressed key regulators in bacterial gene regulatory networks were closely related to phenotypic variations [53]. In the VpaChn25_0734 gene deletion mutant, the secretion of 30S ribosomal protein S1 (RpsA) and DNA-directed RNA polymerase subunit alpha (RpoA) was significantly increased (p < 0.05). Comparative transcriptomic analyses also revealed 13 significantly altered metabolic pathways in the ΔVpaChn25_0734 mutant, including the repressed carbon source transport and utilization, biofilm formation, and T2SS, linked to the defective phenotypes [54]. Additionally, transcriptomic analysis also revealed 15, 14, and 8 significantly altered metabolic pathways in the absence of VpaChn25_RS25055, VpaChn25_0713, and VpaChn25_0714 genes, respectively (p < 0.05) [55]. Most recently, Zhao et al. found that the protein encoded by VpaChn25_RS25055 existed at both poles of the V. parahaemolyticus CHN25 cell [55].
Recently, Li et al. [56] also reported an intact prophage-like element Vaf1 (10,004 bp, GenBank accession no. OP297622) in the genome of the emerging marine pathogen V. alginolyticus AP-1. Vaf1 significantly promoted biofilm formation, swarming motility, and contact-dependent competition of V. alginolyticus AP-1 [56]. The in vivo zebrafish (Danio rerio) infection mode experiment evidenced that Vaf1 contributed to the virulence of V. alginolyticus AP-1 [56]. Most recently, Chen’s research group also reported five prophage regions in V. parahaemolyticus N1-22, N4-46, N8-42, and Q8-15 genomes [47]. For instance, the V. parahaemolyticus N8-42 genome carried two prophage regions with sequence identity to the Vibrio_phage_K139 (33,106 bp, GenBank accession no. NC_003313) and Vibrio_phage_fs2 (8651 bp, GenBank accession no. NC_001956), respectively. The Vibrio_phage_K139 homologue also existed in V. parahaemolyticus N1-22. It contained 46 genes, encoding phage major structure genes that were the same as those in V. parahaemolyticus N8-42, as well as several different accessory genes, implying more recent HGT and genome recombination amongst the V. parahaemolyticus isolates. Moreover, the Pseudomonas_phage_D3 (56,426 bp, GenBank accession no. NC_002484) homologues were found in V. parahaemolyticus N4-46 and Q8-15 genomes, carrying 28 and 29 genes, respectively, indicating possible HGT across different genera of Pseudomonas and Vibrio [47].
Most recently, Qin et al. [57] also reported six prophage gene clusters that contained 248 predicted genes in V. cholerae L1-1, L10-48, and B5-86 genomes, 53.6% of which encoded unknown proteins. For instance, V. cholerae L1-1 carried three prophage regions, which displayed sequence identity to the Escherichia_phage_lys12581Vzw (62,668 bp, GenBank accession no. NC_049917), Vibrio_phage_VHML, and Vibrio_phage_VCY_phi (7103 bp, GenBank accession no. NC_016162), respectively. V. cholerae L10-48 had two prophage regions, with sequence identity to the Burkholderia_cenocepacia_phage_BcepMu (36,748 bp, GenBank accession no. NC_005882) and Escherichia_phage_ArgO145 (62,020 bp, GenBank accession no. NC_049918), respectively. One Escherichia_converting_phage_Stx2a_F451 (64,900 bp, GenBank accession no. NC_049924) homologue existed in V. cholerae B5-86. Notably, the six identified prophages in V. cholerae originated from the distinct genera Burkholderia cenocepacia, Escherichia spp., and Vibrio spp., suggesting that the phage transmission likely occurred across these genera boundaries [57].
Recently, Wang et al. [58] investigated culturable bacteria in the gastric cavity of healthy Galaxea fascicularis, a scleractinian coral prevalent in coral reef areas of the Indo-Pacific Ocean. They reported that temperate phages are key players in the colonization competition in the coral microbiota. Remarkably, Vibrio coralliilyticus outcompeted other coral symbiotic bacteria (e.g., Endozoicomonas spp.), via the LodAB-dependent prophage induction, to obtain a competitive advantage over lysogenic competitors when colonizing corals [58]. Coral mucus also serves as a reservoir of bacteriophages targeting Vibrio pathogens [59].
3.3. Prophages Assist the Superinfection Exclusion in Vibrio spp.
Superinfection exclusion is the ability of bacterial strains carrying temperate phages to resist subsequent infection by the same phage [40]. Superinfection exclusion in the Φ80 in E. coli K-12 and HK97 (39,732 bp, GenBank accession no. NC_002167) in E. coli 594 have been reported [60,61]. However, the underlying mechanism has not been unveiled yet. Kalatzis et al. [62] reported that the H20 (53,224 bp, GenBank accession no. KY658675)-like phage in Vibrio anguillarum A023, T265, BA35, and VaKef strains harbored a repressor gene that genetically and structurally resembled a repressor of the Escherichia_phage_λ (48,502 bp, GenBank accession no. NC_001416). It possibly blocked the gene expression of the lytic pathway and conferred repressor-mediated immunity to other H20-like phages in V. anguillarum [62]. Additionally, it has also been reported that once the VHS1 phage initiated the infection, V. harveyi inhibited and prevented the VHS1 binding receptor from adhering to another phage particle [63].
Pseudolysogens are bacteria that are infected with pseudolysogenic phages that can not became a stable lysogen, e.g., the 493 phage (9300 bp) in V. cholerae and the VHS1 in V. harveyi. Pseudolysogens are classified into two groups: (1) those that contain a defective phage genome and can resist superinfection but do not produce phage particles; (2) those that tolerate superinfection but do not contain the phage genome [40,64]. Recently, a new pre-CTXΦ prophage was found in V. cholera of serogroup O139. The immunity to CTXΦ superinfection triggered by the rstR allele of this pre-CTXΦ is phage-specific [65]. Notably, recently, Pant et al. [66] found that the core genome-encoded RecA helped CTXΦ to bypass V. cholera immunity to replicate in the host, as a similar prophage existed in the host chromosomes [66].
3.4. Prophages Promote the Genome Evolution of Vibrio spp.
As shown in Figure 1, the collected 149 intact prophage gene clusters carrying a number of genes were present in the genomes of 82 Vibrio isolates belonging to seven Vibrio species. They showed sequence similarity to 60 different phages, promoting the genome evolution of Vibrio spp. For instance, the Vibrio_phage_henriette_12B8 (107,218 bp, GenBank accession no. NC_021073) was found in V. alginolyticus and V. parahaemolyticus genomes; the Escherichia_phage_ArgO145 (62,020 bp, GenBank accession no. NC_049918) in V. cholerae and V. penaeicida genomes; the Vibrio_phage_VP882 (38,197 bp, GenBank accession no. NC_009016) in V. gazogenes and V. parahaemolyticus genomes; and the Enterobacteria_phage_mEp235 (37,595 bp, GenBank accession no. NC_019708) in V. nigripulchritudo and V. penaeicida genomes (Figure 1).
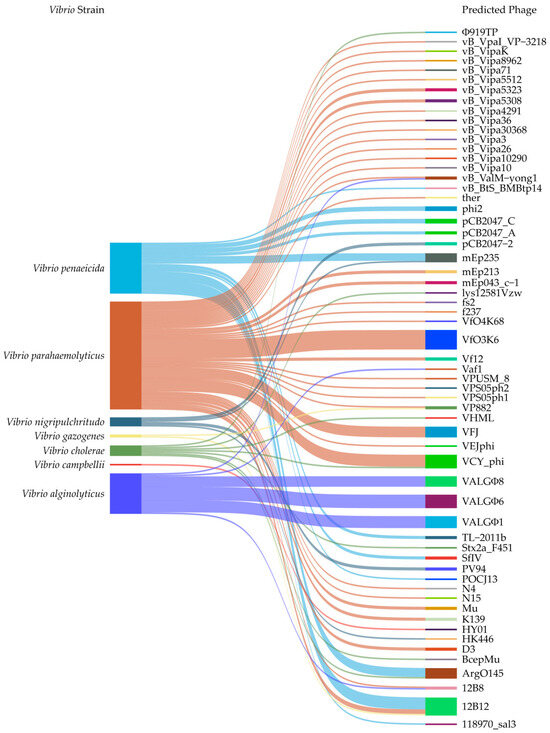
Figure 1.
Sankey diagram showing the relationship of the collected 149 predicted prophage gene clusters (Table S1) identified in genomes of 82 Vibrio isolates in the most recent 5 years. The diagram was plotted and visualized using the SRplot online platform (https://www.bioinformatics.com.cn/srplot, accessed on 15 December 2024).
Studies have evidenced Vibrio spp. undergoing prophage-mediated evolution [67]. For instance, V. cholerae of El Tor biotype can replicate the CTXΦ genome and secrete CTXΦ phage particles. Due to space constraints, readers are referred to reviews on the gene organization of CTXΦ [67]. Recently, Ochi et al. [68] analyzed complete genomes of five clinical V. cholerae strains isolated in Kolkata, India during the period from 2007 to 2011. They found that recent strains had a changed CTXΦ array and could not replicate the CTXΦ genome. Further analysis revealed the identical rstA and intergenic sequence 1 (Ig-1) in all the recent isolates in the altered CTXΦ array [68]. Diarrheal cases caused by non-toxigenic V. cholerae have been reported globally [69]. Lineages L3b and L9, featured as ctxAB-negative and tcpA-positive, pose the most dangerous risk and caused long-term epidemics in different regions in the world [70]. Two waves (2001–2012 and 2013–2018) of cholera were reported in Hangzhou, China, between 2001 and 2018 and were caused by non-toxigenic V. cholerae [71]. Hao et al. [71] analyzed 207 genomes of Hangzhou isolates from these two waves together with 1573 publicly available genomes and found that 21% of L3b and L9 isolates had become CT producers, indicating that the gain of complete CTXΦ-carrying ctxAB genes led to the transition [71]. Recently, Behera et al. [72] reported ctxB, tcpA, and rstR genes in CTX prophages among V. cholerae isolates of serotype O139 (n = 59) in Odisha, India from 1999 to 2017 and found three genotypes, 1, 3, and 4, with at least one copy of CTXCalcΦ in addition to CTXET and CTXCl in the V. cholerae isolates [72]. Additionally, Thong et al. [73] investigated V. cholerae strains of the El Tor biotype (n = 45) related to outbreaks and sporadic cases in Malaysia between 1991 and 2011. They found that the outbreak strains isolated in 1991 carried the El Tor CT gene (ctxB3), whereas sporadic strains from 2004 to 2011 contained the classical ctxB1 gene. Four different CTXΦ arrays existed in the El Tor variants, one of which co-existed with El Tor strains in the 2009 outbreak in Terengganu, Malaysia [73].
The epidemiology of V. parahaemolyticus remarkably changed in America following the Pacific-native lineage called sequence type (ST) 36 in the Atlantic [74]. Most recently, Foxall et al. reported that the major genomic differentiation and competitive success of ST36 included the loss of an inovirus prophage that had been maintained for decades in the endemic north Pacific population [41]. Broader surveys indicated that inoviruses are common and active among the global population of V. parahaemolyticus, and inovirus replacements are infrequent, such as in ST36; however, it is worth noting that they exist in pathogenic lineages that dispersed [41].
Studies have shown that HGT events between different Vibrio spp. (e.g., Vibrio fischeri and V. cholerae) were promoted by mobile genetic elements (MGEs). For example, a CTXΦ phage-like gene cluster was identified in V. fischeri ES114 genome, leading to the virulence genes in CTXΦ from V. cholerae transferred to V. mimicus [75]. Wang et al. [76] identified the two prophages VPS05ph1 (33,915 bp) and VPS05ph2 (39,156 bp) in V. parahaemolyticus S05, showing 38.3% sequence similarity to each other. The majority of the predicted genes (18/24) in VPS05ph1 were similar to the Vibrio phage VPUSM 8 (34,145 bp, GenBank accession no. NC_022747), whereas most genes (19/32) in VPS05ph2 were similar to the Aeromonas phage phiO18P (33,985 bp, GenBank accession no. NC_009542) but showing sequence similarity of only 18.7% [76]. Most recently, Soto et al. [77] searched 4619 Vibrio genomes from 127 species for prophages carrying the zot gene. They found 2030 potential prophages with zot-like genes in 43 Vibrio species. Some prophages, such as CTXΦ or Vf33, were associated with specific species, whereas prophages VCY_phi and VfO3K6 were found in 28 and 20 Vibrio species, respectively [77]. V. campbellii is an emerging pathogen in aquaculture that can cause luminescent vibriosis in farmed shrimp. Nuidate et al. [78] determined the genome sequence of prophage HY01 (41,772 bp, GenBank accession no. MT366580) in V. campbellii, which was distantly associated with the Vibrio phage Va_PF430-3_p42 (51,907 bp, GenBank accession no. MK672805) in V. anguillarum. A total of 60 ORFs of HY01 were predicted, of which 31 had known biological functions. Sequence analysis of clustered regularly interspaced short palindromic repeats (CRISPR) spacers showed two matching sequences between the HY01 genome and viral spacer sequence of Vibrio spp. [78].
The lysogenic phage CTXΦ harbors the precursor genome without the ctxAB, named pre-CTXΦ. Multiple types of the pre-CTXΦ have been found in toxigenic and non-toxigenic V. cholerae strains, based on the transcriptional regulator gene rstR alleles in CTXΦ/pre-CTXΦ [79]. A pre-CTXΦ genome type carrying a new rstR allele, pre-CTXZHJΦ, was identified. This pre-CTXZHJΦ inserted in the small chromosome of V. cholerae and coexisted with a typical CTXETΦ prophage in the large chromosome. RstRZHJ was able to bind to the intergenic sequence 2 (Ig-2) in the rstAB promotor in the pre-CTXZHJΦ genome and inhibited its own rstAB gene expression but did not inhibit this gene in CTXETΦ and CTXclassΦ. These results suggested that V. cholerae isolates harboring the pre-CTXZHJΦ cannot escape from the infection of CTXΦs, thus potentially converting into toxigenic strains [65].
Recently, Garin-Fernandez et al. reported the genome structure of the inducible phage vB_VpaI_VP-3218 (11,082 bp, GenBank accession no. LR595856), a novel filamentous phage in V. parahaemolyticus VN-3218 isolated from the North Sea [80]. This phage can integrate into the host chromosome along with other Vibrio host genomes from the environment. These results suggested that a number of prophage-mediated HGT events occurred during the evolution of Vibrio spp.
3.5. Regulation of Prophages in Vibrio spp.
Studies have shown that CI repressor concentration and cell density regulate prophage induction in V. cholerae and V. anguillarum, respectively [81,82,83,84]. For instance, the phage Φ919TP (33,133 bp, GenBank accession no. KU504502) in V. cholerae was a λ-like phage, harboring a CI repressor that governed the lysis/lysogeny switch in the host cell [81,82]. When the CI repressor concentration was high, it repressed the development of the lytic life cycle of the Φ919TP [82]. Recently, Li et al. [83] analyzed a Φ919TP-deleted variant of V. cholerae and its interaction with a modified lytic variant of the induced prophage (Φ919TP cI−). Comparison of wild-type and Φ919TP cI−-resistant mutant genomes revealed that the Φ919TP cI− selected for phage-resistant mutants in the key steps of lipopolysaccharide (LPS) O-antigen biosynthesis were targeted, resulting in a single-base-pair deletion in the gmd gene. The gmd-mediated O-antigen defect led to pleiotropic phenotypes, e.g., cell auto-aggregation and decreased swarming motility [83]. Tan et al. [84] investigated the effect of quorum sensing (QS) on the interactions between V. anguillarum 90-11-287 and its H20-like prophage p41 (54,432 bp, GenBank accession no. MK672799). They found that the induction of the H20-like prophage was governed by the QS state of the host, and phage particles increased by eight fold per cell in high-cell-density (HCD) cultures of the QS-deficient ΔvanT mutant. Compared to prophage-free strain, H20-like prophage enhanced biofilm formation in low-cell-density (LCD) cultures. Conversely, the HCD state was linked to the decreased prophage induction, the enhanced proteolytic activity, and the inhibited biofilm formation. The intertwined regulation of phage-host interactions and biofilm formation suggested that the increased lysogeny at HCD was not only a successful strategy for phages to utilize in bacterial hosts, but also was a host strategy evolved to govern the lysis–lysogeny switch to benefit the host survival [84,85,86].
Additionally, Xu et al. [87] recently evaluated the potential application of prophages in identifying Vibrio species and strains. They found that prophages in Vibrio strains were highly specific at the strain and species level, implying that prophages could be potentially applied in microbial species, sub-species, and strain-level identification [87].
4. Conclusions
The genus Vibrio includes at least 152 species, 12 of which have been implicated in human severe cholera and vibriosis diseases. These bacteria are frequently detected in aquatic products worldwide, posing a huge danger to food safety and public health. In this review, we searched and collected 149 newly discovered intact prophages located in the genomes of 82 Vibrio spp. in the most recent 5 years. Based on these data, we deciphered the evolutionary relationship between prophages and Vibrio species and highlighted the impact of prophages on the bacterial pathogenicity, environmental fitness, and genome evolution.
Prophages serve as biological reservoirs for the emergence of pandemic, epidemic, or pathogenic clones of Vibrio spp., particularly through the transmission of toxin-encoding genes (e.g., ct, tcp, zot, and ace, as well as T6SS, T3SS, and T2SS-related virulence factors) across genera or species boundaries. However, molecular mechanisms underlying the transmission need to be further investigated. Recent studies also revealed that some prophages (e.g., VHML) carried genes encoding enzymes (e.g., DAM) switching between lytic and lysogenic life cycles to avoid degradation in the host cell; some contributed to Vibrio virulence by up-regulating key virulence-associated genes; and some were pathogenic to aquatic animal hosts as well.
Prophages amplify the ecological persistence of Vibrio spp. Recently, the prophages identified in Vibrio spp. genomes were found to carry a number of genes, endowing the bacteria with additional biological functions (e.g., resistance, competition, and substance metabolism) besides virulence. Notably, in V. parahaemolyticus, some prophage-related genes (e.g., VpaChn25_0724, VpaChn25_0734, VpaChn25_0713, VpaChn25_0714, and VpaChn25_RS25055) have been demonstrated to play essential roles in the bacterial biofilm formation, cell structure integrity, low-temperature survival, and cytotoxicity to the host. In V. alginolyticus, the presence of Vaf1 has been evidenced to significantly increase bacterial biofilm formation, swarming motility, and contact-dependent competition. In V. coralliilyticus, LodAB-dependent prophage induction contributed to a bacterial competitive advantage over lysogenic competitors when colonizing corals.
Prophages promote the genome evolution of Vibrio spp. The collected 149 intact prophages carrying a number of genes were present in the genomes of 82 Vibrio isolates belonging to seven Vibrio species. They showed sequence similarity to 60 different phages, substantially promoting the genome evolution of Vibrio spp. Remarkably, recent studies have evidenced Vibrio spp. undergoing prophage-mediated evolution, such as the CTXΦ in clinical V. cholerae strains and the filamentous prophage in V. parahaemolyticus pathogenic lineages. Moreover, a number of prophage-mediated HGT events likely occurred between different Vibrio spp. (e.g., Vibrio fischeri and V. cholerae) or across genera boundaries (e.g., Burkholderia, Escherichia, and Vibrio). On the other hand, prophages assisted superinfection exclusion in Vibrio spp., leading to the integration into the host genomes, e.g., the H20-like phage in V. anguillarum and VHS1 in V. harveyi. In addition, the intertwined regulation of phage–host interactions, such as between Φ919TP and V. cholerae and between H20-like prophage p41 and V. anguillarum, have recently been reported as well.
Overall, this review facilitates a better understanding of the prophage-promoting evolution of Vibrio spp. and their implications for food safety and public health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods14030403/s1, Table S1. The intact prophage gene clusters identified in Vibrio spp. in the most recent 5 years [80,88,89,90,91,92,93]; Table S2. Information on the 42 articles selected in this review.
Author Contributions
Y.O.: investigation, data curation, and writing—original draft preparation; J.Y.: writing—review and editing; Y.W.: discussion and supervision; L.C.: funding acquisition, conceptualization, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Commission of Shanghai Municipality, grant number 17050502200, and the National Natural Science Foundation of China, grant number 31671946.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to thank Mingjie Chen at Shanghai NewCore Biotechnology Co., Ltd. for the help with Sankey data graphing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lukjancenko, O.; Ussery, D.W. Vibrio chromosome-specifific families. Front. Microbiol. 2014, 5, 73. [Google Scholar] [CrossRef]
- Serratore, P.; Bignami, G.; Ostanello, F.; Lorito, L. Hazard identification related to the presence of Vibrio spp., biogenic amines, and indole-producing bacteria in a non-filter feeding marine gastropod (Tritia mutabilis) commercialized on the Italian market. Foods 2021, 10, 2574. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, C.; Szott, V.; Alter, T.; Huehn-Lindenbein, S.; Fleischmann, S. Prevalence of Vibrio spp. in seafood from German supermarkets and fish markets. Foods 2024, 13, 3987. [Google Scholar] [CrossRef] [PubMed]
- Thompson, F.L.; Iida, T.; Swings, J. Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 2004, 68, 403–431. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primer 2018, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Grimes, D.J. The Vibrios: Scavengers, symbionts, and pathogens from the sea. Microb. Ecol. 2020, 80, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, B.Y.; Waters, C.M. Combating cholera. F1000Research 2019, 8, 589. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.C.; Zahner, V.; Avelar, K.E.S.; Alves, R.M.; Pereira, D.S.G.; Vital, B.J.M.; Freitas, F.S.; Salles, C.A.; Karaolis, D.K.R. Genetic diversity and antibiotic resistance of clinical and environmental suggests that many serogroups are reservoirs of resistance. Epidemiol. Infect. 2004, 132, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Chaguza, C.; Chibwe, I.; Chaima, D.; Musicha, P.; Ndeketa, L.; Kasambara, W.; Mhango, C.; Mseka, U.L.; Bitilinyu-Bangoh, J.; Mvula, B.; et al. Genomic insights into the 2022–2023 Vibrio cholerae outbreak in Malawi. Nat. Commun. 2024, 15, 6291. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, X.; Ni, L.; Xu, D.; Xu, Y.; Ding, Y.; Xie, L.; Chen, L. First experimental evidence for the presence of potentially toxic Vibrio cholerae in snails, and virulence, cross-resistance and genetic diversity of the bacterium in 36 species of aquatic food animals. Antibiotics 2021, 10, 412. [Google Scholar] [CrossRef]
- Yan, L.; Jin, Y.; Zhang, B.; Xu, Y.; Peng, X.; Qin, S.; Chen, L. Diverse aquatic animal matrices play a key role in survival and potential virulence of non-O1/O139 Vibrio cholerae isolates. Front. Microbiol. 2022, 13, 896767. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.; Hasan, N.A.; Huq, A.; Colwell, R.R. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 2013, 3, 97. [Google Scholar] [CrossRef] [PubMed]
- Meza, G.; Majrshi, H.; Tiong, H.K. Recovery of pasteurization-resistant Vibrio parahaemolyticus from seafoods using a modified, two-step enrichment. Foods 2022, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zou, D.; Long, Y.; Xue, L.; Shuai, S.; Tian, F.; Li, M.; Fan, G.; Zheng, Y.; Sun, X.; et al. Contamination of Vibrio parahaemolyticus in crayfish for sale. Front Microbiol. 2024, 15, 1388658. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.J.; Flaherty, E.; Lee, N.; Robbins, A.; Weller, D.L. Severe Vibrio vulnificus infections during heat waves—Three eastern U.S. States, July–August 2023. MMWR Morb. Mortal. Wkly. Rep. 2024, 73, 84–85. [Google Scholar] [CrossRef]
- Yamazaki, K.; Kashimoto, T.; Kado, T.; Yoshioka, K.; Ueno, S. Increased vascular permeability due to spread and invasion of Vibrio vulnificus in the wound infection exacerbates potentially fatal necrotizing disease. Front. Microbiol. 2022, 13, 849600. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, G.; Xu, D.; Ye, J.; Lu, Y. A novel RAA combined test strip method based on dual gene targets for pathogenic Vibrio vulnificus in aquatic products. Foods 2023, 12, 3605. [Google Scholar] [CrossRef] [PubMed]
- Jacobs Slifka, K.M.; Newton, A.E.; Mahon, B.E. Vibrio alginolyticus infections in the USA, 1988-2012. Epidemiol. Infect. 2017, 145, 1491–1499. [Google Scholar] [CrossRef]
- Abdelsalam, M.; Attia, M.M.; Marzouk, M.S.; Korany, R.M.S.; Elgendy, M.Y.; Soliman, A.W.; Prince, A.; Hamada, A.H. Investigating dynamics, etiology, pathology, and therapeutic interventions of Caligus clemensi and Vibrio alginolyticus co-infection in farmed marine fish. Sci. Rep. 2024, 14, 20704. [Google Scholar] [CrossRef]
- Sun, Y.; Yan, Y.; Yan, S.; Li, F.; Li, Y.; Yan, L.; Yang, D.; Peng, Z.; Yang, B.; Sun, J.; et al. Prevalence, antibiotic susceptibility, and genomic analysis of Vibrio alginolyticus isolated from seafood and freshwater products in China. Front Microbiol. 2024, 15, 1381457. [Google Scholar] [CrossRef] [PubMed]
- Morgado, M.E.; Brumfield, K.D.; Mitchell, C.; Boyle, M.M.; Colwell, R.R.; Sapkota, A.R. Increased incidence of vibriosis in Maryland, U.S.A., 2006–2019. Environ. Res. 2024, 244, 117940. [Google Scholar] [CrossRef] [PubMed]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Turkington, C.J.; Hill, C. Mutualistic interplay between bacteriophages and bacteria in the human gut. Nat. Rev. Microbiol. 2022, 20, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Wendling, C.C.; Refardt, D.; Hall, A.R. Fitness benefits to bacteria of carrying prophages and prophage-encoded antibiotic-resistance genes peak in different environments. Evolution 2021, 75, 515–528. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Jacobs-Sera, D.; Bustamante, C.A.G.; Garlena, R.A.; Mavrich, T.N.; Pope, W.H.; Reyes, J.C.C.; Russell, D.A.; Adair, T.; Alvey, R.; et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microbiol. 2017, 2, 16251. [Google Scholar] [CrossRef]
- Garin-Fernandez, A.; Wichels, A. Looking for the hidden: Characterization of lysogenic phages in potential pathogenic Vibrio species from the North Sea. Mar. Genom. 2020, 51, 100725. [Google Scholar] [CrossRef] [PubMed]
- Steensen, K.; Séneca, J.; Bartlau, N.; Yu, X.A.; Hussain, F.A.; Polz, M.F. Tailless and filamentous prophages are predominant in marine Vibrio. ISME J. 2024, 18, wrae202. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Pérez-Reytor, D.; Plaza, N.; Ramírez-Araya, S.; Blondel, C.J.; Corsini, G.; Bastías, R.; Loyola, D.E.; Jaña, V.; Pavez, L.; et al. Exploring the genomic traits of non-toxigenic Vibrio parahaemolyticus strains isolated in Southern Chile. Front. Microbiol. 2018, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Kobakhidze, S.; Koulouris, S.; Kakabadze, N.; Kotetishvili, M. Genetic recombination-mediated evolutionary interactions between phages of potential industrial importance and prophages of their hosts within or across the domains of Escherichia, Listeria, Salmonella, Campylobacter, and Staphylococcus. BMC Microbiol. 2024, 24, 155. [Google Scholar] [CrossRef]
- Molina-Quiroz, R.C.; Dalia, T.N.; Camilli, A.; Dalia, A.B.; Silva-Valenzuela, C.A. Prophage-dependent neighbor predation fosters horizontal gene transfer by natural transformation. mSphere 2020, 5, e00975-20. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Davis, B.M.; Moyer, K.E.; Boyd, E.F.; Waldor, M.K. CTX prophages in classical biotype Vibrio cholerae: Functional phage genes but dysfunctional phage genomes. J. Bacteriol. 2000, 182, 6992–6998. [Google Scholar] [CrossRef] [PubMed]
- Biswas, Q.; Purohit, A.; Kumar, A.; Rakshit, D.; Maiti, D.; Das, B.; Bhadra, R.K. Genetic and mutational analysis of virulence traits and their modulation in an environmental toxigenic Vibrio cholerae non-O1/non-O139 strain, VCE232. Microbiology 2022, 168, 001135. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.E.; Majewski, J.; Faller, R.; Satija, S.; Kuhl, T.L. Cholera toxin assault on lipid monolayers containing ganglioside GM1. Biophys. J. 2004, 86, 3700–3708. [Google Scholar] [CrossRef] [PubMed]
- Krukonis, E.S.; DiRita, V.J. From motility to virulence: Sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 2003, 6, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Reytor, D.; Jaña, V.; Pavez, L.; Navarrete, P.; García, K. Accessory toxins of Vibrio pathogens and their role in epithelial disruption during infection. Front. Microbiol. 2018, 9, 2248. [Google Scholar] [CrossRef] [PubMed]
- Marinaro, M.; Fasano, A.; Magistris, M.T.D. Zonula occludens toxin Acts as an adjuvant through different mucosal routes and induces protective immune responses. Infect. Immun. 2003, 71, 1897–1902. [Google Scholar] [CrossRef]
- Castillo, D.; Kauffman, K.; Hussain, F.; Kalatzis, P.; Rørbo, N.; Polz, M.F.; Middelboe, M. Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Sci. Rep. 2018, 8, 9973. [Google Scholar] [CrossRef]
- Chibani, C.M.; Hertel, R.; Hoppert, M.; Liesegang, H.; Wendling, C.C. Closely related Vibrio alginolyticus strains encode an identical repertoire of caudovirales-like regions and filamentous phages. Viruses 2020, 12, 1359. [Google Scholar] [CrossRef]
- Nawel, Z.; Rima, O.; Amira, B. An overview on Vibrio temperate phages: Integration mechanisms, pathogenicity, and lysogeny regulation. Microb. Pathog. 2022, 165, 105490. [Google Scholar] [CrossRef]
- Foxall, R.L.; Means, J.; Marcinkiewicz, A.L.; Schillaci, C.; DeRosia-Banick, K.; Xu, F.; Hall, J.A.; Jones, S.H.; Cooper, V.S.; Whistler, C.A. Inoviridae prophage and bacterial host dynamics during diversification, succession, and Atlantic invasion of Pacific-native Vibrio parahaemolyticus. mBio 2024, 15, e02851-23. [Google Scholar] [CrossRef] [PubMed]
- Bochow, S.; Elliman, J.; Owens, L. Bacteriophage adenine methyltransferase: A life cycle regulator? Modelled using Vibrio harveyi myovirus like. J. Appl. Microbiol. 2012, 113, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.; Oakey, J.; Bromage, E.; Owens, L. Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Organ. 2003, 54, 187–194. [Google Scholar] [CrossRef]
- Paul, J.H. Prophages in marine bacteria: Dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008, 2, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Santoriello, F.J.; Michel, L.; Unterweger, D.; Pukatzki, S. Pandemic Vibrio cholerae shuts down site-specific recombination to retain an interbacterial defence mechanism. Nat. Commun. 2020, 11, 6246. [Google Scholar] [CrossRef]
- Santoriello, F.J.; Pukatzki, S. When the pandemic opts for the lockdown: Secretion system evolution in the cholera bacterium. Microb. Cell Graz Austria 2021, 8, 69–72. [Google Scholar] [CrossRef]
- Xu, D.; Peng, X.; Xie, L.; Chen, L. Survival and genome diversity of Vibrio parahaemolyticus isolated from edible aquatic animals. Diversity 2022, 14, 350. [Google Scholar] [CrossRef]
- Yu, Z.; Wu, Y.; Chen, M.; Huo, T.; Zheng, W.; Ludtke, S.J.; Shi, X.; Wang, Z. Membrane translocation process revealed by in situ structures of type II secretion system secretins. Nat. Commun. 2023, 14, 4025. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.H.; Teh, C.S.J.; Yap, K.P.; Ung, E.H.; Thong, K.L. Comparative genomic provides insight into the virulence and genetic diversity of Vibrio parahaemolyticus associated with shrimp acute hepatopancreatic necrosis disease. Infect. Genet. Evol. 2020, 83, 104347. [Google Scholar] [CrossRef] [PubMed]
- Khemayan, K.; Prachumwat, A.; Sonthayanon, B.; Intaraprasong, A.; Sriurairatana, S.; Flegel, T.W. Complete genome sequence of virulence-enhancing siphophage VHS1 from Vibrio harveyi. Appl. Environ. Microbiol. 2012, 78, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, H.; Chen, D.; Li, Y. Genomic characterization and comparative genomic analysis of pathogenic Vibrio isolated from aquaculture-grown white-leg shrimp (Penaeus vannamei) in Guangdong and Jiangsu, China. Aquaculture 2024, 580, 740302. [Google Scholar] [CrossRef]
- Mesa, C.A.D.; Mendoza, R.M.; Penir, S.M.U.; de la Peña, L.D.; Amar, E.C.; Saloma, C.P. Genomic analysis of Vibrio harveyi strain PH1009, a potential multi-drug resistant pathogen due to acquisition of toxin genes. Heliyon 2023, 9, e14926. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y.; Yu, P.; Ren, S.; Zhu, Z.; Jin, Y.; Yan, J.; Peng, X.; Chen, L. Prophage-related gene VpaChn25_0724 contributes to cell membrane integrity and growth of Vibrio parahaemolyticus CHN25. Front. Cell. Infect. Microbiol. 2020, 10, 595709. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, L.; Wang, Y.; Zhu, Z.; Yan, J.; Qin, S.; Chen, L. Prophage-encoded gene VpaChn25_0734 amplifies ecological persistence of Vibrio parahaemolyticus CHN25. Curr. Genet. 2022, 68, 267–287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, Y.; Yang, L.; Wang, Y.; Li, M.; Chen, L. Biological function of prophage-related gene cluster ΔVpaChn25_RS25055~ΔVpaChn25_0714 of Vibrio parahaemolyticus CHN25. Int. J. Mol. Sci. 2024, 25, 1393. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Li, R.; Zhang, W.; Wang, L.; Yan, B.; Zhu, T.; Xu, Y.; Tan, D. Characterization of a filamentous Phage, Vaf1, from Vibrio alginolyticus AP-1. Appl. Environ. Microbiol. 2023, 89, e00520-23. [Google Scholar] [CrossRef]
- Qin, X.; Yang, L.; Xu, Y.; Xie, L.; Wang, Y.; Chen, L. Growth and genome features of non-O1/O139 Vibrio cholerae isolated from three species of common freshwater fish. Diversity 2024, 16, 268. [Google Scholar] [CrossRef]
- Wang, W.; Tang, K.; Wang, P.; Zeng, Z.; Xu, T.; Zhan, W.; Liu, T.; Wang, Y.; Wang, X. The coral pathogen Vibrio coralliilyticus kills non-pathogenic holobiont competitors by triggering prophage induction. Nat. Ecol. Evol. 2022, 6, 1132–1144. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Portillo, E.; Robertson, S.; Antón, J. Coral mucus as a reservoir of bacteriophages targeting Vibrio pathogens. ISME J. 2024, 18, wrae017. [Google Scholar] [CrossRef]
- Lewis, J.M.; Janda, K.E.; Kotter, D.B.; Grose, J.H.; McCleary, W.R. Characterization of the attachment of three new coliphages onto the ferrichrome transporter FhuA. J. Virol. 2023, 97, e00667-23. [Google Scholar] [CrossRef] [PubMed]
- Cumby, N.; Reimer, K.; Mengin-Lecreulx, D.; Davidson, A.R.; Maxwell, K.L. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol. Microbiol. 2015, 96, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Kalatzis, P.G.; Rørbo, N.I.; Castillo, D.; Mauritzen, J.J.; Jørgensen, J.; Kokkari, C.; Zhang, F.; Katharios, P.; Middelboe, M. Stumbling across the same phage: Comparative genomics of widespread temperate phages infecting the fish pathogen Vibrio anguillarum. Viruses 2017, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Stalin, N.; Srinivasan, P. Efficacy of potential phage cocktails against Vibrio harveyi and closely related Vibrio species isolated from shrimp aquaculture environment in the south east coast of India. Vet. Microbiol. 2017, 207, 83–96. [Google Scholar] [CrossRef]
- Williamson, S.J.; McLaughlin, M.R.; Paul, J.H. Interaction of the ΦHSIC virus with its host: Lysogeny or pseudolysogeny? Appl. Environ. Microbiol. 2001, 67, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, L.; Gao, H.; Chen, L.; Fan, F.; Li, Z.; Fan, Y.; Li, J.; Liang, W.; Pang, B.; et al. A novel pre-CTX prophage in the Vibrio cholerae serogroup O139 strain. Infect. Genet. Evol. 2020, 81, 104238. [Google Scholar] [CrossRef]
- Pant, A.; Bag, S.; Saha, B.; Verma, J.; Kumar, P.; Banerjee, S.; Kumar, B.; Kumar, Y.; Desigamani, A.; Maiti, S.; et al. Molecular insights into the genome dynamics and interactions between core and acquired genomes of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2020, 117, 23762–23773. [Google Scholar] [CrossRef] [PubMed]
- McLeod, S.M.; Kimsey, H.H.; Davis, B.M.; Waldor, M.K. CTXφ and Vibrio cholerae: Exploring a newly recognized type of phage–host cell relationship. Mol. Microbiol. 2005, 57, 347–356. [Google Scholar] [CrossRef]
- Ochi, K.; Mizuno, T.; Samanta, P.; Mukhopadhyay, A.K.; Miyoshi, S.; Imamura, D. Recent Vibrio cholerae O1 epidemic strains are unable to replicate CTXΦ prophage genome. mSphere 2021, 6, e0033721. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.M.L.; Varkey, J.B.; Petti, C.A.; Liddle, R.A.; Frothingham, R.; Woods, C.W. Non-o1 Vibrio cholerae septicemia: Case report, discussion of literature, and relevance to bioterrorism. Diagn. Microbiol. Infect. Dis. 2004, 49, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, C.; Sun, Z.; Zheng, W.; Zhang, W.; Yu, H.; Wu, Y.; Didelot, X.; Yang, R.; Pan, J.; et al. Genomic epidemiology of Vibrio cholerae reveals the regional and global spread of two epidemic non-toxigenic lineages. PLoS Negl. Trop. Dis. 2020, 14, e0008046. [Google Scholar] [CrossRef]
- Hao, T.; Zheng, W.; Wu, Y.; Yu, H.; Qian, X.; Yang, C.; Zheng, Z.; Zhang, X.; Guo, Y.; Cui, M.; et al. Population genomics implies potential public health risk of two non-toxigenic Vibrio cholerae lineages. Infect. Genet. Evol. 2023, 112, 105441. [Google Scholar] [CrossRef] [PubMed]
- Behera, D.R.; Nayak, A.K.; Nayak, S.R.; Nayak, D.; Swain, S.; Maharana, P.K.; Biswal, B.; Pany, S.; Pati, S.; Pal, B.B. Genomic diversities of ctxB, tcpA and rstR alleles of Vibrio cholerae O139 strains isolated from Odisha, India. Environ. Microbiol. Rep. 2022, 14, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Thong, K.L.; Tham, K.B.L.; Ngoi, S.T.; Tan, S.C.; Wan Yussof, W.N.; Ahmad Hanapi, R.; Mohamad, N.; Teh, C.S.J. Molecular characterization of Vibrio cholerae O1 El Tor strains in Malaysia revealed genetically diverse variant lineages. Transbound. Emerg. Dis. 2022, 69, e693–e703. [Google Scholar] [CrossRef]
- Abanto, M.; Gavilan, R.G.; Baker-Austin, C.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Global expansion of Pacific Northwest Vibrio parahaemolyticus sequence type 36. Emerg. Infect. Dis. 2020, 26, 323–326. [Google Scholar] [CrossRef]
- Boyd, E.F.; Moyer, K.E.; Shi, L.; Waldor, M.K. Infectious CTXΦ; and the Vibrio pathogenicity island prophage in Vibrio mimicus: Evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 2000, 68, 1507–1513. [Google Scholar] [CrossRef]
- Wang, H.; Xie, G.; Huang, J. Genome-based characterization of a novel prophage of Vibrio parahaemolyticus, VPS05ph1, a novel member of Peduoviridae. Virology 2024, 595, 110087. [Google Scholar] [CrossRef] [PubMed]
- Soto, E.; Alegría, M.; Sepúlveda, F.; García, K.; Higuera, G.; Castillo, D.; Fontúrbel, F.; Bastías, R. Prophages carrying Zot toxins on different Vibrio genomes: A comprehensive assessment using multilayer networks. Environ. Microbiol. 2024, 26, e16654. [Google Scholar] [CrossRef]
- Nuidate, T.; Kuaphiriyakul, A.; Surachat, K.; Mittraparp-Arthorn, P. Induction and genome analysis of HY01, a newly reported prophage from an emerging shrimp pathogen Vibrio campbellii. Microorganisms 2021, 9, 400. [Google Scholar] [CrossRef]
- Maiti, D.; Das, B.; Saha, A.; Nandy, R.K.; Nair, G.B.; Bhadra, R.K. Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology 2006, 152, 3633–3641. [Google Scholar] [CrossRef] [PubMed]
- Garin-Fernandez, A.; Glöckner, F.O.; Wichels, A. Genomic characterization of filamentous phage vB_VpaI_VP-3218, an inducible prophage of Vibrio parahaemolyticus. Mar. Genom. 2020, 53, 100767. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, J.; Xu, J.; Du, P.; Pang, B.; Li, J.; Kan, B. The resistance of Vibrio cholerae O1 El Tor strains to the typing phage 919TP, a member of K139 phage family. Front. Microbiol. 2016, 7, 726. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in bacteriophage Lambda development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zeng, Y.; Hu, B.; Zhu, T.; Svenningsen, S.L.; Middelboe, M.; Tan, D. Interactions between the prophage 919TP and its Vibrio cholerae host: Implications of gmd mutation for phage resistance, cell auto-aggregation, and motility. Viruses 2021, 13, 2342. [Google Scholar] [CrossRef]
- Tan, D.; Hansen, M.F.; de Carvalho, L.N.; Røder, H.L.; Burmølle, M.; Middelboe, M.; Lo Svenningsen, S. High cell densities favor lysogeny: Induction of an H20 prophage is repressed by quorum sensing and enhances biofilm formation in Vibrio anguillarum. ISME J. 2020, 14, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Alvise, P.D.; Xu, R.; Zhang, F.; Middelboe, M.; Gram, L. Comparative genome analyses of Vibrio anguillarum strains reveal a link with pathogenicity traits. mSystems 2017, 2, e00001-17. [Google Scholar] [CrossRef]
- Castillo, D.; Andersen, N.; Kalatzis, P.G.; Middelboe, M. Large phenotypic and genetic diversity of prophages induced from the fish pthogen Vibrio anguillarum. Viruses 2019, 11, 983. [Google Scholar] [CrossRef]
- Xu, M.; Xu, M.; Tu, Q. Comparative evaluation of Vibrio delineation methodologies in post-genomic era. Environ. Microbiol. Rep. 2021, 13, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Li, D.; Xu, L.; Lin, W.; Tong, Y. Complete genome analysis of an active prophage of Vibrio alginolyticus. Arch. Virol. 2021, 166, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Baby, B.; Vijay, D.; Philip, P.S.; Alnuaimi, A.A.; Almansoori, H.M.; Areidat, S.O.; Khan, G.; Vijayan, R.; Akhtar, M.K. Complete genome sequence of Vibrio gazogenes PB1: An estuarine bacterium capable of producing prodigiosin from starch or cellulose. Front. Mar. Sci. 2023, 10, 1028319. [Google Scholar] [CrossRef]
- Rathnapala, J.M.S.N.; Ragab, W.; Kawato, S.; Furukawa, M.; Nozaki, R.; Kondo, H.; Hirono, I. Genomic characterization and identification of virulence-related genes in Vibrio nigripulchritudo isolated from white leg shrimp Penaeus vannamei. J. Fish. Dis. 2023, 46, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yu, P.; Wang, J.; Zhao, T.; Zhao, Y.; Pan, Y.; Chen, L. Genomic and transcriptomic analyses reveal multiple strategies for Vibrio parahaemolyticus to tolerate sub-lethal concentrations of three antibiotics. Foods 2024, 13, 1674. [Google Scholar] [CrossRef] [PubMed]
- Ragab, W.; Kawato, S.; Nozaki, R.; Kondo, H.; Hirono, I. Comparative genome analyses of five Vibrio penaeicida strains provide insights into their virulence-related factors. Microb. Genom. 2022, 8, 000766. [Google Scholar] [CrossRef] [PubMed]
- Zago, V.; Veschetti, L.; Patuzzo, C.; Malerba, G.; Lleo, M.M. Resistome, mobilome and virulome analysis of Shewanella algae and Vibrio spp. strains isolated in Italian aquaculture centers. Microorganisms 2020, 8, 572. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).