Comprehensive Characterization of Metabolites in Multiplier Onion Bulbs and Identification of Regulatory Genes for Nutritional Improvement
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Metabolite Extraction
2.3. UPLC-MS/MS Detection
2.4. Differentially Accumulated Metabolites Analysis and KEGG Enrichment
2.5. RNA Extraction and Illumina RNA Sequencing
2.6. Gene Differential Expression Analysis and Gene Ontology Enrichment
2.7. Integration Analysis of Metabolomics and Transcriptomics
3. Results
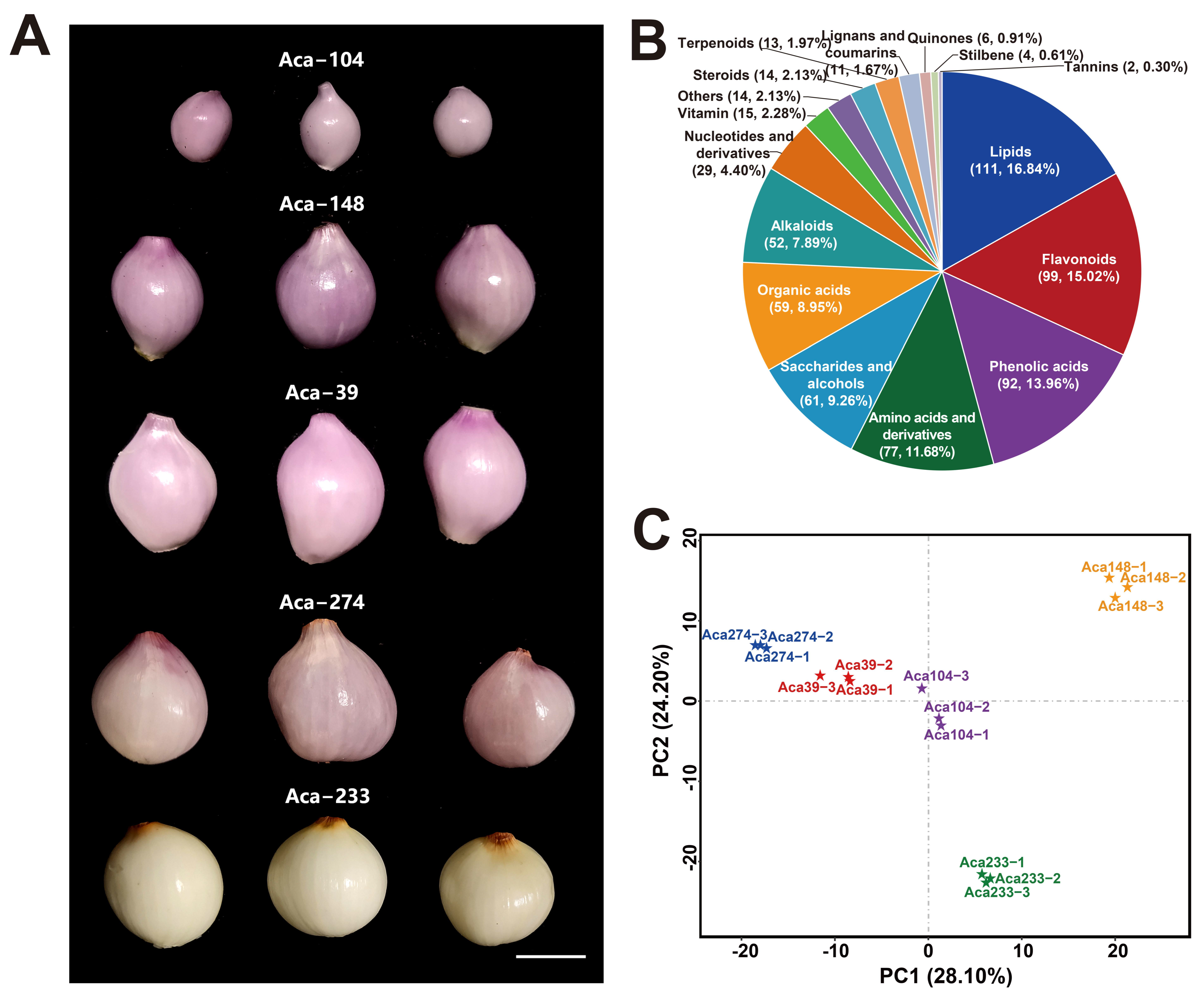
3.1. Metabolic Profiling Analysis
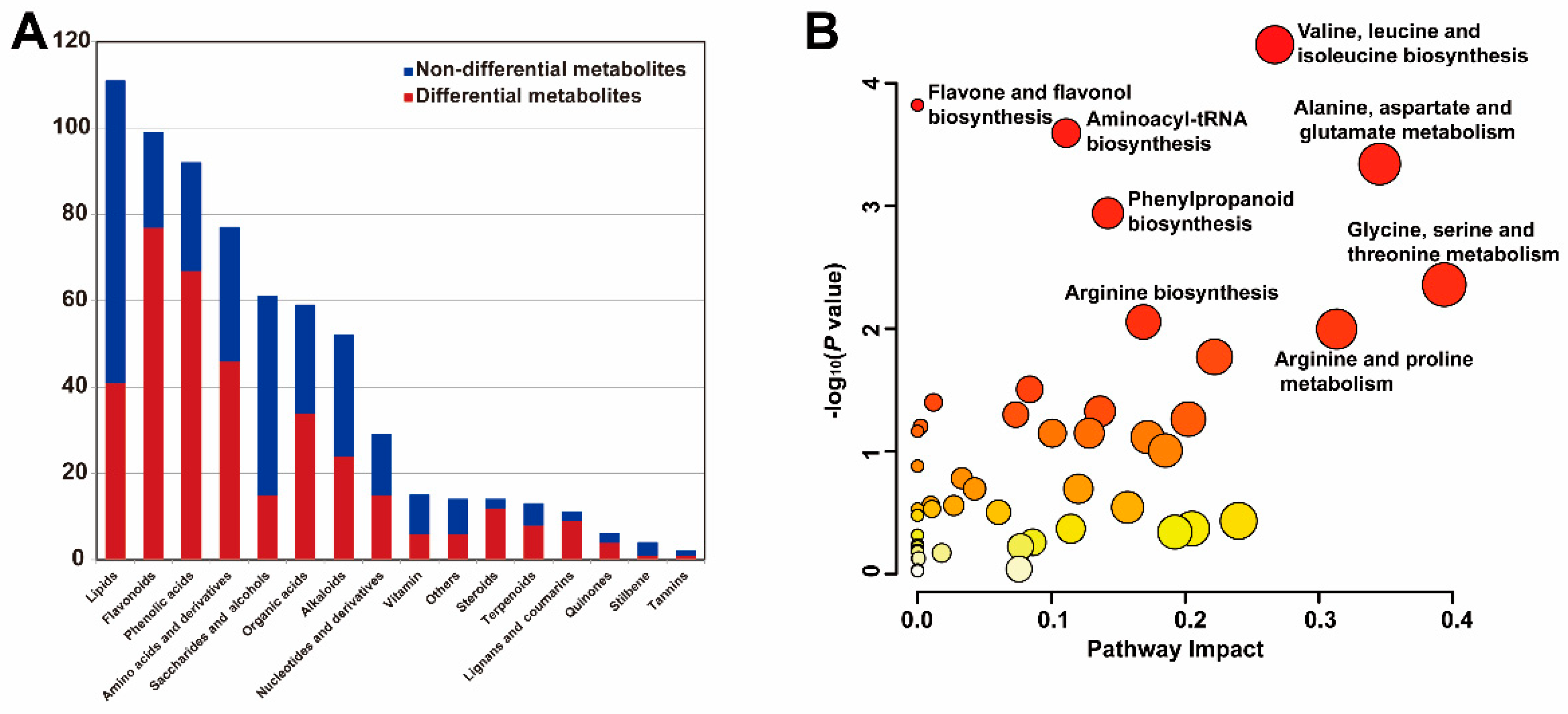
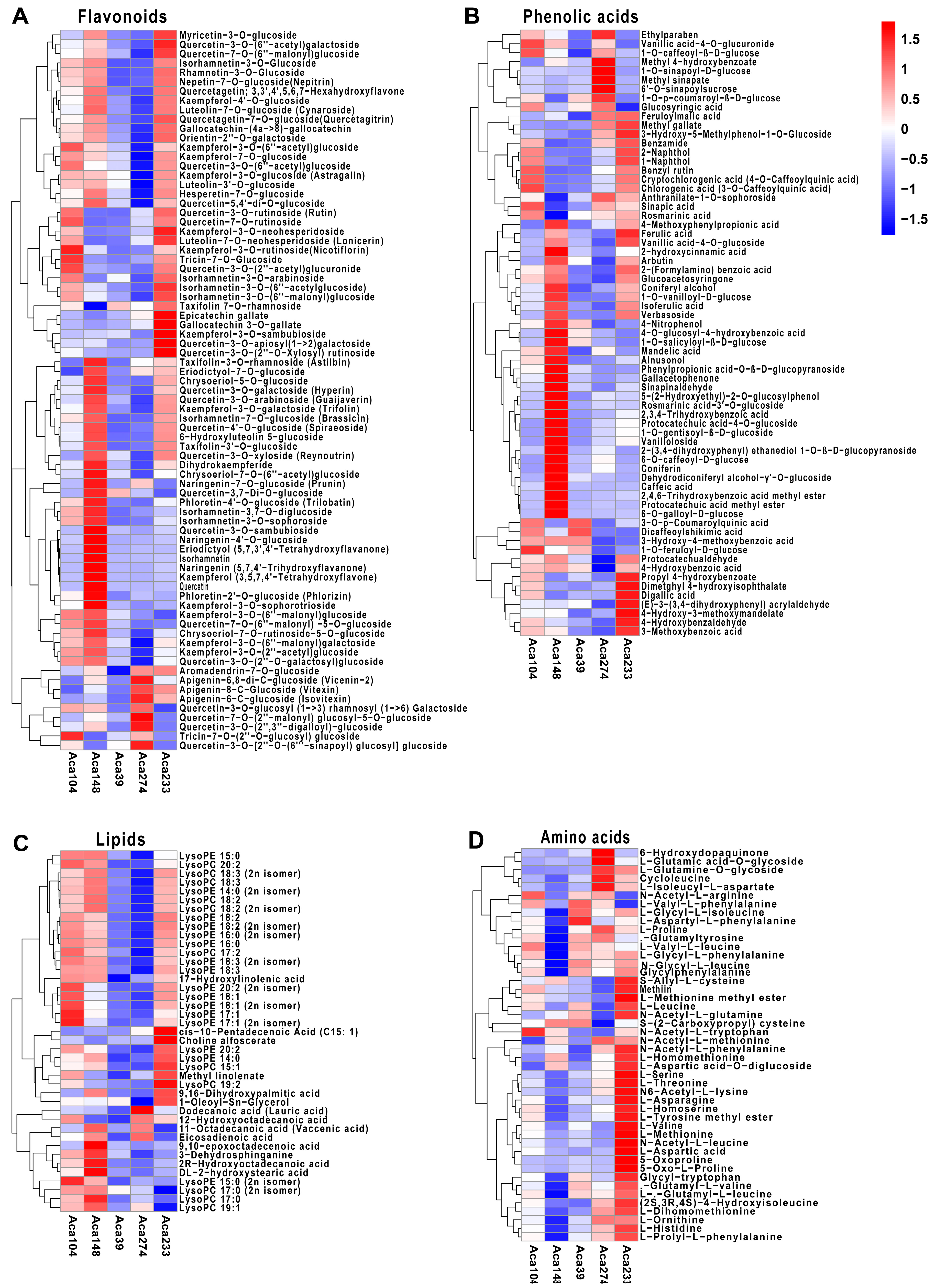
3.2. Differentially Accumulated Metabolites Analysis
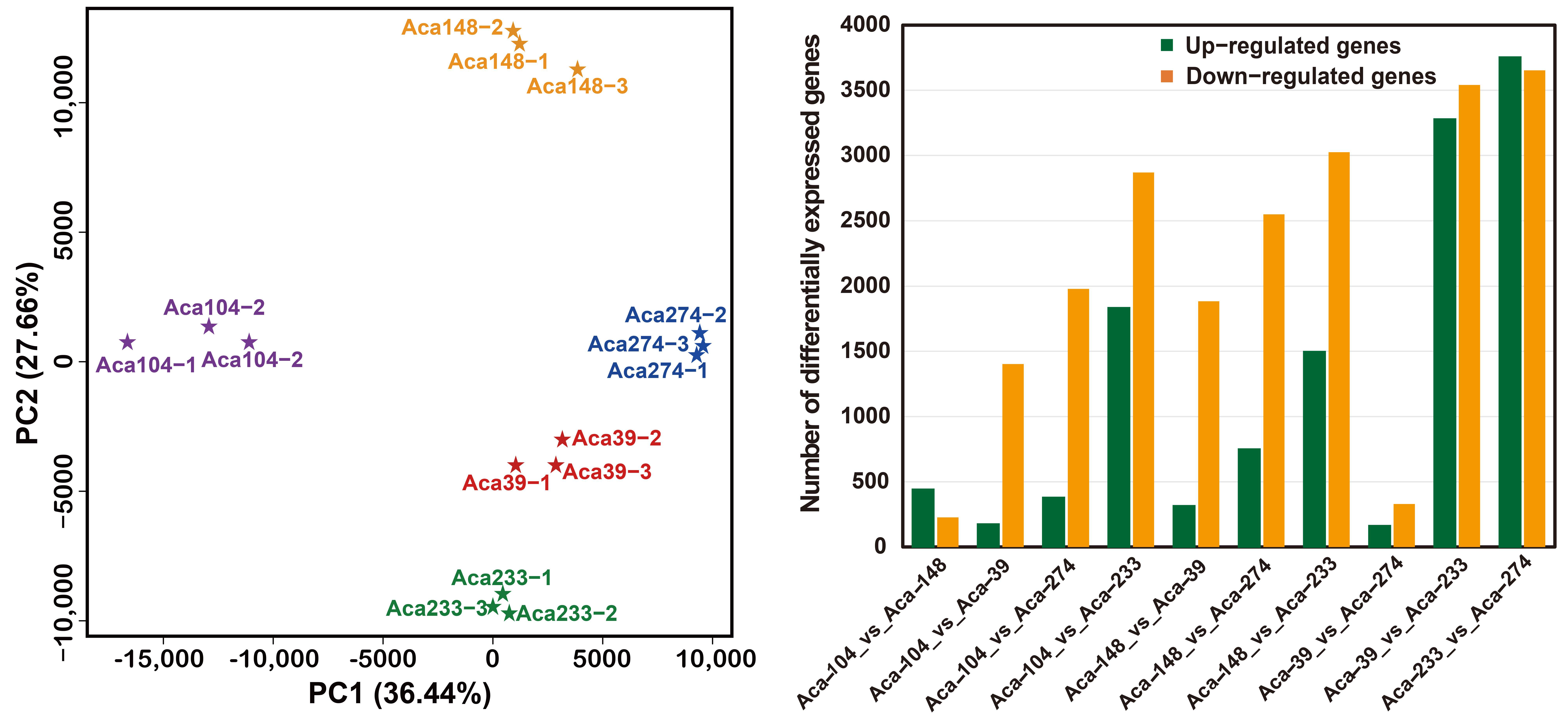
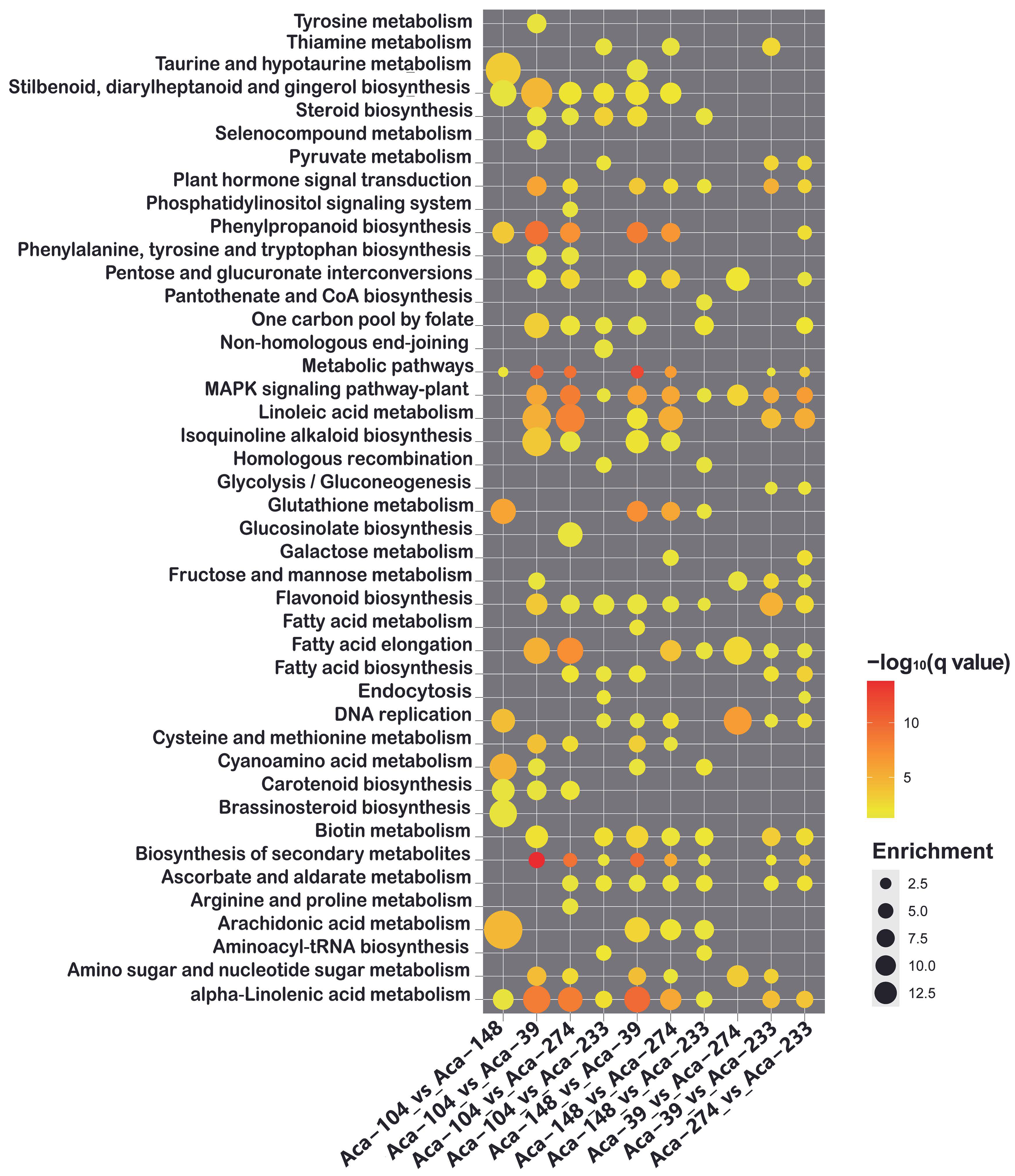
3.3. Transcriptome Sequencing and Differential Expression Gene Analysis
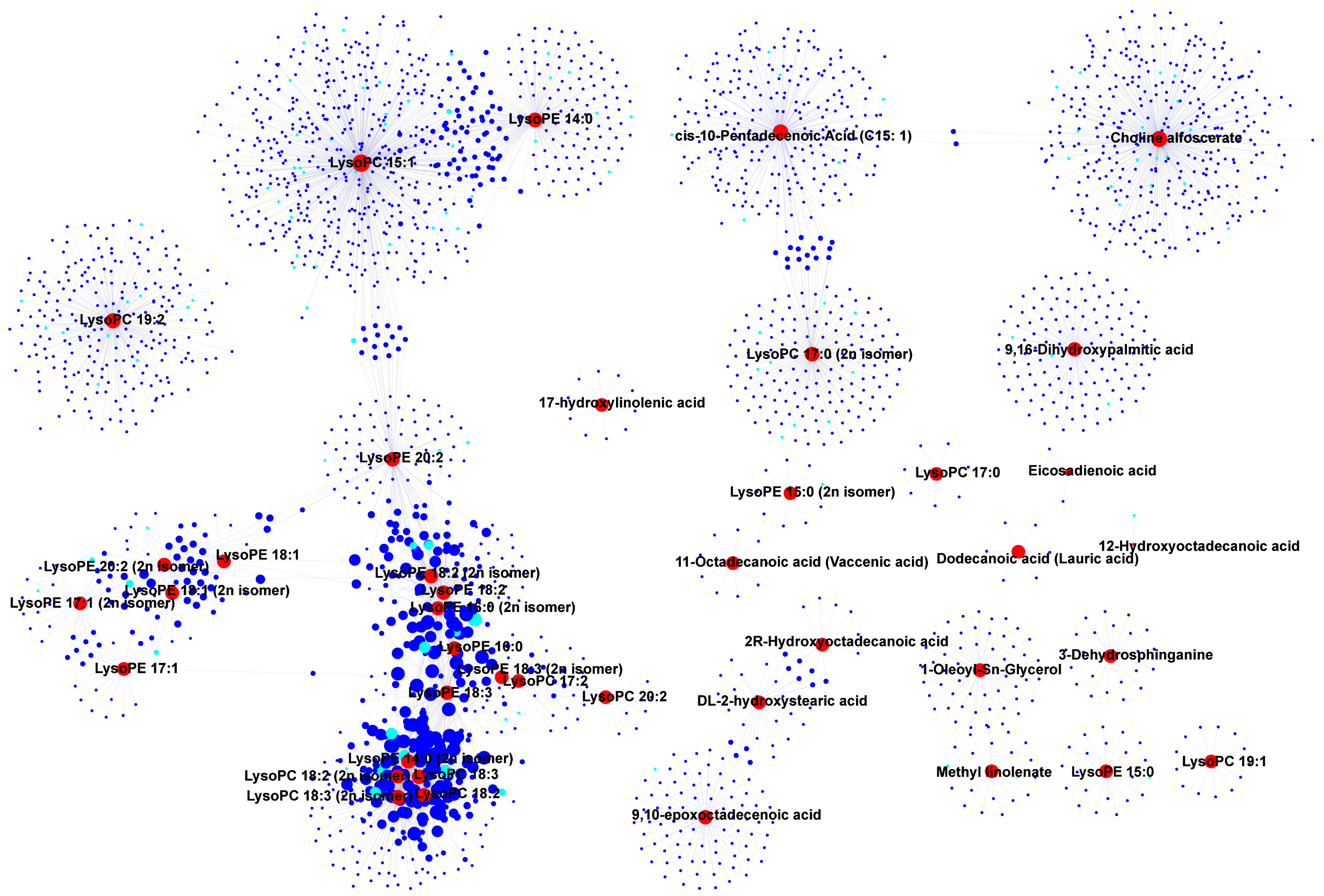
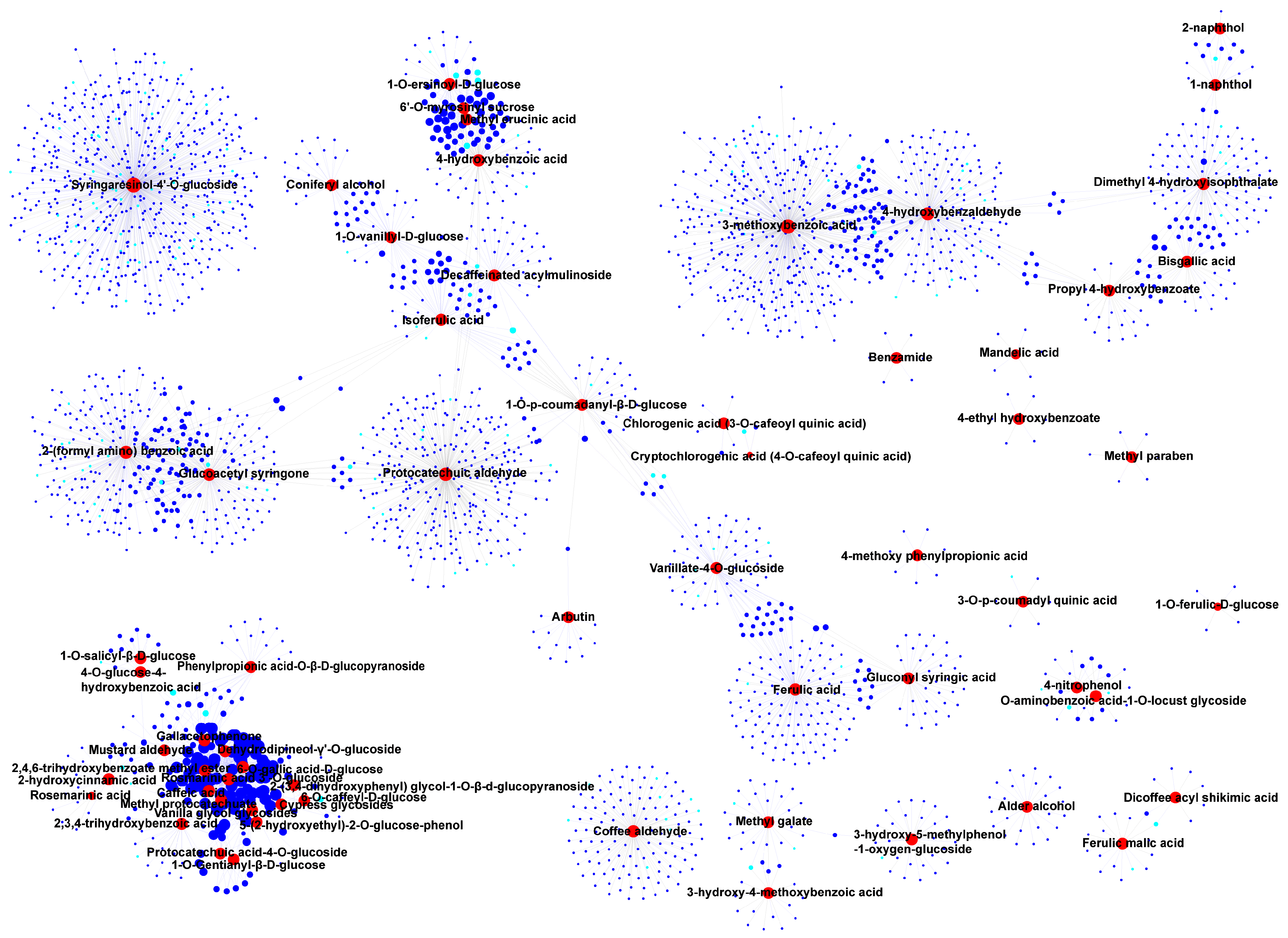
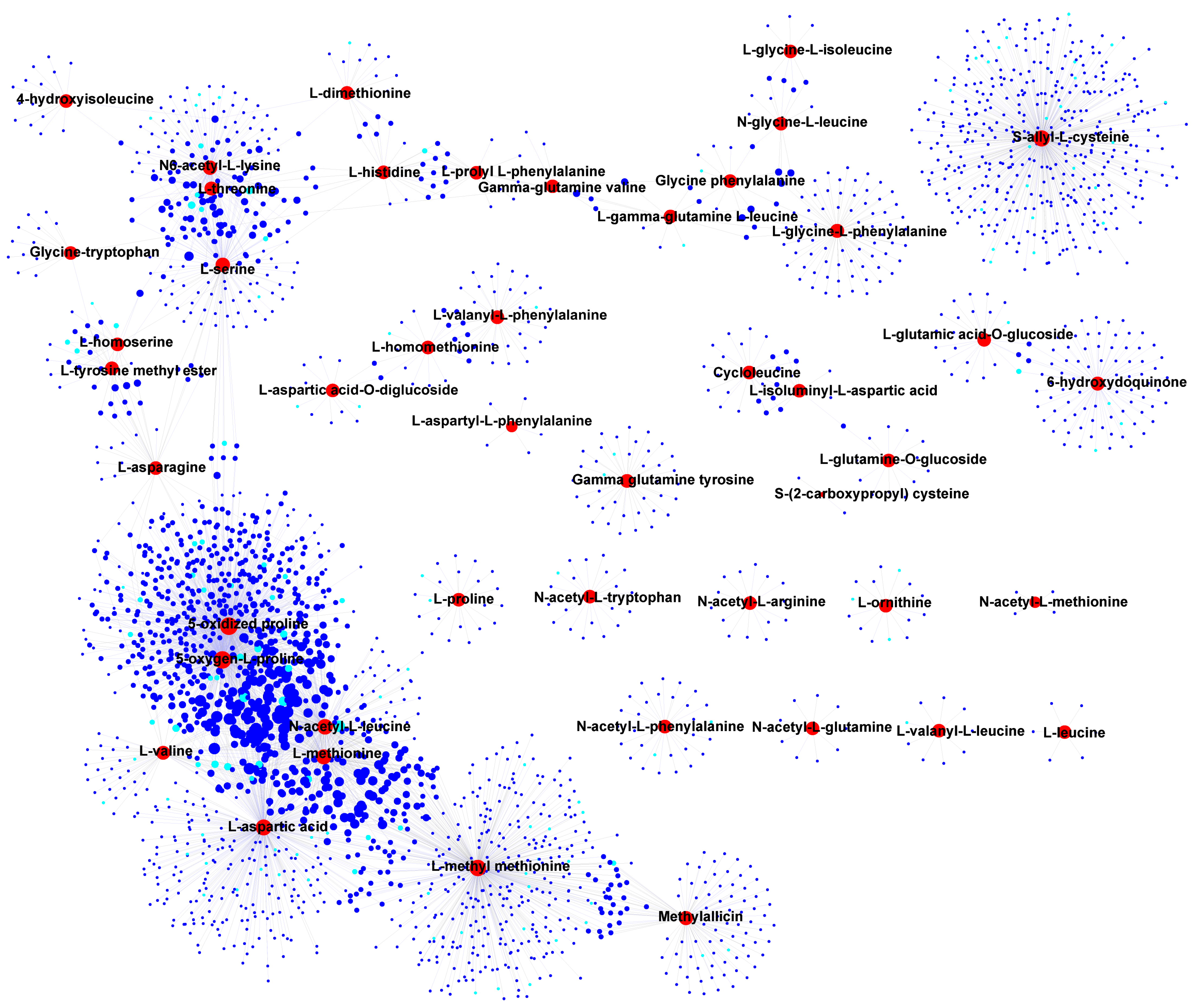
3.4. Correlation and Network Analysis of DAMs and DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Getachew, T. Development of seed propagated shallot (Allium cepa L var. aggregatum) varieties in Ethiopia. Sci. Hortic. 2018, 240, 89–93. [Google Scholar] [CrossRef]
- Poovamma, C.; Devi, A.B.K.; Singh, K.J. Effect of different levels of planting time and spacing on quality and economics of multiplier onion (Allium cepa L. var. aggregatum Don.) Cv. Meitei Tilhou. Int. J. Curr. Microbiol. App. Sci. 2021, 10, 1–8. [Google Scholar] [CrossRef]
- Wang, M.R.; Hamborg, Z.; Slimestad, R.; Elameen, A.; Blystad, D.R.; Haugslien, S.; Skjeseth, G.; Wang, Q.C. Assessments of rooting, vegetative growth, bulb production, genetic integrity and biochemical compounds in cryopreserved plants of shallot. Plant Cell Tissue Organ Cult. 2021, 144, 123–131. [Google Scholar] [CrossRef]
- Metrani, R.; Singh, J.; Acharya, P.; Jayaprakasha, G.K.; Patil, B.S. Comparative metabolomics profiling of polyphenols, nutrients and antioxidant activities of two red onion (Allium cepa L.) cultivars. Plants 2020, 9, 1077. [Google Scholar] [CrossRef]
- Celano, R.; Docimo, T.; Piccinelli, A.L.; Gazzerro, P.; Tucci, M.; Sanzo, R.D.; Di Sanzo, R.; Carabetta, S.; Campone, L.; Russo, M.; et al. Onion peel: Turning a food waste into a resource. Antioxidants 2021, 10, 304. [Google Scholar] [CrossRef]
- Kiplagat, E.; Ramezani, M.; Malla, S.; Cisneros-Zevallos, L.; Joshi, V.; Castillo, A. Factors affecting growth and survival of salmonella in onion extracts and onion bulbs. Foods 2025, 14, 1. [Google Scholar] [CrossRef]
- Esuyawukal-Moges, K.; Biruk-Masrie, Z. Growth and yield response of shallot (Allium cepa var. aggregatum) varieties to intra-row spacing in Eastern Amhara, Ethiopia. Curr. Res. Agric. Sci. 2019, 6, 157–168. [Google Scholar] [CrossRef]
- Nile, S.H.; Park, S.W. Total phenolics, antioxidant and xanthine oxidase inhibitory activity of three colored onions (Allium cepa L.). Front. Life Sci. 2013, 7, 224–228. [Google Scholar] [CrossRef]
- Kumari, N.; Kumar, M.; Radha; Lorenzo, J.M.; Sharma, D.; Puri, S.; Pundi, A.; Dhumal, S.; Bhuyan, D.J.; Jayanthy, G.; et al. Onion and garlic polysaccharides: A review on extraction, characterization, bioactivity, and modifications. Int. J. Biol. Macromol. 2022, 219, 1047–1061. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Zhan, Z.; Liu, B.; Chen, Z.; Liang, Y. Transcriptome analysis of sucrose metabolism during bulb swelling and development in onion (Allium cepa L.). Front. Plant Sci. 2016, 7, 1425. [Google Scholar] [CrossRef]
- Zhang, C.; Li, X.; Zhan, Z.; Cao, L.; Zeng, A.; Chang, G.; Liang, Y. Transcriptome sequencing and metabolism analysis reveals the role of cyanidin metabolism in dark-red onion (Allium cepa L.) bulbs. Sci. Rep. 2018, 8, 14109. [Google Scholar] [CrossRef]
- Böttcher, C.; Krähmer, A.; Stürtz, M.; Widder, S.; Schulz, H. Comprehensive metabolite profiling of onion bulbs (Allium cepa) using liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. Metabolomics 2017, 13, 35. [Google Scholar] [CrossRef]
- El-Din, M.S.; Fahmy, H.A.; Ragab, N.A.; Khalifa, M.R.; Zahran, H.; Soliman, N.R.; Farag, M.A. Comprehensive metabolome classification of four onion (Allium cepa) cultivars via GC–MS and UPLC-MS: Insights into chemical diversity and remote antimicrobial activity against foodborne pathogens. Food Res. Int. 2025, 221, 117367. [Google Scholar] [CrossRef]
- Böttcher, C.; Krähmer, A.; Stürtz, M.; Widder, S.; Schulz, H. Effect of Cultivar and Cultivation Year on the Metabolite Profile of Onion Bulbs (Allium cepa L.). J. Agric. Food Chem. 2018, 66, 3229–3238. [Google Scholar] [CrossRef]
- Ferioli, F.; D’Antuono, L.F. Evaluation of phenolics and cysteine sulfoxides in local onion and shallot germplasm from Italy and Ukraine. Genet. Resour. Crop Evol. 2016, 63, 601–614. [Google Scholar] [CrossRef]
- Tian, P.; Zhan, P.; Tian, H.; Wang, P.; Lu, C.; Zhao, Y.; Ni, R.; Zhang, Y. Analysis of volatile compound changes in fried shallot (Allium cepa L. var. aggregatum) oil at different frying temperatures by GC–MS, OAV, and multivariate analysis. Food Chem. 2021, 345, 128748. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the fruit metabolome in tomato breeding. Cell 2018, 172, 249–261. [Google Scholar] [CrossRef]
- Zhou, X.; Obel, H.O.; Liu, S.; Yang, Y.; Liu, J.; Zhuang, Y. Comparative analysis of metabolic variation in eggplant fruit of different varieties reveals metabolites important for quality traits. Foods 2023, 12, 4383. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; Morais, D.; Chang, L.; Barrette, M.; Michel, B.; Carol, G.; Jacques, P.; Li, S.; et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.V.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids—Biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Hirata, S.; Sawada, Y.; Hirai, M.Y.; Sato, S.; Hirakawa, H.Y.; Mine, Y.; Tanaka, K.; Shigyo, M. Widely targeted metabolome and transcriptome landscapes of Allium fistulosum-A. cepa chromosome addition lines revealed a flavonoid hot spot on chromosome 5A. Sci. Rep. 2019, 9, 3541. [Google Scholar] [CrossRef]
- Rohani, A.S.; Yuandani; Sitorus, P.; Andrianto, D.; Dalimunthe, A. Anti-hypercholesterolemic activity of standardized fermented Allium cepa L. var aggregatum extract: In vitro and in vivo studies. J. Herbmed Pharmacol. 2022, 11, 383–388. [Google Scholar] [CrossRef]
- Galili, G.; Amir, R.; Fernie, A.R. The regulation of essential amino acid synthesis and accumulation in plants. Annu. Rev. Plant Biol. 2016, 67, 153–178. [Google Scholar] [CrossRef]
- Tuncel, A.; Qi, Y. CRISPR/Cas mediated genome editing in potato: Past achievements and future directions. Plant Sci. 2022, 325, 111474. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Sharma, M.; Chai, C.; Morohashi, K.; Grotewold, E.; Snook, M.E.; Chopra, S. Expression of flavonoid 3′-hydroxylase is controlled by P1, the regulator of 3-deoxyflavonoid biosynthesis in maize. BMC Plant Biol. 2012, 12, 196. [Google Scholar] [CrossRef]
- Wang, L.; Lui, A.C.W.; Lam, P.Y.; Liu, G.; Godwin, I.D.; Lo, C. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol. J. 2020, 18, 2170–2172. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Bai, Y.; Li, S.; Chen, H.; Han, X.; Zhao, H.; Shao, J.; Park, S.U.; Wu, Q. Cloning, characterization, and activity analysis of a flavonol synthase gene FtFLS1 and its association with flavonoid content in tartary buckwheat. J. Agric. Food Chem. 2012, 60, 5161–5168. [Google Scholar] [CrossRef]
- Manzoor, M.A.; Sabir, I.A.; Shah, I.H.; Riaz, M.W.; Rehman, S.; Song, C.; Li, G.; Malik, M.S.; Ashraf, G.A.; Haider, M.S.; et al. Flavonoids: A review on biosynthesis and transportation mechanism in plants. Funct. Integr. Genom. 2023, 23, 315. [Google Scholar] [CrossRef]
- Yang, J.; Li, H.; Ma, R.; Chang, Y.; Qin, X.; Xu, J.; Fu, Y. Genome-wide transcriptome analysis and characterization of the cytochrome P450 flavonoid biosynthesis genes in pigeon pea (Cajanus cajan). Planta 2022, 255, 120. [Google Scholar] [CrossRef]
- Liu, X.; Gong, Q.; Zhao, C.; Wang, D.; Ye, X.; Zheng, G.; Wang, Y.; Cao, J.; Sun, C. Genome-wide analysis of cytochrome P450 genes in Citrus clementina and characterization of a CYP gene encoding flavonoid 3′-hydroxylase. Hortic. Res. 2022, 10, uhac283. [Google Scholar] [CrossRef] [PubMed]
- Sanders, A.; Collier, R.; Trethewy, A.; Gould, G.; Sieker, R.; Tegeder, M. AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 2009, 59, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.R.; Reinders, A.; Ward, J.M. Transport function of rice Amino Acid Permeases (AAPs). Plant Cell Physiol. 2015, 56, 1355–1363. [Google Scholar] [CrossRef]
- Cantor, J.R.; Stone, E.M.; Chantranupong, L.; Georgiou, G. The human asparaginase-like protein 1 hASRGL1 is an Ntn hydrolase with beta-aspartyl peptidase activity. Biochemistry 2009, 48, 11026–11031. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Song, J.; Zhang, T.; Tan, Y.; Wang, M.; Zang, J.; Zhang, X.; Yang, W.; Pang, Y.; Yang, Y.; et al. Comprehensive Characterization of Metabolites in Multiplier Onion Bulbs and Identification of Regulatory Genes for Nutritional Improvement. Foods 2025, 14, 3290. https://doi.org/10.3390/foods14193290
Jia H, Song J, Zhang T, Tan Y, Wang M, Zang J, Zhang X, Yang W, Pang Y, Yang Y, et al. Comprehensive Characterization of Metabolites in Multiplier Onion Bulbs and Identification of Regulatory Genes for Nutritional Improvement. Foods. 2025; 14(19):3290. https://doi.org/10.3390/foods14193290
Chicago/Turabian StyleJia, Huixia, Jiangping Song, Tingting Zhang, Yumin Tan, Mengzhen Wang, Jiyan Zang, Xiaohui Zhang, Wenlong Yang, Yanhui Pang, Yanfei Yang, and et al. 2025. "Comprehensive Characterization of Metabolites in Multiplier Onion Bulbs and Identification of Regulatory Genes for Nutritional Improvement" Foods 14, no. 19: 3290. https://doi.org/10.3390/foods14193290
APA StyleJia, H., Song, J., Zhang, T., Tan, Y., Wang, M., Zang, J., Zhang, X., Yang, W., Pang, Y., Yang, Y., & Wang, H. (2025). Comprehensive Characterization of Metabolites in Multiplier Onion Bulbs and Identification of Regulatory Genes for Nutritional Improvement. Foods, 14(19), 3290. https://doi.org/10.3390/foods14193290







