Integrated Enzyme-Mediated One-Step Sample Processing and Duplex Amplification System for Rapid Detection of Carpione rhabdovirus in Aquaculture-Derived Food Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Sample Collection and RNA Preparation
2.2. Construction of Plasmid for Standard Quantification
2.3. EmDEA Reaction System Development
2.3.1. Design of EmDEA Primers
2.3.2. Screening of EmDEA Reaction and Primers
2.4. Development of RT-EmDEA Reaction System
2.4.1. Preparation of RNA Standard Samples
2.4.2. Reverse Transcription Primer Design and RT-EmDEA Reaction
2.4.3. Screening of Compatible Primer Pairs for RT-EmDEA of CAPRV2023
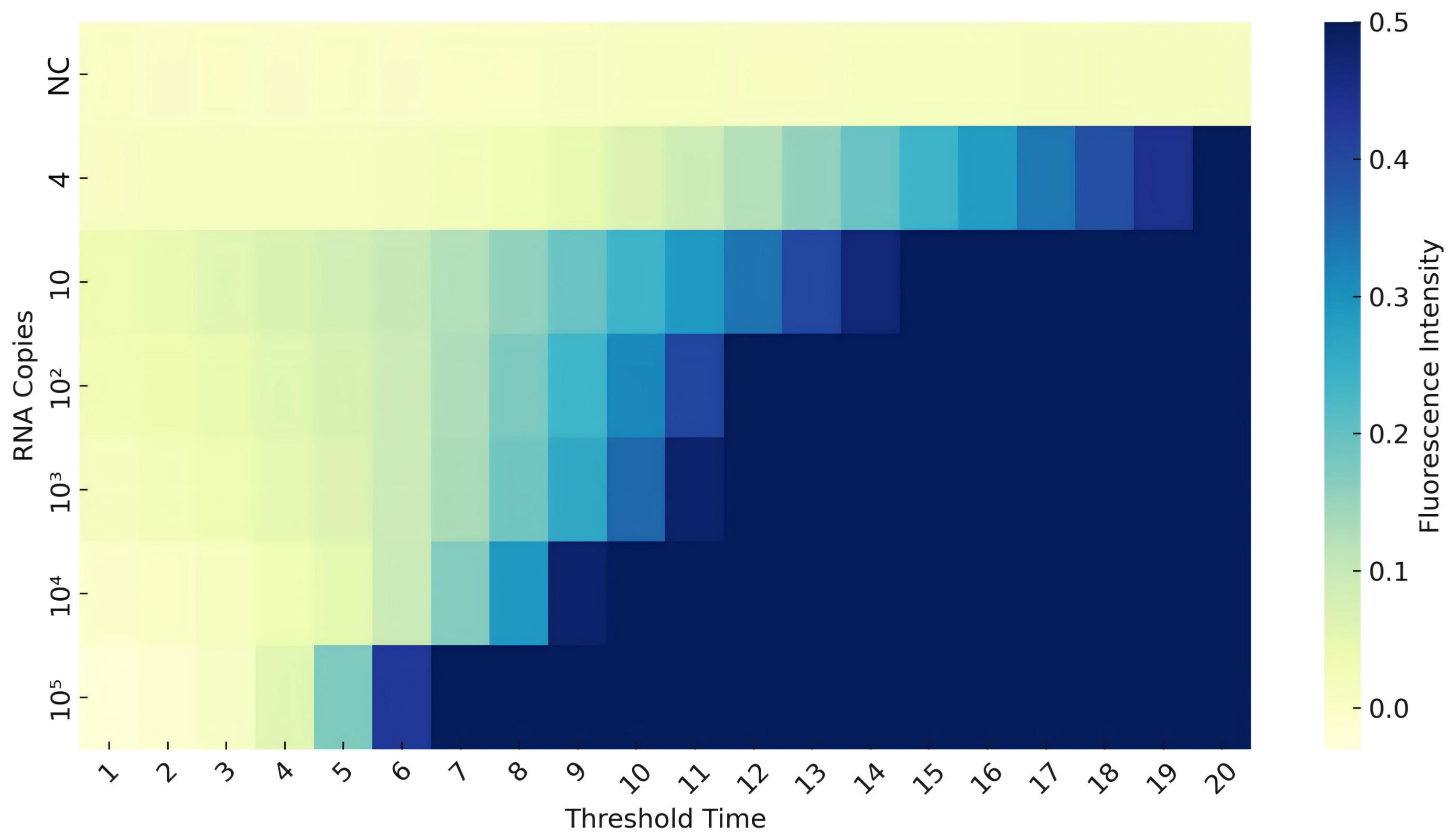
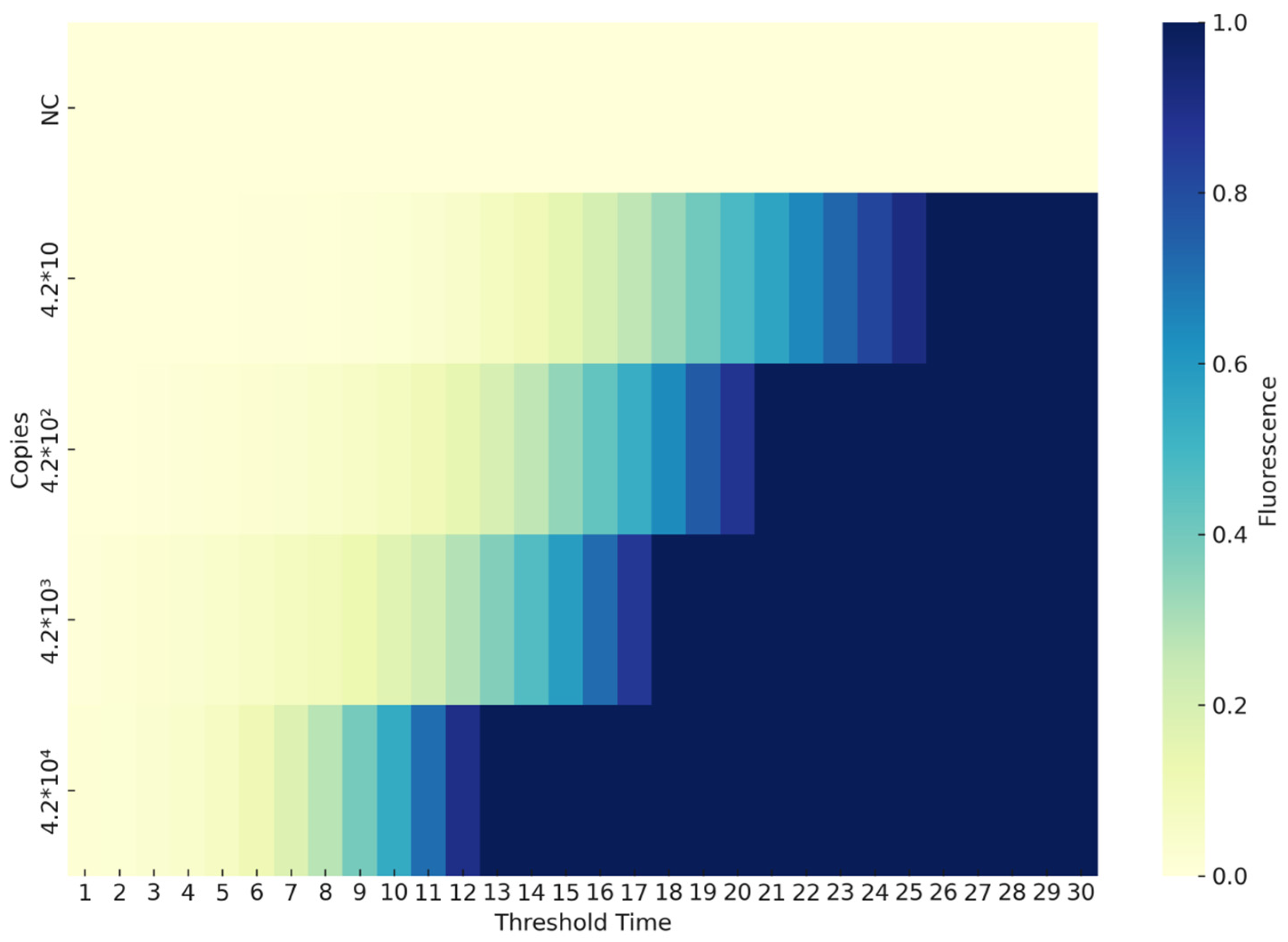
2.4.4. Sensitivity of the CAPRV2023 RT-EmDEA Reaction
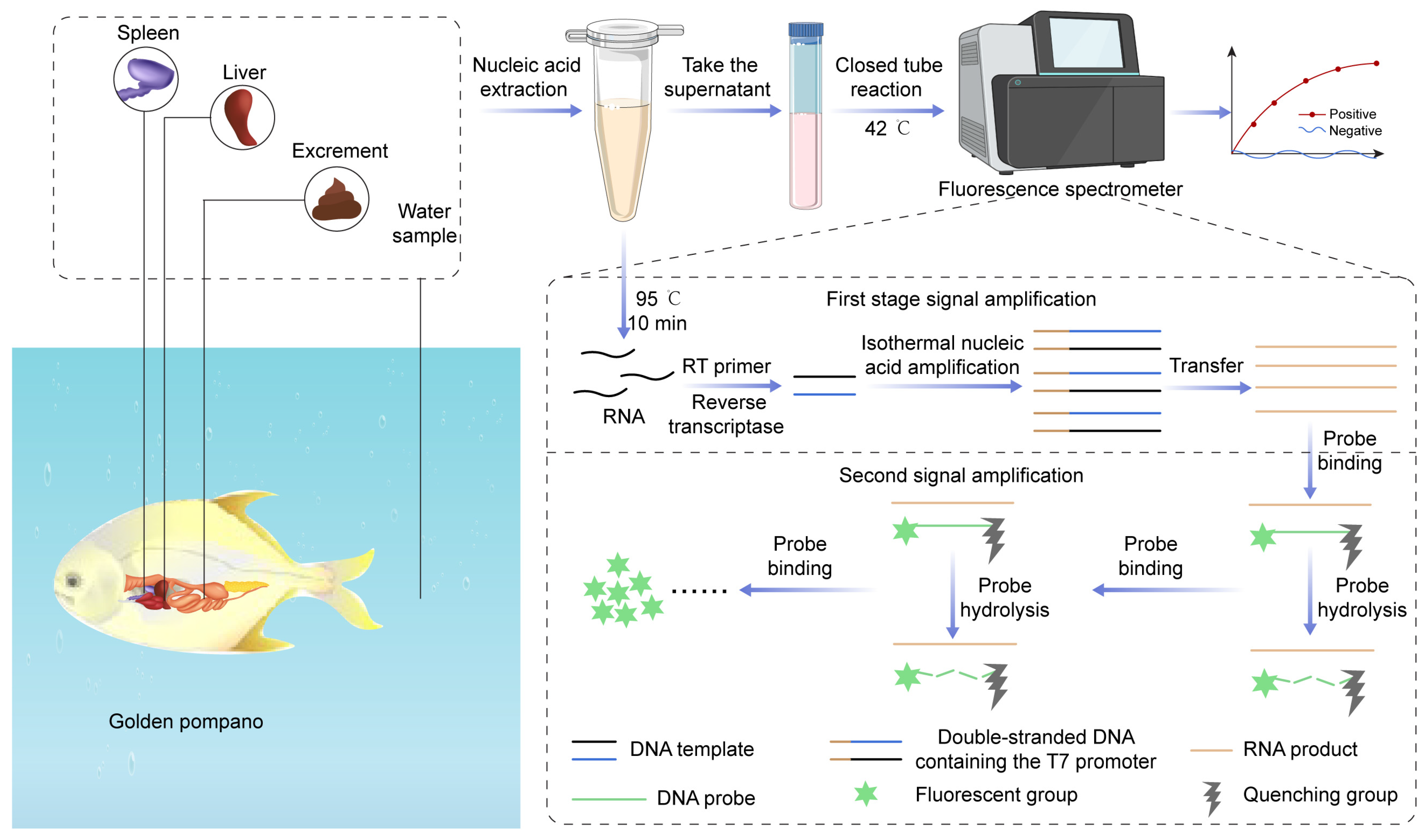
2.5. Enzyme-Mediated One-Step Sample Processing Technique
2.5.1. Sample Processing Method
2.5.2. Minimally Invasive Sample Extraction
2.5.3. Invasive Sample Extraction
2.5.4. Environmental Sample Extraction
2.5.5. Optimization of EmOSP for Environmental Samples
2.6. EmOSP-RT-EmDEA Detection System Development for CAPRV2023
2.6.1. Integrating RT-EmDEA System for CAPRV2023
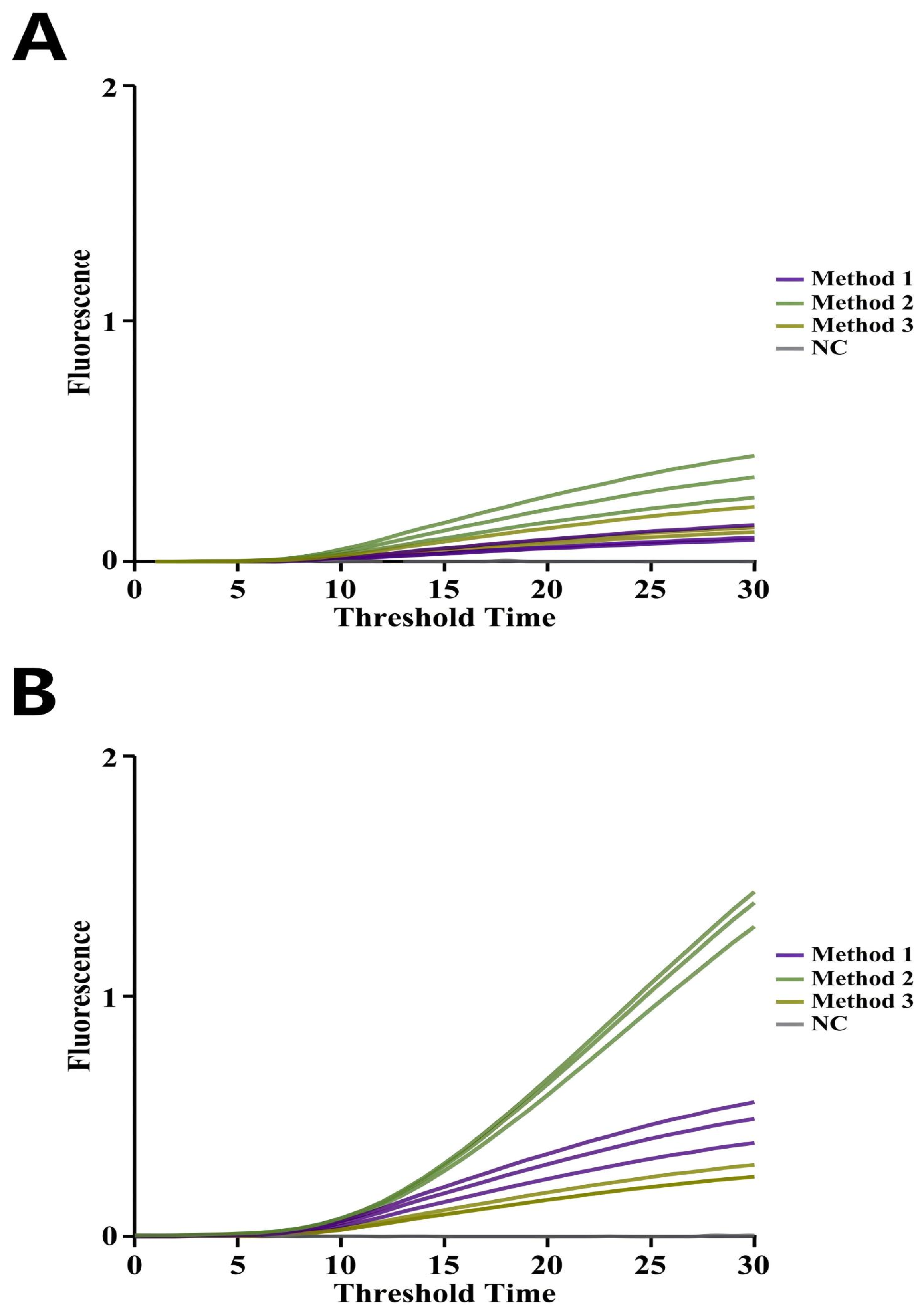
2.6.2. Specificity of the EmOSP-RT-EmDEA Detection System for CAPRV2023
2.6.3. Repeatability of the EmOSP-RT-EmDEA Detection System for CAPRV2023
2.6.4. Application of the CAPRV2023 EmOSP-RT-EmDEA Detection System
2.7. Clinical Samples and Ethics Statement
3. Results
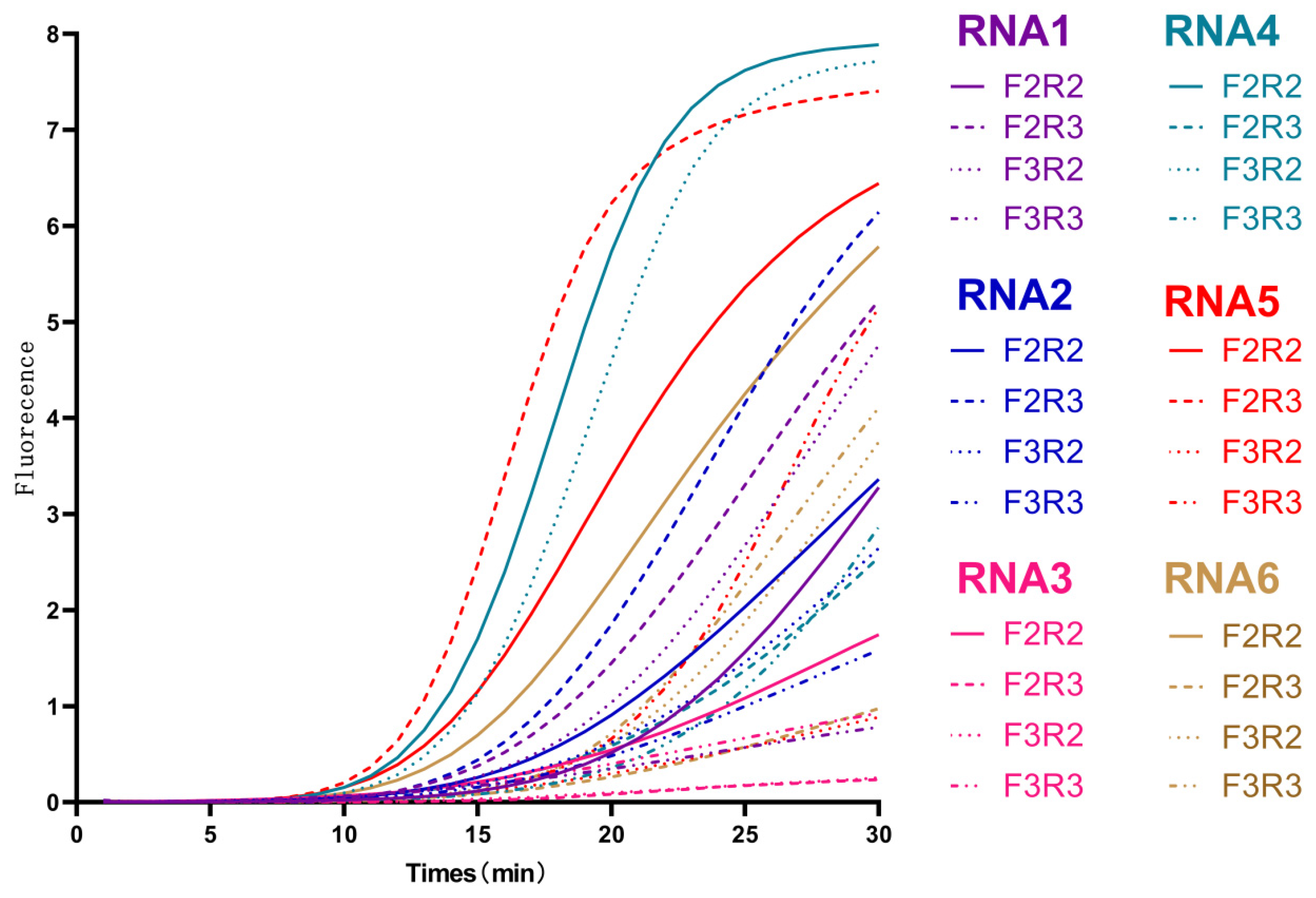
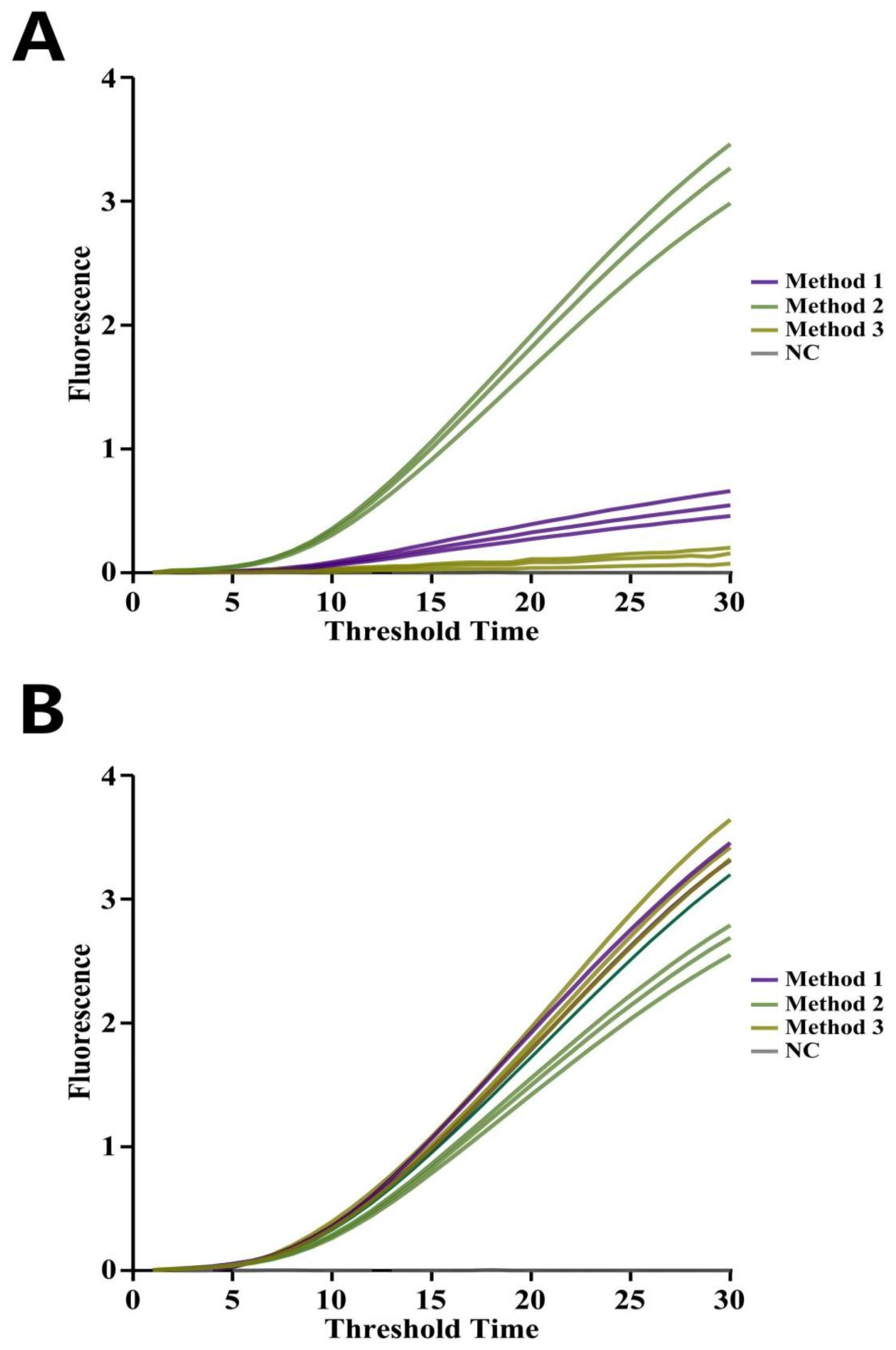
3.1. Screening of RNA and EmDEA Primers
3.1.1. Screening of RNA Primer
3.1.2. Screening of EmDEA Primer
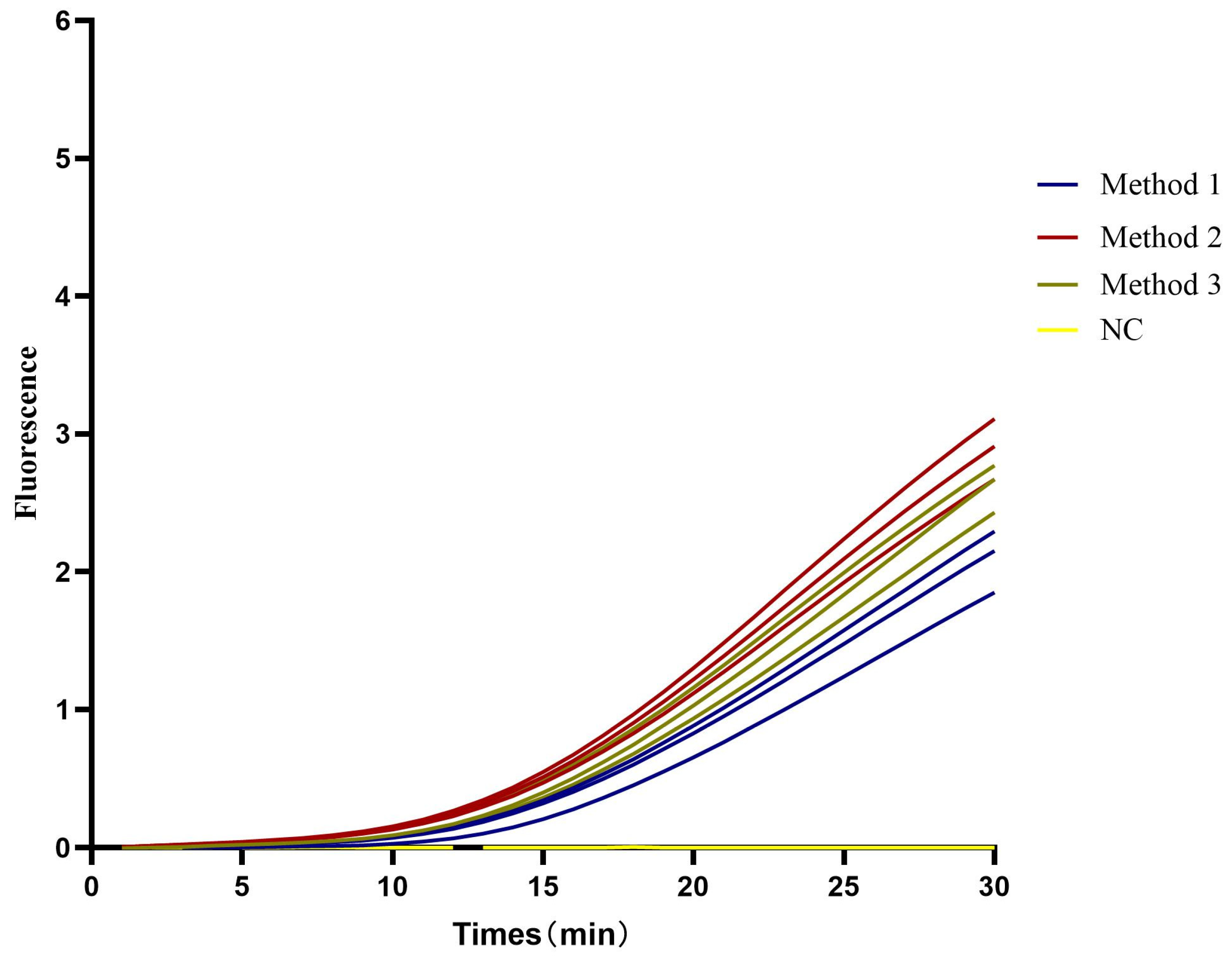
3.2. CAPRV-RT-EmDEA
3.2.1. RT-EmDEA Primer
3.2.2. Sensitivity of the CAPRV2023-RT-EmDEA
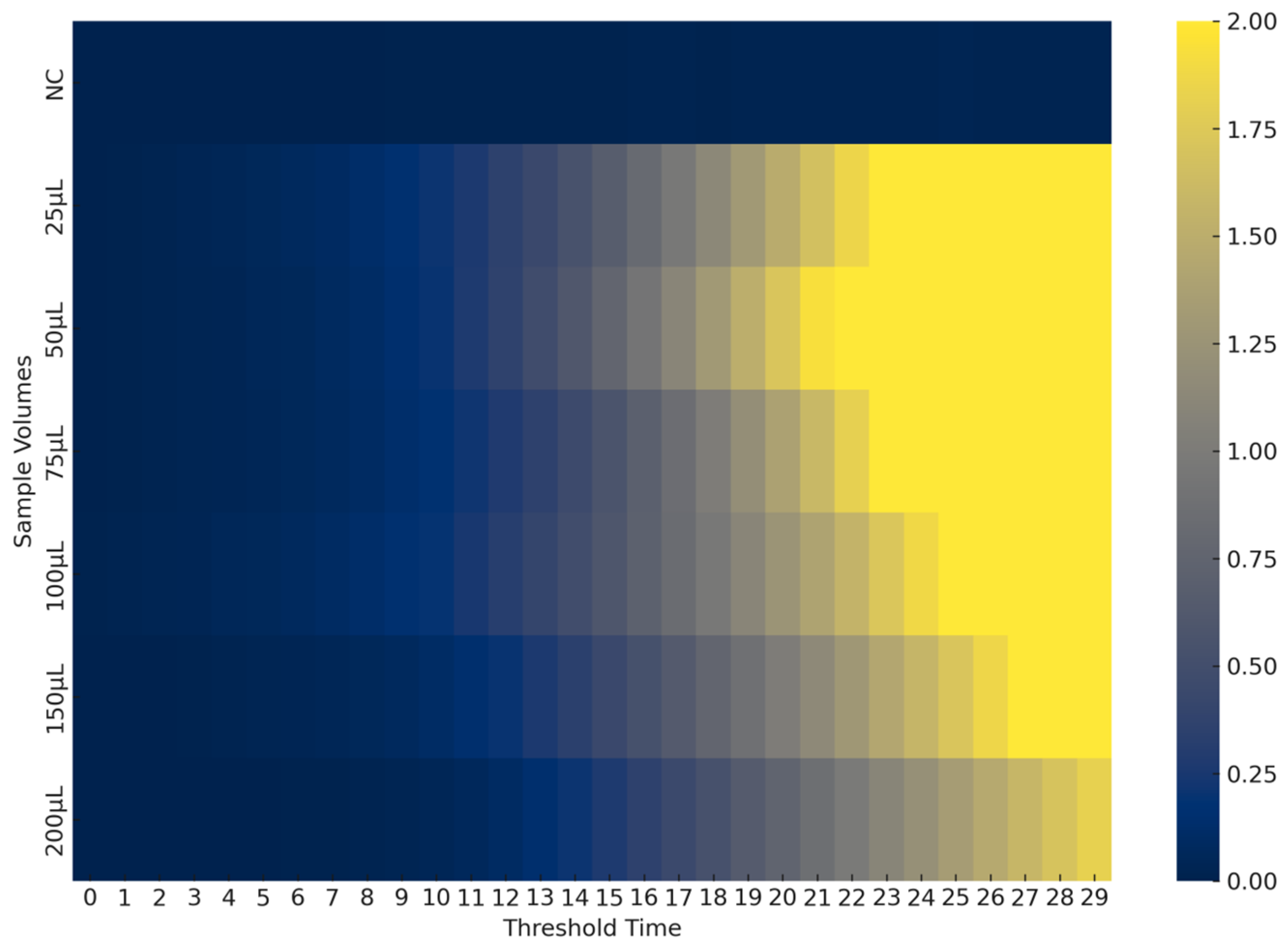
3.3. Enzyme-Mediated One-Step Sample Processing
3.3.1. Minimally Invasive Sample
3.3.2. Invasive Sample
3.3.3. Environmental Sample
3.4. CAPRV2023 EmOSP-RT-EmDEA Detection System
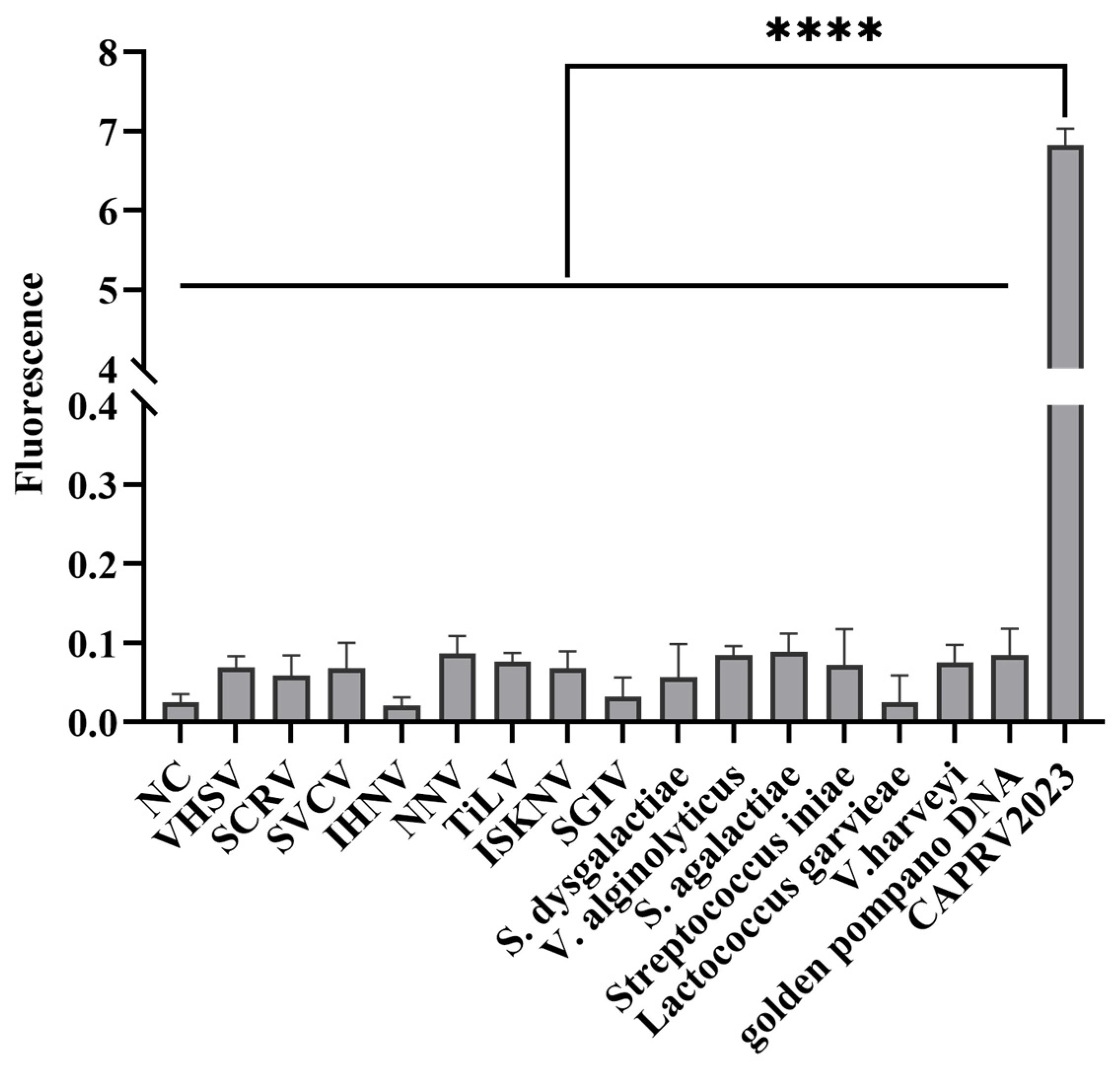
3.4.1. Specificity of the CAPRV2023 EmOSP-RT-EmDEA Detection System
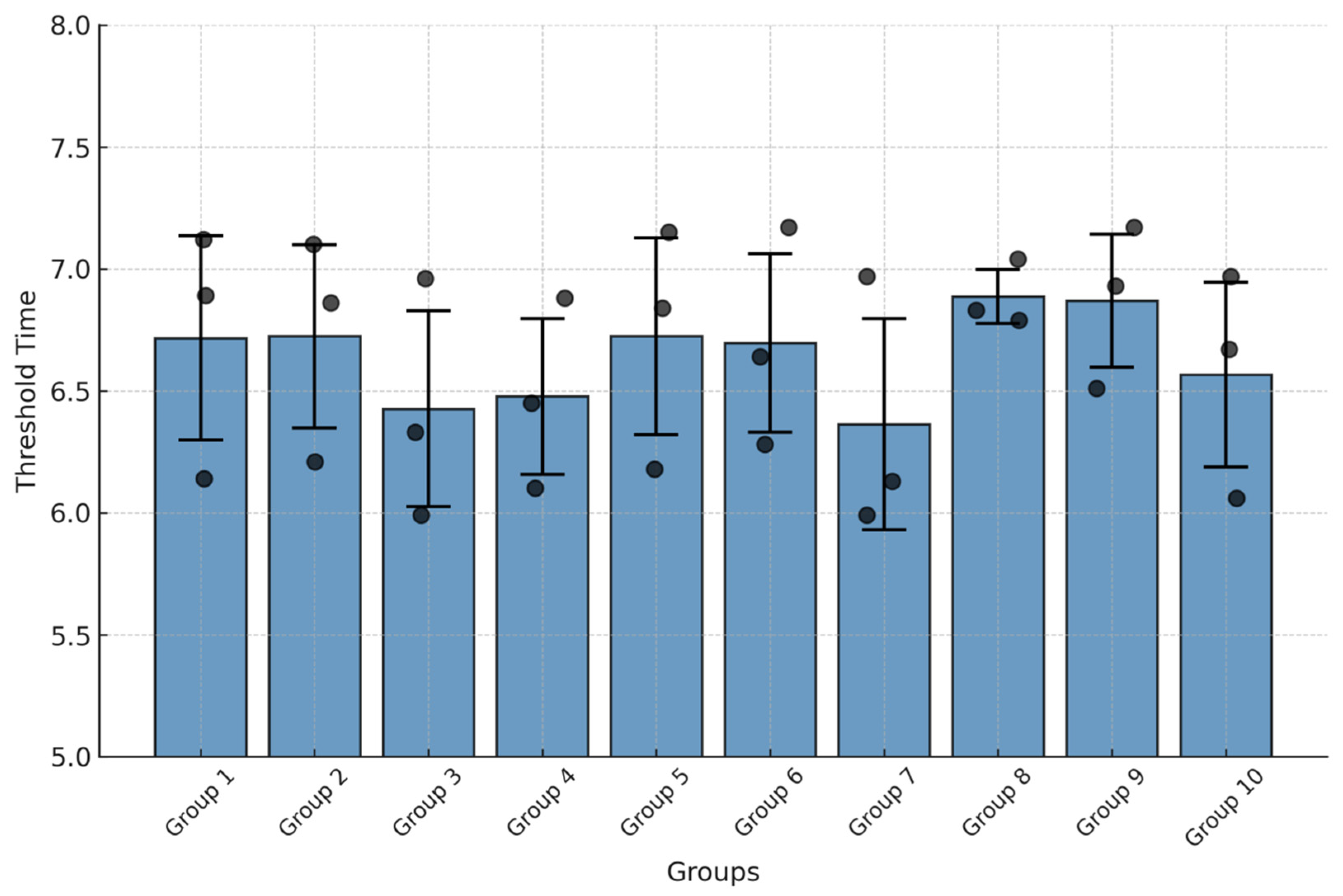
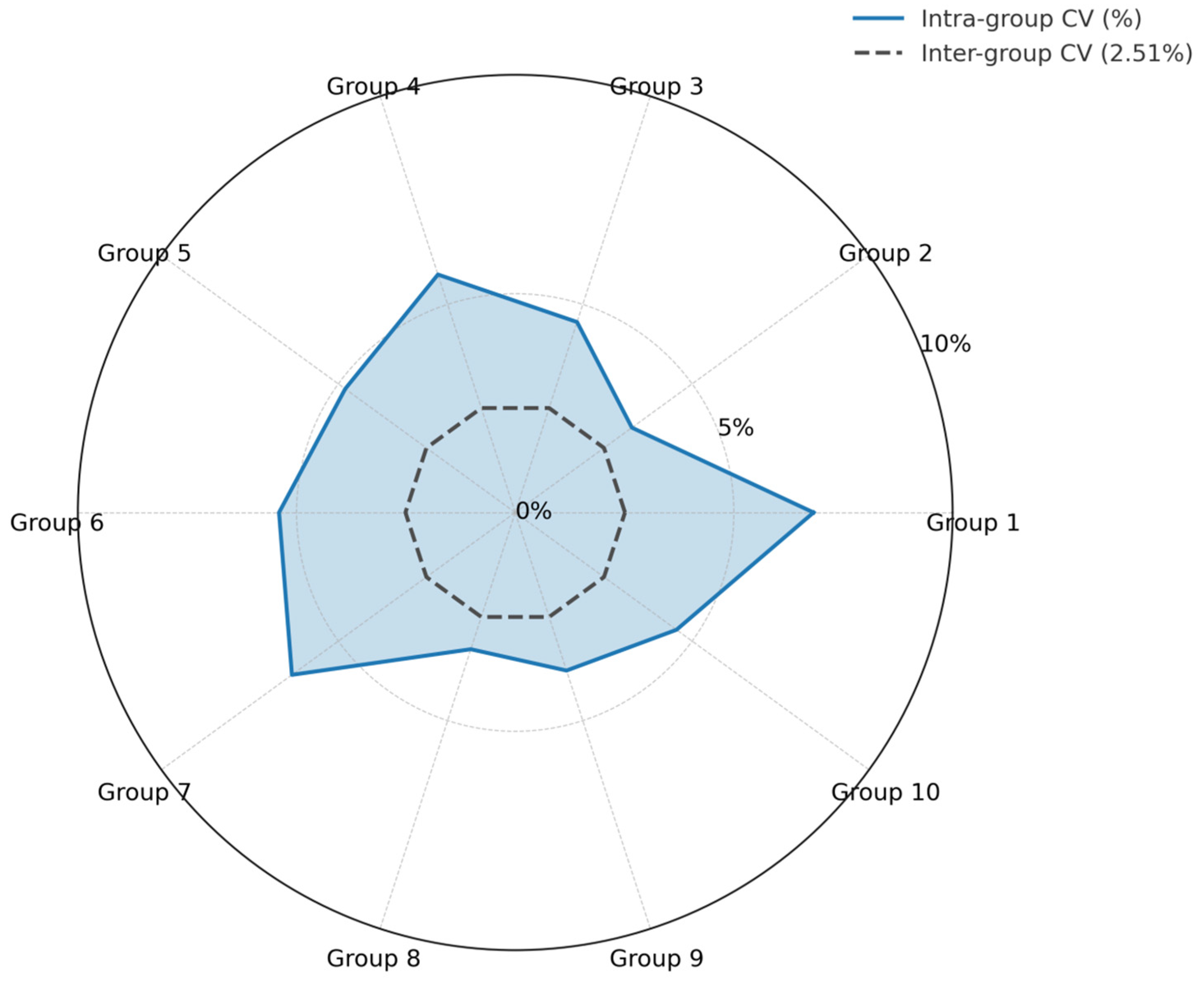
3.4.2. Repeatability of the CAPRV2023 EmOSP-RT-EmDEA Detection System
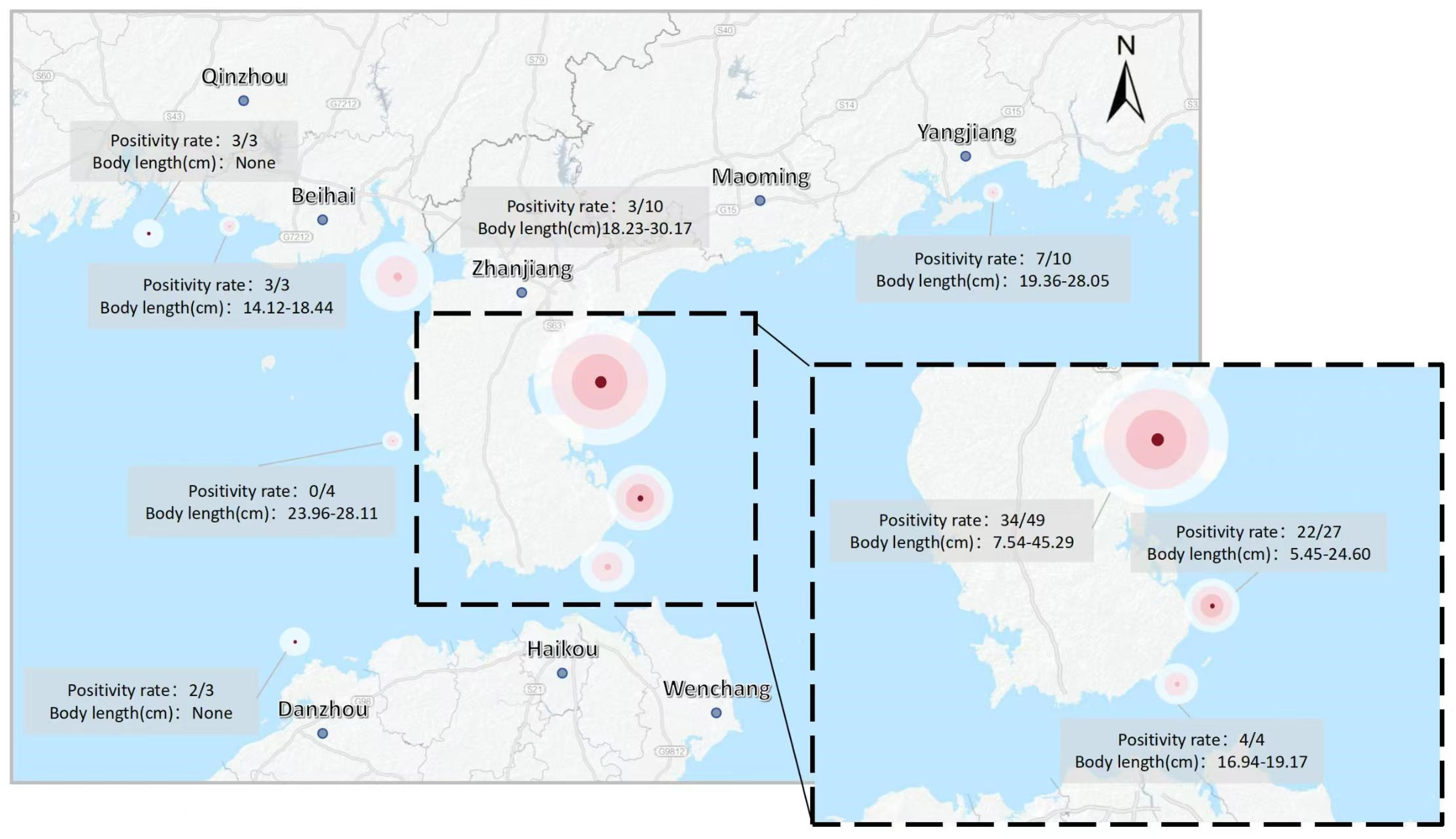
3.4.3. Application
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, H.; Fred, B.; Wang, H.; Huang, J.; Wu, Z.; Wu, D.; Lu, Y.; Jian, J.; Huang, Y. One-step and two-step qPCR assays for CAPRV2023: Development and application in full-cycle epidemiological surveillance of golden pompano. Front. Vet. Sci. 2025, 12, 1620997. [Google Scholar] [CrossRef]
- Sun, H.; Huang, J.; Wang, H.; Zhang, Y.; Fei, Q.; Zhou, J.; Yang, L.; Li, Y.; Jian, J.; Lu, Y.; et al. Mass mortality associated with Carpione rhabdovirus in golden pompano (Trachinotus ovatus) in China: First report. Aquaculture 2025, 595, 741638. [Google Scholar] [CrossRef]
- PRC. 2024 China Fishery Products Report; FAS China Staff, Ed.; PRC: Beijing, China, 2024. [Google Scholar]
- Bovo, G.; Olesen, N.; Jørgensen, P.; Ahne, W.; Winton, J. Characterization of a rhabdovirus isolated from carpione Salmo trutta carpio in Italy. Dis. Aquat. Org. 1995, 21, 115–122. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Balea, R.; Pollak, N.M.; Hobson-Peters, J.; Macdonald, J.; McMillan, D.J. Development and pre-clinical evaluation of a Zika virus diagnostic for low resource settings. Front. Microbiol. 2023, 14, 1214148. [Google Scholar] [CrossRef] [PubMed]
- Zyrina, N.V.; Antipova, V.N. Nonspecific Synthesis in the Reactions of Isothermal Nucleic Acid Amplification. Biochemistry 2021, 86, 887–897. [Google Scholar]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Sun, Z.; Lin, K.-F.; Zhao, Z.-H.; Wang, Y.; Hong, X.-X.; Guo, J.-G.; Ruan, Q.-Y.; Lu, L.-Y.; Li, X.; Zhang, R.; et al. An automated nucleic acid detection platform using digital microfluidics with an optimized Cas12a system. Sci. China Chem. 2022, 65, 630–640. [Google Scholar] [CrossRef]
- Wang, B.; Wang, R.; Wang, D.; Wu, J.; Li, J.; Wang, J.; Liu, H.; Wang, Y. Cas12aVDet: A CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 2019, 91, 12156–12161. [Google Scholar] [CrossRef]
- Habib, S.; Azmai, M.N.A.; Yasin, I.-S.M.; Masdor, N.A.; Said, N.A.M.; Yasid, N.A. Streamlined boiling lysis DNA extraction for Gram-positive aquaculture pathogen Streptococcus agalactiae. Arch. Microbiol. 2024, 206, 435. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, G.; Nguyen, V.G.; Kim, M.C.; Noh, J.Y.; Jang, S.S.; Yoon, S.-W.; Jeong, D.G.; Le Huynh, T.M.; Le, V.P.; Song, D.; et al. Magnetic-bead-based DNA-capture-assisted real-time polymerase chain reaction and recombinase polymerase amplification for the detection of African swine fever virus. Arch. Virol. 2023, 168, 21. [Google Scholar] [CrossRef] [PubMed]
- World Organization for Animal Health (WOAH). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organization for Animal Health: Paris, France, 2016. [Google Scholar]
- Coll, J.M. The glycoprotein G of rhabdoviruses. Arch. Virol. 1995, 140, 827–851. [Google Scholar] [CrossRef]
- Chico, V.; Martinez-Lopez, A.; Ortega-Villaizan, M.; Falco, A.; Perez, L.; Coll, J.M.; Estepa, A. Pepscan Mapping of Viral Hemorrhagic Septicemia Virus Glycoprotein G Major Lineal Determinants Implicated in Triggering Host Cell Antiviral Responses Mediated by Type I Interferon. J. Virol. 2010, 84, 7140–7150. [Google Scholar] [CrossRef]
- Albertini, A.A.V.; Baquero, E.; Ferlin, A.; Gaudin, Y. Molecular and Cellular Aspects of Rhabdovirus Entry. Viruses 2012, 4, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Laing, K.J.; Winton, J.R. Immunity to fish rhabdoviruses. Viruses 2012, 4, 140–166. [Google Scholar] [CrossRef]
- Ahmadivand, S.; Palić, D.; Weidmann, M. Molecular Epidemiology of Novirhabdoviruses Emerging in Iranian Trout Farms. Viruses 2021, 13, 448. [Google Scholar] [CrossRef]
- Abdulgani, N.; Sudaryatma, P.E.; Berliana, N.P.S. (Eds.) Comparison of RT-PCR and rRT-PCR Methods in Detection of Viral Hemorrhagic Septicemia Virus (VHSV) in Marine Ornamental Fish. BIO Web Conf. 2024, 89, 02007. [Google Scholar]
- Chan, K.G.; Ang, G.Y.; Yu, C.Y.; Yean, C.Y. Harnessing CRISPR-Cas to Combat COVID-19: From Diagnostics to Therapeutics. Life 2021, 11, 1210. [Google Scholar] [CrossRef]
- Chen, L.; Hu, M.; Zhou, X. Trends in developing one-pot CRISPR diagnostics strategies. Trends Biotechnol. 2025, 43, 98–110. [Google Scholar] [CrossRef]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.; Drew, T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef]
- Dong, J.; Feng, W.; Lin, M.; Chen, S.; Liu, X.; Wang, X.; Chen, Q. Comparative Evaluation of PCR-Based, LAMP and RPA-CRISPR/Cas12a Assays for the Rapid Detection of Diaporthe aspalathi. Int. J. Mol. Sci. 2024, 25, 5773. [Google Scholar] [CrossRef] [PubMed]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Li, Z.; Feng, W.; Zhu, Z.; Lu, S.; Lin, M.; Dong, J.; Wang, Z.; Liu, F.; Chen, Q. Cas-OPRAD: A one-pot RPA/PCR CRISPR/Cas12 assay for on-site Phytophthora root rot detection. Front. Microbiol. 2024, 15, 1390422. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, F.; Xue, B.; Zhou, X. Field-deployable assay based on CRISPR-Cas13a coupled with RT-RPA in one tube for the detection of SARS-CoV-2 in wastewater. J. Hazard. Mater. 2023, 459, 132077. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Yu, G.; Sun, W.; Aizaz, M.; Yang, G.; Chen, L. One-tube RPA-CRISPR Cas12a/Cas13a rapid detection of methicillin-resistant Staphylococcus aureus. Anal. Chim. Acta 2023, 1278, 341757. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mukama, O.; Wu, W.; Li, Z.; De Dieu Habimana, J.; Zhang, Y.; Zeng, R.; Nie, C.; Zeng, L. A CRISPR/Cas12a Based Universal Lateral Flow Biosensor for the Sensitive and Specific Detection of African Swine-Fever Viruses in Whole Blood. Biosensors 2020, 10, 203. [Google Scholar] [CrossRef]
- Bhardwaj, P.; Dhangur, P.; Kalichamy, A.; Singh, R. RT-RPA Assisted CRISPR/Cas12a Based One-Pot Rapid and Visual Detection of the Pan-Dengue Virus. J. Med. Virol. 2025, 97, e70219. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Othman, I.; Helmi, A.; Slama, I.; Hamdi, R.; Mastouri, M.; Aouni, M. Evaluation of three viral concentration methods for detection and quantification of SARS-CoV-2 in wastewater. J. Water Health 2023, 21, 354–360. [Google Scholar] [CrossRef] [PubMed]


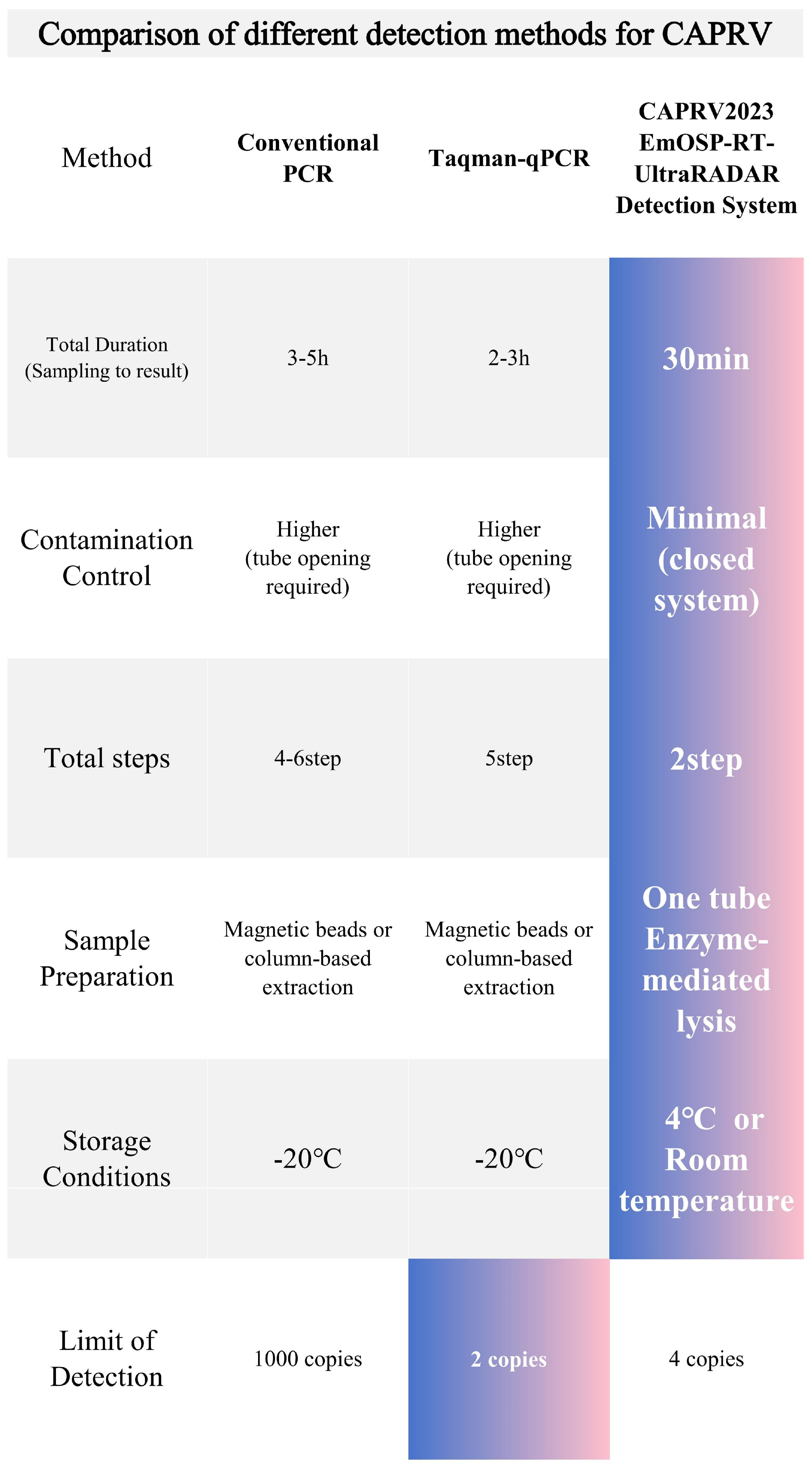
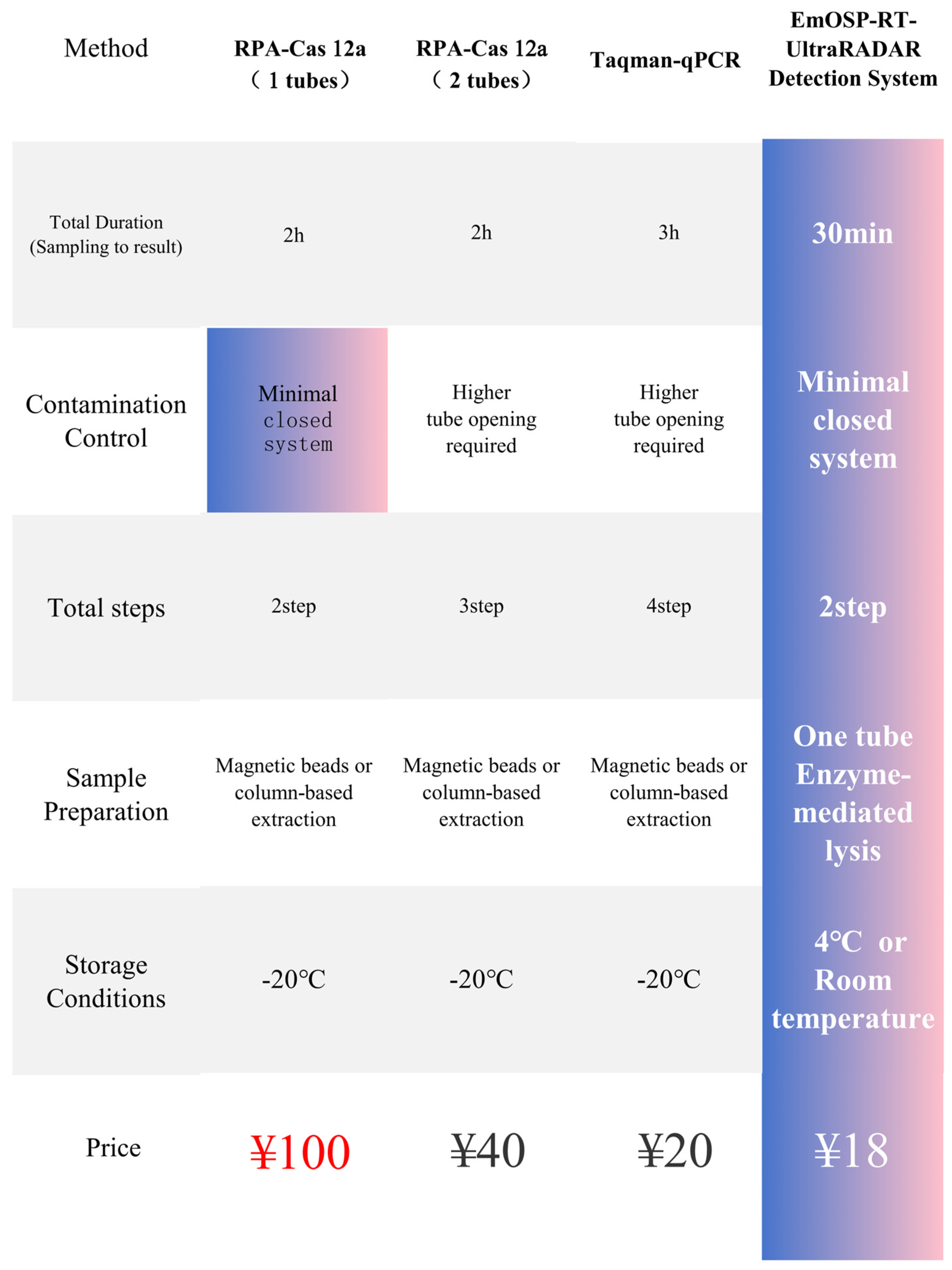
| Types of Sequence | Primer Names | Sequence |
|---|---|---|
| Forward primer sequence | CAP-ULTRA-F1 | 5’-CAACCTATCCCACCAAGAACACTGCTGA-3’ |
| CAP-ULTRA-F2 | 5’-ATCCCACCAAGAACACTGCTGAGGGACG-3’ | |
| CAP-ULTRA-F3 | 5’-CCAAGAACACTGCTGAGGGACGAATCTT-3’ | |
| CAP-ULTRA-F4 | 5’-ACACTGCTGAGGGACGAATCTTTACAAG-3’ | |
| CAP-ULTRA-F5 | 5’-CTGAGGGACGAATCTTTACAAGAAAGGC-3’ | |
| CAP-ULTRA-F6 | 5’-GACGAATCTTTACAAGAAAGGCAGATCA-3’ | |
| RNA probe sequences | 2-GD-RNA1 | 5’6-FAM-rApp -CACAAUCAUAAAGGCGGUUGCGCCUGGA-3`BHQ1 |
| 2-GD-RNA2 | 5’6-FAM-rApp -CAUAAAGGCGGUUGCGCCUGGACACCAU-3`BHQ1 | |
| 2-GD-RNA3 | 5’6-FAM-rApp -GGCGGUUGCGCCUGGACACCAUCCAUGG-3`BHQ1 | |
| 2-GD-RNA4 | 5’6-FAM-rApp -UGCGCCUGGACACCAUCCAUGGGGACUU-3`BHQ1 | |
| 2-GD-RNA5 | 5’6-FAM-rApp -UGGACACCAUCCAUGGGGACUUACCAAG-3`BHQ1 | |
| 2-GD-RNA6 | 5’6-FAM-rApp -CCAUCCAUGGGGACUUACCAAGGCCUGC-3`BHQ1 | |
| Reverse primer sequence | CAP-ULTRA-R1 | 5’-AAGCTAATACGACTCACTATAGGGTCCATTGTTCTCCAGCAAACTGCAGTGT-3’ |
| CAP-ULTRA-R2 | 5’-AAGCTAATACGACTCACTATAGGGTCCTGATCCATTGTTCTCCAGCAAACTG-3’ | |
| CAP-ULTRA-R3 | 5’-AAGCTAATACGACTCACTATAGGGGGTCTGTCCTGATCCATTGTTCTCCAGC-3’ | |
| CAP-ULTRA-R4 | 5’-AAGCTAATACGACTCACTATAGGGCCCCGAGGTCTGTCCTGATCCATTGTTC-3’ | |
| CAP-ULTRA-R5 | 5’-AAGCTAATACGACTCACTATAGGGTGAGGTCCCCGAGGTCTGTCCTGATCCA-3’ | |
| CAP-ULTRA-R6 | 5’-AAGCTAATACGACTCACTATAGGGTTTGGATGAGGTCCCCGAGGTCTGTCCT-3’ | |
| Taq Man-qPCR primer | CAP-GQF | 5’-TGCATGATCGGTCCATGGACTG-3’ |
| CAR-GQR | 5’-GTTTGGACTCTCTAGCTAGCTAG-3’ | |
| CAR-GProbe | 5’6-FAM-GTGGGGTACGATCGCATCCCAGATGATCGATG-3`BHQ1 | |
| CAPRV-G-Clone | CAP-G-COPY-F | 5’-CGCAACCGCCTTTATGATTG-3’ |
| CAP-G-COPY-R | 5’-GTCCCCTTCCTGGGTGATGAGG-3’ |
| Types of Primers | Primer Names | Sequence |
|---|---|---|
| RT-Primer | CAP-RT1 | CCACCA |
| CAP-RT2 | AACTTT | |
| CAP-RT3 | AATGAG | |
| CAP-RT4 | CATATT | |
| CAP-RT5 | CCCATC | |
| CAP-RT6 | TTGGCA | |
| CAP-RT7 | TGACCC | |
| CAP-RT8 | TCGTCC | |
| CAP-RT9 | TAACCA | |
| CAP-RT10 | TGATTG | |
| CAP-RT11 | CACAAT | |
| CAP-RT12 | TTTCAT | |
| CAP-RT13 | CTCCCG | |
| CAP-RT14 | AAGGCA | |
| CAP-RT15 | TGTGAG | |
| CAP-RT16 | CAATCT | |
| CAP-RT17 | TCTGAG | |
| CAP-RT18 | GAGACT | |
| CAP-RT19 | GGTAGG | |
| CAP-RT20 | TGACAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Wang, H.; Huang, J.; Wu, Y.; Hu, Z.; Huang, Y. Integrated Enzyme-Mediated One-Step Sample Processing and Duplex Amplification System for Rapid Detection of Carpione rhabdovirus in Aquaculture-Derived Food Products. Foods 2025, 14, 3929. https://doi.org/10.3390/foods14223929
Sun H, Wang H, Huang J, Wu Y, Hu Z, Huang Y. Integrated Enzyme-Mediated One-Step Sample Processing and Duplex Amplification System for Rapid Detection of Carpione rhabdovirus in Aquaculture-Derived Food Products. Foods. 2025; 14(22):3929. https://doi.org/10.3390/foods14223929
Chicago/Turabian StyleSun, Heng, Haoyu Wang, Jie Huang, Yao Wu, Zhenxin Hu, and Yucong Huang. 2025. "Integrated Enzyme-Mediated One-Step Sample Processing and Duplex Amplification System for Rapid Detection of Carpione rhabdovirus in Aquaculture-Derived Food Products" Foods 14, no. 22: 3929. https://doi.org/10.3390/foods14223929
APA StyleSun, H., Wang, H., Huang, J., Wu, Y., Hu, Z., & Huang, Y. (2025). Integrated Enzyme-Mediated One-Step Sample Processing and Duplex Amplification System for Rapid Detection of Carpione rhabdovirus in Aquaculture-Derived Food Products. Foods, 14(22), 3929. https://doi.org/10.3390/foods14223929






