Abstract
Clostridium perfringens is responsible for various diseases. Foodborne outbreaks (FBOs) result from the in situ production of C. perfringens enterotoxin (CPE) by type F strains during sporulation. The cpe gene can be plasmidic (p-cpe) or chromosomal (c-cpe). Strains (c-cpe) exhibit greater heat resistance and are frequently associated with FBO. Strains cpe-negative are considered heat-sensitive. This study investigates the sporulation abilities and heat resistance of eight C. perfringens strains isolated from French foodborne outbreaks. Whole-genome sequencing classified the strains into two clades: the “chromosomal cpe clade,”, mainly composed of cpe-positive strains with c-cpe and some cpe-negative strains, and the “plasmidic cpe clade,”, primarily containing cpe-negative strains and a few with plasmid-borne cpe. Sporulation assays and thermal inactivation kinetics were performed on FBO strains to evaluate the influence of genetic variability on sporulation abilities and heat resistance. Experimental analyses revealed that strains within the “chromosomal cpe clade” exhibited the highest sporulation abilities, regardless of cpe presence, while those in the “plasmidic cpe clade” had low sporulation ability. Moreover, heat-resistant spores were produced exclusively by strains of the “chromosomal cpe clade,” with c-cpe strains exhibiting the highest heat resistance (δ95 °C ≈ 49 min), followed by cpe-negative strains (δ95 °C ≈ 9.5 min). p-cpe strains exhibited a heat-sensitive phenotype, with δ85 °C values of 12 min. A key finding of this study is the identification of a group with intermediate heat resistance, distinct from the highly heat-resistant (c-cpe) and heat-sensitive (p-cpe) strains. This intermediate heat-resistance phenotype, observed in cpe-negative strains within the “chromosomal cpe clade,” offers a new perspective on C. perfringens stress adaptation, suggesting their potential for persistence in food. Their heat resistance, along with the potential for cpe gene transfer, could make these strains a relevant hazard for cooked, cooled, and re-heated meat products.
1. Introduction
Clostridium perfringens is a Gram-positive, spore-forming anaerobic bacterium found in various environments and is responsible for a variety of human and animal diseases. The ability of C. perfringens to cause disease in various hosts is primarily attributed to its arsenal of virulence factors, including toxins and enzymes [1,2]. The production patterns of six toxins (α-toxin, β-toxin, ε-toxin, ι-toxin, CPE, and NetB) among different strains form the basis for a recently revised classification scheme [3]. This new classification scheme categorizes C. perfringens isolates into one of seven toxinotypes from A to G (Table 1), which improves epidemiological and diagnostic value in infections in both humans and animals [3]. The toxin-based typing method for C. perfringens has shown that different toxinotype strains have distinct host preferences and are linked to specific diseases (Table 1) [2,3]. C. perfringens type F produces the α-toxin and, upon sporulation, also produces the C. perfringens enterotoxin (CPE), which is responsible for food poisoning (FP).
CPE is considered the main toxin responsible for symptoms in foodborne illnesses caused by C. perfringens [4]. This enterotoxin is produced by all type F strains and by some type C, D, and E strains [2,3].

Table 1.
C. perfringens toxinotype classification scheme and disease association.
Table 1.
C. perfringens toxinotype classification scheme and disease association.
| Toxinotype | α-Toxin, CPA (plc or cpa) | β-Toxin, CPB (cpb) | ε-Toxin, ETX (etx) | ι-Toxin, ITX (iap and ibp) | CPE (cpe) | NetB (netB) | Main Diseases and Affected Species |
|---|---|---|---|---|---|---|---|
| A | + | − | − | − | − | − | Gas gangrene in humans and several animals; possible involvement in enterotoxemia and GI disease in ruminants, horses, and pigs; hemorrhagic gastroenteritis in dogs and horses |
| B | + | + | + | − | − | − | Lamb dysentery |
| C | + | + | − | − | +/− | − | Hemorrhagic and necrotizing enteritis in several neonatal animals; struck; enteritis necroticans (pig-bel, Darmbrand) in humans |
| D | + | − | + | − | +/− | − | Enterotoxemia in sheep, goats, and cattle; enterocolitis in goats |
| E (*) | + | − | − | + | +/− | − | Possible involvement in gastroenteritis of cattle and rabbits |
| F | + | − | − | − | + | − | Human food poisoning, antibiotic-associated diarrhea and sporadic diarrhea |
| G | + | − | − | − | − | + | Necrotic enteritis in poultry |
(*) Type E strains either carry a functional or a silent cpe gene [5]; (+) toxin produced; (−) toxin not produced.
Food poisoning can occur when food contaminated with a high concentration of vegetative cells (i.e., >106 to 107 vegetative cells/gram of food) from a type F strain of C. perfringens is ingested [6]. Many of these bacteria are destroyed by gastric acids but some survive and reach the intestines. After a growth phase, C. perfringens undergoes sporulation and synthesizes CPE. The exact triggers for in vivo sporulation are not fully understood, but may involve exposure to stomach acidity, bile salts, or phosphate in the intestines [6,7].
Approximately 5% of global C. perfringens isolates carry the cpe gene and produce CPE, making the isolation of cpe-positive strains relatively uncommon [4,8,9,10,11]. The cpe gene can be located either on plasmids or on a transposable element integrated into the chromosome, and both plasmid-mediated (p-cpe) and chromosomal (c-cpe) strains can cause foodborne outbreaks (FBOs) [9,10]. Approximately 70% of type F human food poisoning isolates carry their cpe gene on the chromosome. The remaining 30% of food poisoning strains, and virtually all non-foodborne human gastrointestinal (GI) diseases cpe-positive strains, carry their cpe gene on plasmids [4,12]. A recent study [1] conducted on a collection of 141 strains of C. perfringens detected in 42 FBOs in the Paris region have shown that 79 strains were assigned to C. perfringens type F (56%), 61 strains were assigned to C. perfringens type A (43%), and one strain was assigned to C. perfringens type E (1%). Genomic analysis performed on 58 strains revealed that C. perfringens strains carrying the cpe gene on the chromosome (24/58) are more abundant compared to strains with plasmid-mediated cpe (3/58), confirming the prominent implication of chromosomal cpe (c-cpe) strains in human food poisoning [1,12]. Many studies suggest that the specific association between chromosomal cpe isolates and C. perfringens type F food poisoning is partly due to the significantly greater heat resistance of chromosomal cpe isolates compared to plasmid cpe isolates [8,9]. This enhanced heat resistance allows chromosomal cpe isolates to survive better in cooked meat products, which are the major vehicle implicated in C. perfringens type F outbreaks [1,13,14].
Several known mechanisms contribute to the heat resistance of spores within Clostridia species, particularly C. perfringens, which share similarities with the spores of Bacillus subtilis [15,16,17]. The main factor responsible for spore heat resistance is the low water content of the protoplast or the spore core [18,19,20]. In addition to the water content of the protoplast, other factors may contribute to spore resistance: (i) The accumulation of dipicolinic acid (DPA) and its chelated bivalent cations (Ca2+) in the spore core helps reduce its water content during sporulation and consequently increases resistance to wet heat [21,22,23]. (ii) The nature of the mineral cation associated with DPA. In fact, spores formed in the presence of Ca2+ exhibit greater protection against moist heat compared to those formed with other divalent cations (Mg2+, Mn2+) or monovalent cations (K+, Na+) [21,24]. (iii) The saturation of DNA with small acid-soluble spore proteins (SASPs) which protect DNA from heat [19,20]. SASPs are a class of low-molecular weight proteins that bind to spore DNA and provide protection from various environmental stresses. For C. perfringens food poisoning strains, a variant allele of small acid-soluble proteins ssp4 was associated with spore’s heat resistance [12,25]. However, a recent study has reported a heat-resistant phenotype for a c-cpe strain without the presence of heat resistance-associated allele of Ssp4 [9].
Although the sporulation and heat resistance of C. perfringens have been investigated in previous studies, none have compared sporulation abilities between cpe-negative strains and cpe-positive strains (c-cpe and p-cpe). Moreover, in the reviewed literature, it seems that cpe-negative strains have generally been considered as heat-sensitive, with studies mainly focusing on heat resistance comparison between c-cpe and p-cpe strains [9,26,27]. Accordingly, the present study aims to examine the sporulation abilities and the heat-resistant characteristics of eight strains of C. perfringens detected in French FBO (cpe-negative and cpe-positive strains). A comparison was made with other studies to examine the link between sporulation, heat resistance, and the presence or absence of the cpe gene and its location (on the chromosome or on the plasmid).
The last sentence of the paragraph refers exclusively to C. perfringens.
2. Materials and Methods
2.1. Bacterial Strains
A total of eight C. perfringens strains isolated from foods involved in French FBOs from 2016 to 2017 were selected and used for sporulation and heat resistance assays (Table 2). Strains were classified and selected according to their affiliation with either the “chromosomal cpe clade” or to the “plasmidic cpe clade”. The “chromosomal cpe clade” is a clade that contains mainly cpe-positive strains carrying the cpe gene on a chromosome (c-cpe) and few strains in which the cpe gene is absent [1]. The “plasmidic cpe clade” is a clade that contains mainly cpe-negative strains and few cpe-positive strains with plasmid-mediated cpe (p-cpe) [1]. The phylogenetic tree was published in 2019 [1]. C. perfringens NCTC 8239 (ATCC 12917; CIP 104880), originally isolated from boiled salted beef, was purchased from the Pasteur Institute collection and used in this study as a control strain for sporulation and heat resistance essays. This strain carries a chromosomal cpe (c-cpe) [7,15].

Table 2.
Bacterial strains of C. perfringens used in this study.
2.2. Media Preparation for Growth and Sporulation
Tryptone–Peptone–Glucose (TPG) supplemented with sodium thioglycolate was used as liquid growth media for all strains of C. perfringens. Tryptone and peptone were purchased from Oxoid (Lyon, France). Glucose and sodium thioglycolate were purchased from Oxoid (France) and Merck (Lyon, France), respectively. For sporulation of C. perfringens, modified Tryptone–Peptone–Glucose (m-TPG) medium was used. Tryptone–Peptone–Glucose used for sporulation was supplemented with sodium thioglycolate (1.0 g L −1, MW 114.10, Merck, France), MnSO4 (5.0 mg L−1, MW 169.02, Sigma-Aldrich, Fallavier, France) and CaCl2 (5.0 mg L−1, MW 110.99, Thermoscientific, Illkirch, France). The pH was adjusted to 7.5 with NaOH (1 M) in the presence of phosphate-buffered saline (KCl 2.7 mM, NaCl 140 mM, Phosphate 10 mM, PanReac AppliChem, Darmstadt, Germany). Glass jars containing sporulation media (m-TPG) were sealed and autoclaved at 121 °C for 15 min. Finally, sodium taurocholate (MW 537, Merck, France) was added after sterilization at a final concentration of 500 µM.
2.3. Spore Production
For all experiments, a first pre-culture was obtained from a vegetative cell stock of cryogenic beads (CRYOBEADS, bioMérieux, Marcy-l’Étoile, France). This first pre-culture was carried out by adding a cryogenic bead to 9 mL of TPG contained in a sealed jar in the presence of an anaerobic generator (GENbox anae, bioMérieux, France) at 37 °C for 24 h to reach an approximate concentration of 7.0 log10 CFU/mL. An anaerobe indicator (Anaer Indicator, bioMéreux, France) was introduced in sealed jars with media flasks to check anaerobiosis. A second pre-culture was carried out by inoculation of 9 mL of TPG with 100 µL of the first pre-culture, followed by anaerobic incubation at 37 °C for 6 h. The second pre-culture (300 µL) was used to inoculate 30 mL of sporulation media; then, sporulation was conducted for three days at 37 °C under anaerobic conditions. Spores were harvested by centrifugation, washed three times (3000 rpm for 15 min) in sterile ultrapure water (Millipore Simplicity Water Purification System, Merck, France) and kept at 4 °C. Sporulation was verified using a phase-contrast microscope (Olympus BX51, Olympus, Rungis, France, 10 × 100 Oil). To deactivate vegetative cells and ensure that only spores were recovered and counted, the produced spore suspension was heat-treated in a water bath at 75 °C for 15 min. Spore concentration was evaluated after the heat treatment by plating on Columbia agar + 5% sheep blood (bioMérieux, France).
2.4. Heat Resistance Assays
Inactivation kinetics were performed at 85 °C or 95 °C according to the capillary tube method, allowing heat treatment in isothermal conditions. Glass capillary tubes with a 200 µL capacity (Hirschman Laborgerate, Germany) were filled with 100 µL of spore suspension and then sealed at both ends with a gas burner flame. The capillary tubes were heated at 85 °C or 95 °C in a thermostated glycerol/water bath with a circulating-water pump to maximize heat exchange. Immediately after heat treatment, the glass capillary tubes were placed in melting ice. The glass tubes were washed, broken, and poured into 0.9 mL of tryptone salt broth [28,29]. The heat-treated spores were counted by plating on Columbia agar + 5% sheep blood after anaerobic incubation at 37 °C for 24 h.
2.5. Heat Resistance Estimation
Survival curves (log CFU/mL vs. heating time) were fitted by the model based on the Weibull distribution proposed by Mafart et al. (2002) [30]:
where N is the population size at time t (CFU/mL), N0 is the initial population size (CFU/mL), δ is the first decimal reduction time that led to a 10-fold reduction (min), and p is the shape parameter of the survivor curve. For p > 1, the curve is convex; for p < 1, the curve is concave; and for p = 1, the curve is log-linear. A single p value was estimated for the whole set of data. The initial population size, as well as the first decimal reduction time, was estimated for each survival curve.
2.6. Statistical Analysis
A non-linear fitting function (lsqcurvefit function, Optimization Toolbox, MATLAB R2024b, The MathWorks, Natick, MA, USA) was used to estimate δ-values. Goodness of fit and the performance of the inactivation model was evaluated by calculating the following indicators: the coefficient of determination (R2), the mean squared error (MSE), the root mean square error (RMSE) and the corrected Akaike Information Criterion (AICc). The closer R2 is to 1, the better the prediction. For MSE and RMSE, a value close to zero indicates a smaller random error component. The corrected Akaike Information Criterion (AICc) was calculated to evaluate the fitting performance of the model used for heat resistance estimation. A lower AICc reveals an appropriate model for prediction [31]. A multiple comparison procedure (anova1 and multcompare function, Statistical Toolbox, Matlab R2024b, The MathWorks, Natick, MA, USA) was implemented to assess the impact of strain variability on spore production ability and heat resistance abilities.
3. Results
3.1. Sporulation Abilities of C. perfringens Strains
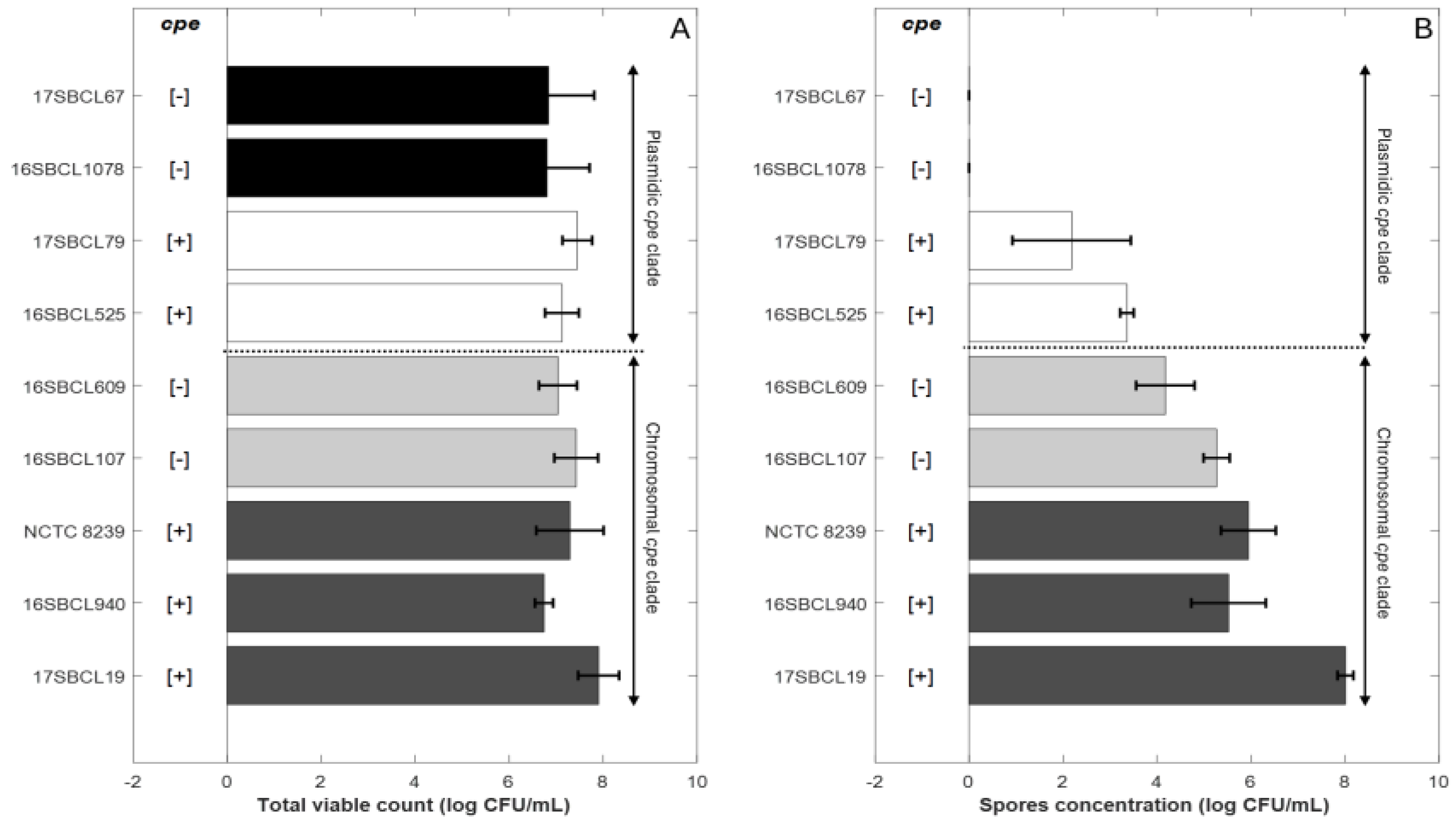
The spore quantities produced by the control strains C. perfringens NCTC 8239 and the FBO strains were estimated. The final spore concentration obtained for each strain following heat treatment at 75 °C for 15 min (Figure 1B) was compared to the total viable count (Figure 1A) obtained after three days of sporulation without heat treatment.
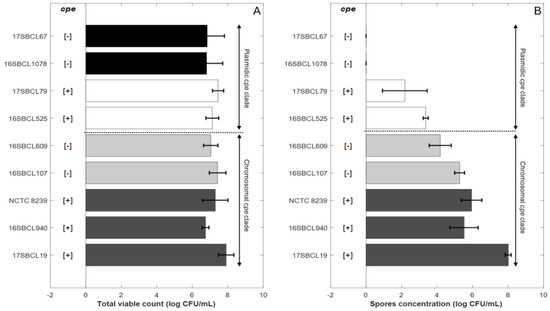
Figure 1.
Sporulation abilities of Clostridium perfringens strains. Control strains NCTC 8239 (ATCC 12917/CIP 104880) and eight FBO strains were included in the study. The “chromosomal cpe clade” is a clade that contains mainly cpe-positive strains carrying the cpe gene on a chromosome (c-cpe) and few strains in which the cpe gene is absent [1]. The “plasmidic cpe clade” is a clade that contains mainly cpe-negative strains and few cpe-positive strains with plasmid-mediated cpe (p-cpe) [1]. (A) represents the total viable count obtained after three days of sporulation without heat treatment for the different strains. (B) represents the spore concentration for each strain following heat treatment at 75 °C for 15 min. Dark grey bars (c-cpe strains), light grey bars (cpe-negative strains belonging to chromosomal cpe clade), white bars (p-cpe strains), black bars (cpe-negative strains belonging to plasmidic cpe clade). [+]: presence of cpe gene. [−]: absence of cpe gene.The plot was generated in MATLAB R2024b.
The strains were classified based on their belonging to the “chromosomal cpe clade” or to the “plasmidic cpe clade” [1]. The details of the classification are provided in the Section 2.
For strains belonging to the “chromosomal cpe clade”, final mean spore concentrations were about 6.24 (±1.11) log10 CFU/mL for chromosomal cpe-positive strains (17SBCL19, 16SBCL940, NCTC 8239) and about 4.72 (±0.73) log10 CFU/mL for the cpe-negative strains (16SBCL107, 16SBCL609) (Figure 1.B). For strains belonging to the “plasmidic cpe clade”, final mean spore concentrations were about 2.77 (±1.02) log10 CFU/mL for plasmid-mediated cpe-positive strains (16SBCL525, 17SBCL79). No spores were detected in our experimental conditions for cpe-negative strains belonging to the “plasmidic cpe clade” (16SBCL1078, 17SBCL67).
The results show that the greatest spore production was generally observed for strains belonging to “chromosomal cpe clade” whether or not they carried the cpe gene. In comparison, strains belonging to “plasmidic cpe clade” exhibited a low ability to produce spores under our experimental conditions. A significant difference in spore production was observed between cpe-positive strains carrying the cpe gene on a chromosome (c-cpe) compared to (p-cpe) strains (p value < 0.05). Furthermore, within the “chromosomal cpe clade”, cpe-negative strains did not show a significant difference in spore production compared to cpe-positive strains, except for the strain 17SBSC19, which produced the highest spore concentration (7.9 ± 0.43 log10 CFU/mL).
3.2. Heat Resistance Characterization of C. perfringens Strains
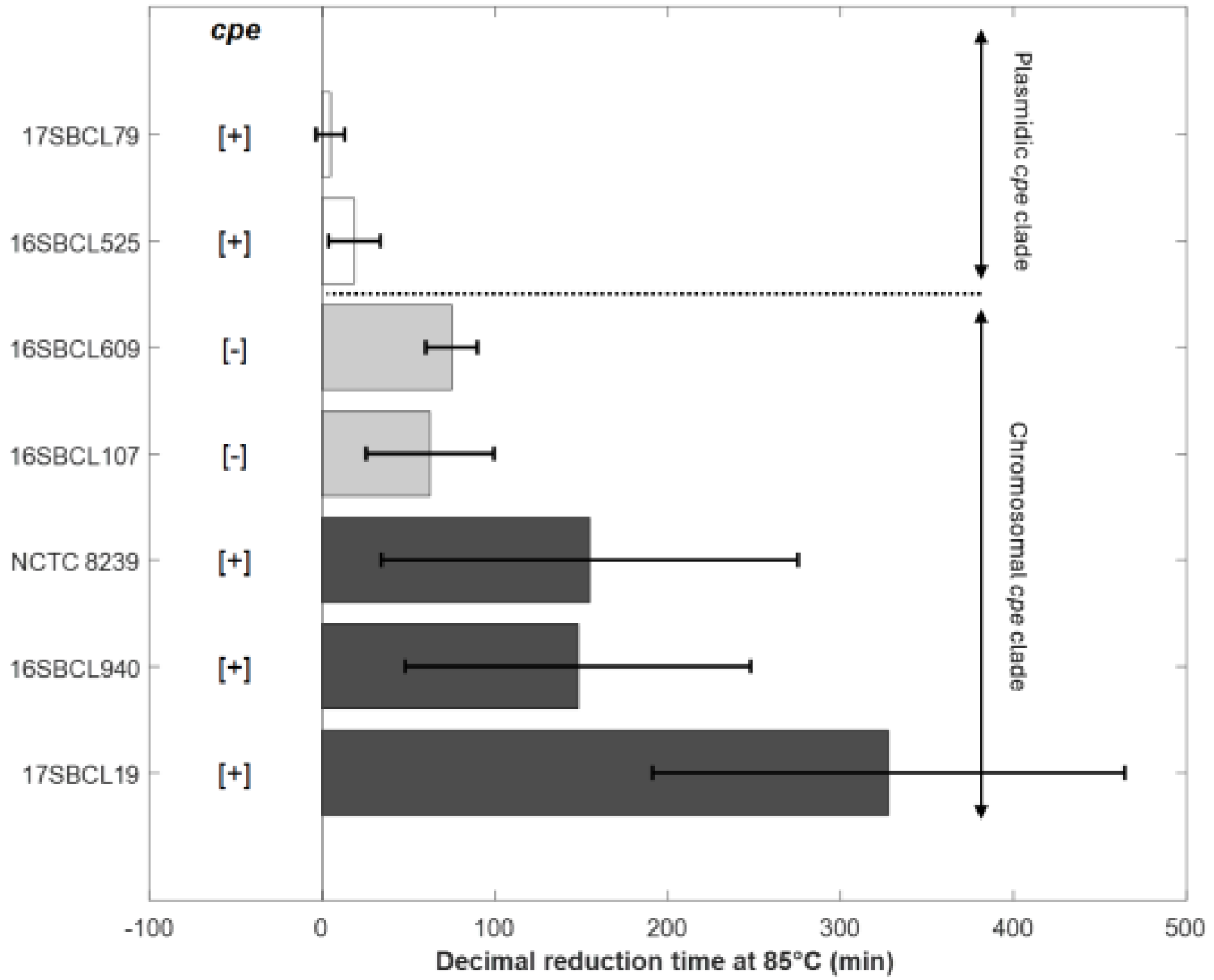
To investigate the effect of strain genetic variability on heat resistance properties, thermal inactivation kinetics were performed at 85 °C or 95 °C on spore suspensions produced as described previously. Triplicates were performed for each strain from three independent spore batches. Typical survivals curves were obtained and then fitted to the Weibull model. The spore heat resistance for each strain was estimated using the first decimal reduction time (δ) (Table 3). The goodness of fit was evaluated by calculating the RMSE, AICc, and other statistical indicators (Table 3). The results show that the model provided a good fit to the data (34 survival curves) and accurately describes the thermal inactivation kinetics of C. perfringens spores. The thermal inactivation curves exhibited a concave shape, with a p-value estimated at 0.68 (Table 3).

Table 3.
Estimation of heat resistance parameters (δ and p-value) after a heat treatment at 85 °C and 95 °C for the control strains NCTC 8239 (ATCC 12917/CIP 104880) and six FBO strains. The mean values of heat resistance parameters for each strain, standard deviation, and goodness of fit indicators were calculated and evaluated using MATLAB R2024b.
To characterize the heat resistance phenotypes associated with the two clades established by Abdelrahim et al. (2019) [1], spores produced from the sequenced strains underwent heat resistance assays. As shown in Figure 2 and Table 3, C. perfringens strains carrying a chromosomal cpe gene (c-cpe) produced highly heat-resistant spores with δ85 °C values over 100 min and an average of 210 min. However, strains with a plasmid cpe gene (p-cpe) produced heat-sensitive spores with δ85 °C values ranging from 5 to 19 min (Table 3). A significant difference (p value < 0.05) was observed between c-cpe strain (17SBCL19) and two p-cpe strains (16SBCL525, 17SBCL79) (Figure 2).
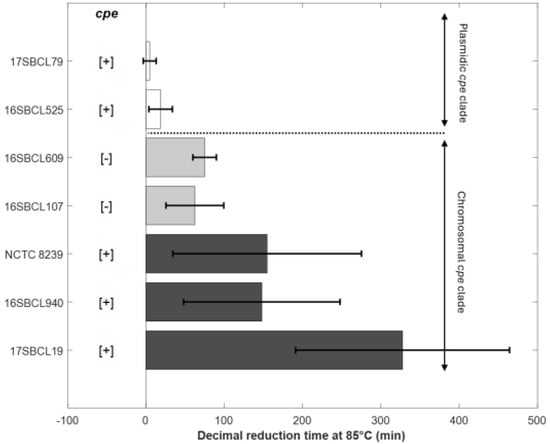
Figure 2.
Decimal reduction time (min) obtained at 85 °C for the control strains NCTC 8239 (ATCC 12917/CIP 104880) and six FBO strains. The error bars indicate standard deviation. The “chromosomal cpe clade” is a clade that contains mainly cpe-positive strains carrying the cpe gene on a chromosome (c-cpe) and few strains in which the cpe gene is absent [1]. The “plasmidic cpe clade” is a clade that contains mainly cpe-negative strains and few cpe-positive strains with plasmid-mediated cpe (p-cpe) [1]. Dark grey bars (c-cpe strains), light grey bars (cpe-negative strains belonging to chromosomal cpe clade), white bars (p-cpe strains). [+]: presence of cpe gene. [−]: absence of cpe gene. The plot was generated in MATLAB R2024b.
Interestingly, cpe-negative strains belonging to the “chromosomal cpe clade” produced relatively heat-resistant spores with δ85 °C values ranging from 62 to 75 min (Table 3). To further investigate the differences between cpe-positive and cpe-negative strains within the “chromosomal cpe clade”, we compared their δ95 °C values (Table 3). Indeed, a heat treatment at this temperature is more suitable for studying the decimal reduction time of these heat-resistant strains. No significant difference was observed among the strains within the “chromosomal cpe clade,” regardless of the presence or absence of the cpe gene (p value < 0.05). Our results show that cpe-negative strains belonging to the “chromosomal cpe clade” form a group with an intermediate level of heat resistance, positioned between the highly heat-resistant strains (c-cpe) and the heat-sensitive strains (p-cpe).
We also acknowledge that under our experimental conditions, no spores were recovered after a heat treatment at 75 °C for 15 min intended to eliminate vegetative cells of cpe-negative strains from the “plasmidic cpe clade”. As a result, we were unable to assess their heat resistance.
4. Discussion
C. perfringens is a well-known spore-forming bacterium and a significant contributor to bacterial food poisoning, particularly in cooked meat products. However, it exhibits a notoriously poor ability to form spores in most laboratory media [32]. In our study, preliminary experiments revealed that the control strain’s sporulation yield (NCTC 8239) was significantly higher (approximately 3 log10 units greater) in the newly developed medium (m-TPG) compared to the commonly used Duncan–Strong medium. For this reason, this sporulation medium was adopted for subsequent experiments. However, we acknowledge at this stage that there is no evidence indicating that this medium is suitable for the sporulation of all C. perfringens strains. Many sporulation media have been developed to enhance and optimize the sporulation of C. perfringens [32,33,34]; but, to our knowledge, none of these media can be considered as the most suitable for the sporulation of all strains. All we can conclude is that sporulation initiation requires inorganic phosphate (Pi) in the environment and that Pi neutralizes the inhibitory effect of glucose and induces the expression of spo0A [35]. Another parameter that might regulate the formation of C. perfringens spores is the pH. Sporulation efficiency can be enhanced at a pH of 7.5–8 in the medium [32,34]. Finally, sodium taurocholate and thioglycolate have been reported to stimulate sporulation, although this effect varies depending on the strain and experimental conditions [33,36]. The significant variability observed in the sporulation conditions of C. perfringens strains remains fascinating and calls for further investigation.
In our experimental conditions, we observed that the greatest spore production was generally associated with strains belonging to “chromosomal cpe clade” regardless of the presence or absence of the cpe gene. The “chromosomal cpe clade” is a clade that contains mainly cpe-positive strains carrying the cpe gene on a chromosome (c-cpe) and few strains in which the cpe gene is absent [1]. The clonal genomic relationship of chromosomal cpe strains was reported and described recently [7,37]. The authors associated this clade to phylogroup I, which contains c-cpe strains, including our control strain C. perfringens NCTC 8239 and other strains in which the cpe gene is absent. Phylogroup I presents a distinctive feature, as its isolates exhibit a markedly smaller genome size compared to other phylogroups [7]. For cpe-negative strains belonging to the phylogroup I (“chromosomal cpe clade”), the possibility of cpe gene loss was suggested by the authors in [7,37]. In addition to their high sporulation ability, strains belonging to this clade have demonstrated the ability to produce heat-resistant spores. Some authors have identified two genes potentially associated with spore heat resistance in c-cpe strains: a specific allele of GrpE and the presence of CoA-disulfide reductase NaoX [9]. A variant allele of the small acid-soluble protein Ssp4 has also been linked to spore heat resistance [12,25]. However, a recent study reported a heat-resistant phenotype in a c-cpe strain lacking the heat-resistance-associated allele of Ssp4 [9]. Regarding the “plasmidic cpe clade”, it contains mainly cpe-negative strains and few cpe-positive strains with plasmid-mediated cpe [1]. For this clade, strains exhibited limited ability to produce spores, and when formed, these spores are generally heat-sensitive [9,27].
In fact, our results are in agreement with the literature and the findings obtained by other authors. Table 4 presents a non-exhaustive review of heat resistance value obtained for different C. perfringens strains by different authors. When log-linear model was used for heat resistance estimation, results were expressed as D-value in min. When model based on Weibul distribution was used for heat resistance estimation, results were expressed as δ-value in min. By comparing the results obtained for the reference strain NCTC 8239 (c-cpe), we found that the heat resistance in our study (δ95 °C = 46 min) was similar to that reported by Mehdizadeh Gohari’s (2024) study (D100 °C = 43.3 min) [27], despite differences in the sporulation medium (Table 4). Indeed, we used the m-TPG medium instead of the commonly used Duncan–Strong medium employed by Mehdizadeh Gohari, since our developed medium yielded more spores.

Table 4.
Review of heat resistance value obtained for different C. perfringens strains by different authors. The results obtained in this study have been included in the table.
Our results confirm previous studies showing that strains carrying cpe chromosomally exhibit high heat resistance, enabling them to survive more effectively in improperly cooked or stored food, which also explains their significant involvement in FBO [9]. The most heat-resistant strain was 17SBSC19 (c-cpe), which exhibited a δ95 °C value of 47 min, along with the highest sporulation ability (7.9 ± 0.43 log10 CFU/mL). These traits make this strain a significant hazard due to its ability to withstand cooking. On the other hand, strains carrying cpe on plasmid exhibit a low heat resistance with an average of 2.4 ± 2.44 min at 89 °C [9], 1.77 ± 0.73 min at 100 °C [27] and 12 ± 13 min at 85 °C (our study, Table 4). This heat-sensitive phenotype may explain the low prevalence of these strains (p-cpe) in food poisoning cases associated with C. perfringens toxinotype F.
The main finding in this study is the identification of a third group with intermediate heat resistance values, which lies between the highly heat-resistant strains (c-cpe) and the heat-sensitive strains (p-cpe). Strains exhibiting intermediate heat-resistance phenotype are cpe-negative and phylogenetically located within the “chromosomal cpe clade”. As shown previously, these strains are genetically very close to strains that carry cpe chromosomally [7,37]. To our knowledge, our study is the first one to provide heat resistance values for this group of strains, with an average of 69 ± 26 min at 85 °C (Table 4). Indeed, the reviewed literature suggests that cpe-negative strains are given less importance since they generally exhibit a heat-sensitive phenotype with D89 °C-value of 1.5 ± 0.63 min [9] (Table 4) and D95 °C-value of 2.18 ± 0.46 min [26].
This article highlights the existence of an intermediate heat-resistant group of cpe-negative strains and challenges the traditional binary classification (heat-resistant vs. heat-sensitive). It also suggests a new layer of variability in heat stress adaptation. This observation is corroborated by Orsburn et al. (2008) [16], who noted that the distinction between cpe-positive and cpe-negative strains was not always marked in terms of heat resistance, but without being able to provide an explanation. Other Clostridium species are also known to exhibit variable thermal resistance. In C. botulinum, heat resistance varies both between and within groups: proteolytic Group I strains produce the most heat-resistant spores (D121.1 °C = 0.21 min), whereas non-proteolytic Group II strains are the most heat-sensitive (D80 °C = 0.6–1.25 min). Group III strains display D104 °C values ranging from 0.1 to 0.9 min, while Group IV strains exhibit D104 °C values between 0.8 and 1.12 min [37].
Although cpe-negative Type A strains are not generally associated with food poisoning, they possess other virulence factors [1]. For example, nagH gene has been identified in cpe-negative strains associated with food poisoning and may contribute to their enterotoxigenic potential [1]. This gene encodes a hyaluronidase that increases tissue permeability and facilitates the spread of alpha-toxins [38]. Furthermore, the possibility of their conversion into Type F strains through cpe plasmid transfer cannot be ruled out [6]. Pangenome analysis revealed a high degree of variability and genomic plasticity in C. perfringens, reflecting frequent events of gene gain and loss [7,39]. Their heat resistance, along with the potential for cpe gene transfer, could make these strains a relevant hazard for cooked, cooled, and re-heated meat products.
5. Conclusions
This study provides new insights into the phenotypic and genetic diversity of Clostridium perfringens regarding its sporulation ability and heat resistance. The results show that strains belonging to “the chromosomal cpe clade”, whether carrying the cpe gene or not, exhibit high sporulation ability and produce heat-resistant spores. In contrast, strains within the “plasmidic cpe clade” demonstrate low sporulation abilities and are heat-sensitive. A key finding of this work is the identification of a group of cpe-negative strains with intermediate heat resistance, a phenotype not previously described. This challenges the traditional classification, which strictly opposes heat-resistant c-cpe strains and heat-sensitive p-cpe strains. It also suggests the existence of unknown mechanisms of stress adaptation that are potentially associated with the genomic and molecular environment of the “chromosomal cpe clade” (mostly cpe-positive, with some cpe-negative), and that are independent of the cpe gene.
Author Contributions
O.F.: Writing—review and editing, conceptualization, supervision, funding acquisition. V.M.: methodology, investigation. W.B.: methodology, investigation. M.F.: review, validation, funding acquisition. C.T.: review, validation. N.M.: writing—original draft preparation, visualization, methodology, data curation, formal analysis, supervision, funding acquisition, conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. This study was funded by the National Veterinary School (EnvA) of Alfort and the French Agency for Food, Environmental and Occupational Health & Safety (ANSES).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| FBO | Food-Borne outbreaks |
| CPE | Clostridium perfringens enterotoxin |
| c-cpe | chromosomal cpe |
| p-cpe | plasmidic cpe |
References
- Mahamat Abdelrahim, A.; Radomski, N.; Delannoy, S.; Djellal, S.; Le Négrate, M.; Hadjab, K.; Fach, P.; Hennekinne, J.-A.; Mistou, M.-Y.; Firmesse, O. Large-Scale Genomic Analyses and Toxinotyping of Clostridium perfringens Implicated in Foodborne Outbreaks in France. Front. Microbiol. 2019, 10, 777. [Google Scholar] [CrossRef] [PubMed]
- Mehdizadeh Gohari, I.; Navarro, M.A.; Li, J.; Shrestha, A.; Uzal, F.; McClane, B.A. Pathogenicity and Virulence of Clostridium perfringens. Virulence 2021, 12, 723–753. [Google Scholar] [CrossRef] [PubMed]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G.; et al. Expansion of the Clostridium perfringens Toxin-Based Typing Scheme. Anaerobe 2018, 53, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.C.; Shrestha, A.; McClane, B.A. Clostridium perfringens Enterotoxin: Action, Genetics, and Translational Applications. Toxins 2016, 8, 73. [Google Scholar] [CrossRef]
- Shrestha, A.; Mehdizadeh Gohari, I.; Li, J.; Navarro, M.; Uzal, F.A.; McClane, B.A. The Biology and Pathogenicity of Clostridium perfringens Type F: A Common Human Enteropathogen with a New(Ish) Name. Microbiol. Mol. Biol. Rev. 2024, 88, e00140-23. [Google Scholar] [CrossRef]
- Shrestha, A.; Uzal, F.A.; McClane, B.A. Enterotoxic Clostridia: Clostridium perfringens Enteric Diseases. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Abdel-Glil, M.Y.; Thomas, P.; Linde, J.; Busch, A.; Wieler, L.H.; Neubauer, H.; Seyboldt, C. Comparative in Silico Genome Analysis of Clostridium perfringens Unravels Stable Phylogroups with Different Genome Characteristics and Pathogenic Potential. Sci. Rep. 2021, 11, 6756. [Google Scholar] [CrossRef]
- Brynestad, S.; Sarker, M.R.; McClane, B.A.; Granum, P.E.; Rood, J.I. Enterotoxin Plasmid from Clostridium perfringens Is Conjugative. Infect. Immun. 2001, 69, 3483–3487. [Google Scholar] [CrossRef]
- Jaakkola, K.; Virtanen, K.; Lahti, P.; Keto-Timonen, R.; Lindström, M.; Korkeala, H. Comparative Genome Analysis and Spore Heat Resistance Assay Reveal a New Component to Population Structure and Genome Epidemiology Within Clostridium perfringens Enterotoxin-Carrying Isolates. Front. Microbiol. 2021, 12, 717176. [Google Scholar] [CrossRef]
- Miyamoto, K.; Fisher, D.J.; Li, J.; Sayeed, S.; Akimoto, S.; McClane, B.A. Complete Sequencing and Diversity Analysis of the Enterotoxin-Encoding Plasmids in Clostridium perfringens Type A Non-Food-Borne Human Gastrointestinal Disease Isolates. J. Bacteriol. 2006, 188, 1585–1598. [Google Scholar] [CrossRef]
- Tran, C.; Poezevara, T.; Maladen, V.; Guillier, L.; Mtimet, N.; Malayrat, C.; Coadou, T.; Jambou, L.; Rouxel, S.; Le Bouquin, S.; et al. Isolation Rate, Genetic Diversity, and Toxinotyping of Clostridium perfringens Isolated from French Cattle, Pig or Poultry Slaughterhouses. Food Microbiol. 2026, 133, 104898. [Google Scholar] [CrossRef]
- Li, J.; Paredes-Sabja, D.; Sarker, M.R.; McClane, B.A. Further Characterization of Clostridium perfringens Small Acid Soluble Protein-4 (Ssp4) Properties and Expression. PLoS ONE 2009, 4, e6249. [Google Scholar] [CrossRef]
- Grass, J.E.; Gould, L.H.; Mahon, B.E. Epidemiology of Foodborne Disease Outbreaks Caused by Clostridium perfringens, United States, 1998–2010. Foodborne Pathog. Dis. 2013, 10, 131–136. [Google Scholar] [CrossRef]
- SPF. Surveillance des Toxi-Infections Alimentaires Collectives. Données de la Déclaration Obligatoire. 2022. Available online: https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-infectieuses-d-origine-alimentaire/toxi-infections-alimentaires-collectives/documents/bulletin-national/surveillance-des-toxi-infections-alimentaires-collectives.-donnees-de-la-declaration-obligatoire-2022 (accessed on 20 August 2024).
- Novak, J.S.; Juneja, V.K.; McClane, B.A. An Ultrastructural Comparison of Spores from Various Strains of Clostridium perfringens and Correlations with Heat Resistance Parameters. Int. J. Food Microbiol. 2003, 86, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Orsburn, B.; Melville, S.B.; Popham, D.L. Factors Contributing to Heat Resistance of Clostridium Perfringens Endospores. Appl. Environ. Microbiol. 2008, 74, 3328–3335. [Google Scholar] [CrossRef] [PubMed]
- Setlow, P.; Christie, G. New Thoughts on an Old Topic: Secrets of Bacterial Spore Resistance Slowly Being Revealed. Microbiol. Mol. Biol. Rev. 2023, 87, e0008022. [Google Scholar] [CrossRef] [PubMed]
- Leggett, M.J.; McDonnell, G.; Denyer, S.P.; Setlow, P.; Maillard, J.-Y. Bacterial Spore Structures and Their Protective Role in Biocide Resistance. J. Appl. Microbiol. 2012, 113, 485–498. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus Endospores to Extreme Terrestrial and Extraterrestrial Environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef]
- Setlow, P. I Will Survive: DNA Protection in Bacterial Spores. Trends Microbiol. 2007, 15, 172–180. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their Resistance to and Killing by Radiation, Heat and Chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Coleman, W.H.; Chen, D.; Li, Y.; Cowan, A.E.; Setlow, P. How Moist Heat Kills Spores of Bacillus subtilis. J. Bacteriol. 2007, 189, 8458–8466. [Google Scholar] [CrossRef]
- Zhang, P.; Kong, L.; Setlow, P.; Li, Y. Characterization of Wet-Heat Inactivation of Single Spores of Bacillus Species by Dual-Trap Raman Spectroscopy and Elastic Light Scattering. Appl. Environ. Microbiol. 2010, 76, 1796–1805. [Google Scholar] [CrossRef] [PubMed]
- Bender, G.R.; Marquis, R.E. Spore Heat Resistance and Specific Mineralization. Appl. Environ. Microbiol. 1985, 50, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; McClane, B.A. A Novel Small Acid Soluble Protein Variant Is Important for Spore Resistance of Most Clostridium perfringens Food Poisoning Isolates. PLOS Pathog. 2008, 4, e1000056. [Google Scholar] [CrossRef]
- Ando, Y.; Tsuzuki, T.; Sunagawa, H.; Oka, S. Heat Resistance, Spore Germination, and Enterotoxigenicity of Clostridium perfringens. Microbiol. Immunol. 1985, 29, 317–326. [Google Scholar] [CrossRef]
- Mehdizadeh Gohari, I.; Li, J.; Shivers, R.; Sparks, S.G.; McClane, B.A. Heat Resistance Differences Are Common between Both Vegetative Cells and Spores of Clostridium perfringens Type F Isolates Carrying a Chromosomal vs Plasmid-Borne Enterotoxin Gene. Appl. Environ. Microbiol. 2024, 90, e0091424. [Google Scholar] [CrossRef]
- Mtimet, N.; Guégan, S.; Durand, L.; Mathot, A.-G.; Venaille, L.; Leguérinel, I.; Coroller, L.; Couvert, O. Effect of pH on Thermoanaerobacterium thermosaccharolyticum DSM 571 Growth, Spore Heat Resistance and Recovery. Food Microbiol. 2016, 55, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Mtimet, N.; Trunet, C.; Mathot, A.-G.; Venaille, L.; Leguérinel, I.; Coroller, L.; Couvert, O. Modeling the Behavior of Geobacillus stearothermophilus ATCC 12980 throughout Its Life Cycle as Vegetative Cells or Spores Using Growth Boundaries. Food Microbiol. 2015, 48, 153–162. [Google Scholar] [CrossRef]
- Mafart, P.; Couvert, O.; Gaillard, S.; Leguerinel, I. On Calculating Sterility in Thermal Preservation Methods: Application of the Weibull Frequency Distribution Model. Int. J. Food Microbiol. 2002, 72, 107–113. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Tsai, C.L. Model Selection for Extended Quasi-Likelihood Models in Small Samples. Biometrics 1995, 51, 1077–1084. [Google Scholar] [CrossRef]
- Ushijima, T.; Sugitani, A.; Ozaki, Y. A Pair of Semisolid Media Facilitate Detection of Spore and Enterotoxin of Clostridium perfringens. J. Microbiol. Methods 1987, 6, 145–152. [Google Scholar] [CrossRef]
- De Jong, A.E.I.; Beumer, R.R.; Rombouts, F.M. Optimizing Sporulation of Clostridium perfringens. J. Food Prot. 2002, 65, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Cui, X.; Li, M.; Zhu, Y.; Zhao, L.; Liu, S.; Zhao, G.; Wang, N.; Ma, Y.; Xu, L. Effects of Sporulation Conditions on the Growth, Germination, and Resistance of Clostridium perfringens Spores. Int. J. Food Microbiol. 2023, 396, 110200. [Google Scholar] [CrossRef] [PubMed]
- Philippe, V.A.; Méndez, M.B.; Huang, I.-H.; Orsaria, L.M.; Sarker, M.R.; Grau, R.R. Inorganic Phosphate Induces Spore Morphogenesis and Enterotoxin Production in the Intestinal Pathogen Clostridium perfringens. Infect. Immun. 2006, 74, 3651–3656. [Google Scholar] [CrossRef]
- Liggins, M.; Ramírez Ramírez, N.; Abel-Santos, E. Comparison of Sporulation and Germination Conditions for Clostridium perfringens Type A and G Strains. Front. Microbiol. 2023, 14, 1143399. [Google Scholar] [CrossRef]
- Anses. Hazard Datasheet: Clostridium botulinum, Clostridium Neurotoxinogènes. 2019. Available online: https://www.anses.fr/fr/system/files?file=BIORISK2016SA0074Fi.pdf (accessed on 29 October 2025).
- Camargo, A.; Ramírez, J.D.; Kiu, R.; Hall, L.J.; Muñoz, M. Unveiling the pathogenic mechanisms of Clostridium perfringens toxins and virulence factors. Emerg. Microbes Infect. 2024, 13, 2341968. [Google Scholar] [CrossRef]
- Kiu, R.; Caim, S.; Painset, A.; Pickard, D.; Swift, C.; Dougan, G.; Mather, A.E.; Amar, C.; Hall, L.J. Phylogenomic Analysis of Gastroenteritis-Associated Clostridium perfringens in England and Wales over a 7-Year Period Indicates Distribution of Clonal Toxigenic Strains in Multiple Outbreaks and Extensive Involvement of Enterotoxin-Encoding (CPE) Plasmids. Microb. Genom. 2019, 5, e000297. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).