Genomic Landscape and Antimicrobial Resistance of Listeria monocytogenes in Retail Chicken in Qingdao, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
2.2. Antibiotic Susceptibility Testing
2.3. Preparation of Genome DNA
2.4. Whole-Genome Sequencing of L. monocytogenes Isolates
2.5. In Silico Subtyping and Phylogenetic Analysis
2.6. Genome Annotation
3. Results
3.1. Prevalence and Antimicrobial Susceptibility of L. monocytogenes in Chicken Meats
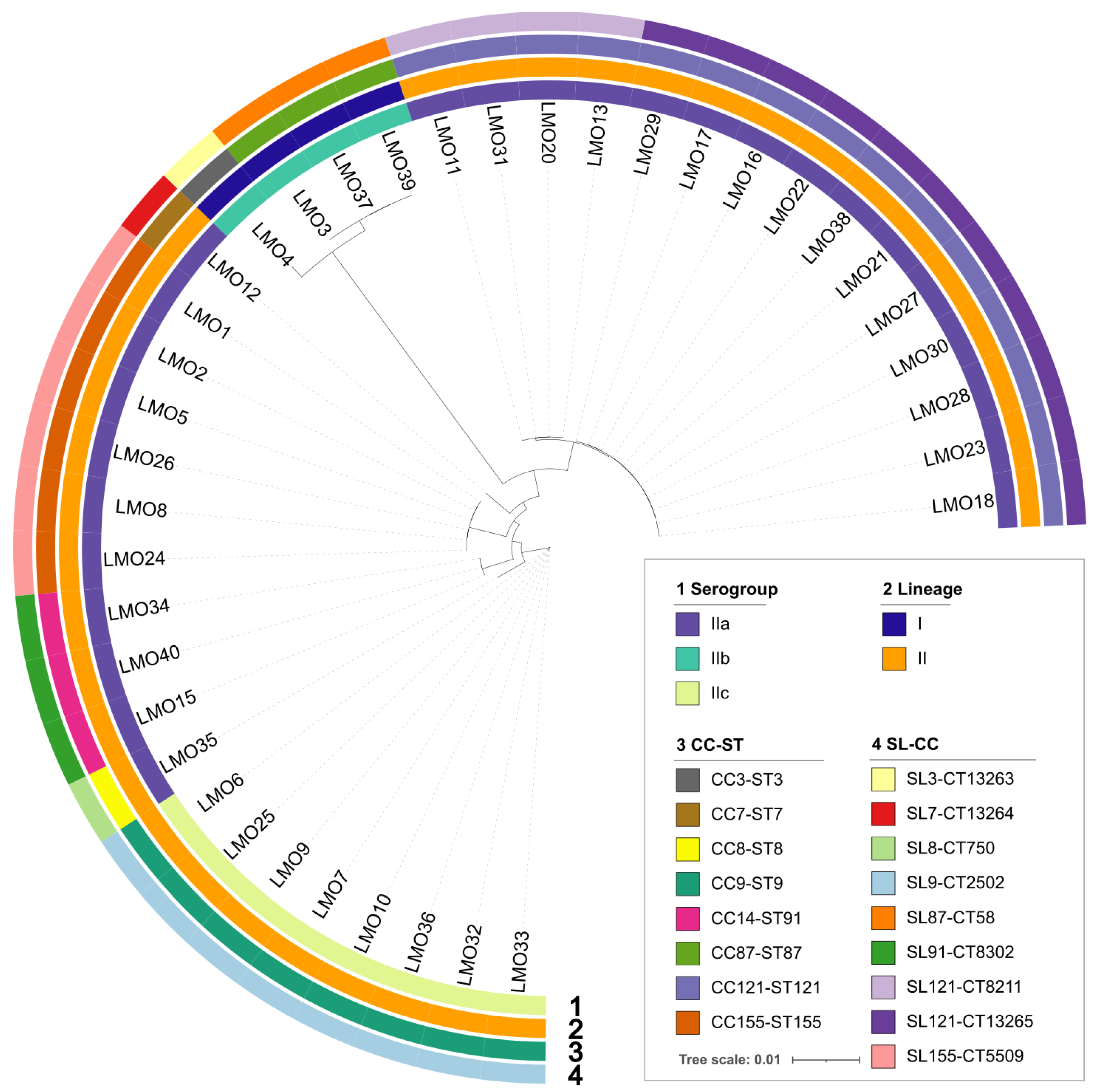
3.2. L. monocytogenes Population Structure
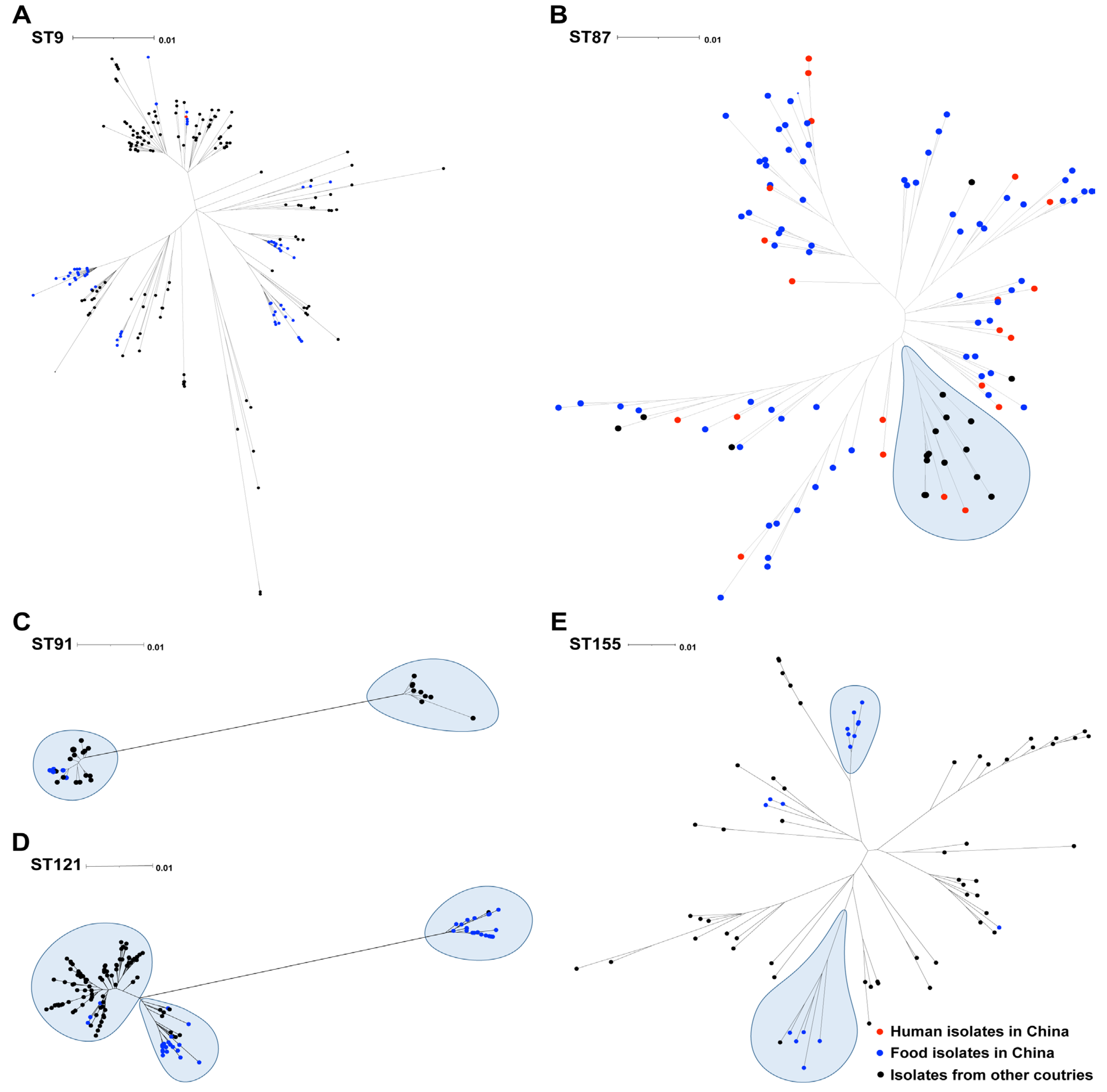
3.3. The Distribution of the Prevalent Sequence Types of L. monocytogenes Isolates from Food in China in the Context of Global Isolates
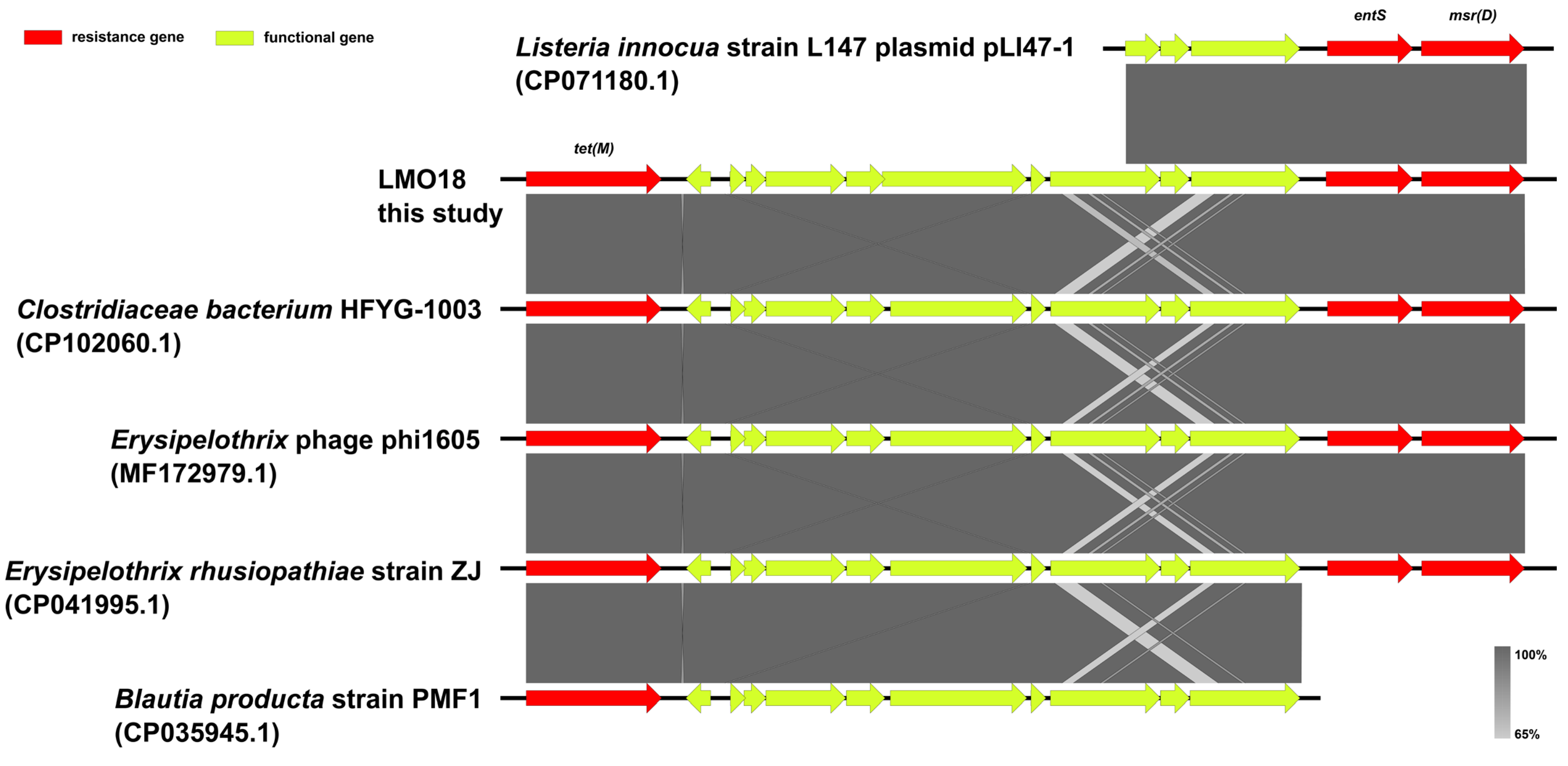
3.4. Antimicrobial Resistance Genes in the Studied L. monocytogenes Isolates
3.5. Assessment of Virulence Factor Profiles and Stress Islands
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meloni, D. Presence of Listeria monocytogenes in mediterranean-style dry fermented sausages. Foods 2015, 4, 34–50. [Google Scholar] [CrossRef]
- Hernandez-Milian, A.; Payeras-Cifre, A. What is new in listeriosis? Biomed Res. Int. 2014, 2014, 358051. [Google Scholar] [CrossRef]
- CDC Listeria (Listeriosis)|Listeria. Available online: https://www.cdc.gov/Listeria/outbreaks/index.html (accessed on 13 September 2022).
- European Food Safety Authority; European Centre for Disease Prevention Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Li, W.; Bai, L.; Fu, P.; Han, H.; Liu, J.; Guo, Y. The Epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 2018, 15, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Beresford, M.R.; Andrew, P.W.; Shama, G. Listeria monocytogenes adheres to many materials found in food-processing environments. J. Appl. Microbiol. 2001, 90, 1000–5. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, T.; Cervantes-Huamán, B.R.H.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes biofilms in the food industry: Is the current hygiene program sufficient to combat the persistence of the pathogen? Microorganisms 2021, 9, 181. [Google Scholar] [CrossRef] [PubMed]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef]
- Cruz, C.D.; Pitman, A.R.; Harrow, S.A.; Fletcher, G.C. Listeria monocytogenes associated with New Zealand seafood production and clinical cases: Unique sequence types, truncated InlA, and attenuated invasiveness. Appl. Environ. Microbiol. 2014, 80, 1489–1497. [Google Scholar] [CrossRef]
- Manuel, C.S.; Van Stelten, A.; Wiedmann, M.; Nightingale, M.; Orsi, R.H. Prevalence and distribution of Listeria monocytogenes inlA alleles prone to phase variation and inlA alleles with premature stop codon mutations among human, food, animal, and environmental isolates. Appl. Environ. Microbiol. 2015, 81, 8339–8345. [Google Scholar] [CrossRef]
- Leclercq, A.; Chenal-Francisque, V.; Dieye, H.; Cantinelli, T.; Drali, R.; Brisse, S.; Lecuit, M. Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb-v1. Int. J. Food Microbiol. 2011, 147, 74–77. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Author Correction: Hypervirulent Listeria monocytogenes clones’ adaptation to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 3619. [Google Scholar] [CrossRef]
- Matle, I.; Mbatha, K.R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, e1–e20. [Google Scholar] [CrossRef]
- Abdeen, E.E.; Mousa, W.S.; Harb, O.H.; Fath-Elbab, G.A.; Nooruzzaman, M.; Gaber, A.; Alsanie, W.F.; Abdeen, A. Prevalence, antibiogram and genetic characterization of Listeria monocytogenes from food products in Egypt. Foods 2021, 10, 1381. [Google Scholar] [CrossRef]
- Singh, R.; Kusalik, A.; Dillon, J.R. Bioinformatics tools used for whole-genome sequencing analysis of Neisseria gonorrhoeae: A literature review. Brief. Funct. Genom. 2022, 21, 78–89. [Google Scholar] [CrossRef]
- Truchado, P.; Gil, M.I.; Querido-Ferreira, A.P.; Capón, C.L.; Álvarez-Ordoñez, A.; Allende, A. Frozen vegetable processing plants can harbour diverse Listeria monocytogenes populations: Identification of critical operations by WGS. Foods 2022, 11, 1546. [Google Scholar] [CrossRef]
- Tirloni, E.; Bernardi, C.; Pomilio, F.; Torresi, M.; De Santis, E.P.L.; Scarano, C.; Stella, S. Occurrence of Listeria spp. and Listeria monocytogenes isolated from PDO Taleggio production plants. Foods 2020, 9, 1636. [Google Scholar] [CrossRef]
- Oliveira, M.; Antunes, W.; Mota, S.; Madureira-Carvalho, Á.; Dinis-Oliveira, R.J.; Dias da Silva, D. An overview of the recent advances in antimicrobial resistance. Microorganisms 2024, 12, 1920. [Google Scholar] [CrossRef] [PubMed]
- Poyart-Salmeron, C.; Carlier, C.; Trieu-Cuot, P.; Courtieu, A.L.; Courvalin, P. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 1990, 335, 1422–1426. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, Y.; Yamada, F.; Batmunkh, O.; Mochizuki, M.; Takano, T.; Hondo, R.; Ueda, F. Prevalence of Listeria monocytogenes in retailed meat in the Tokyo metropolitan area. J. Food Prot. 2010, 73, 1688–1693. [Google Scholar] [CrossRef]
- GB 4789.30-2016; National Food Safety Standard of China-Food Microbiological Examination, Listeria monocytogenes. Standards Press of China: Beijing, China, 2016.
- Liu, P.; Mizue, H.; Fujihara, K.; Kobayashi, H.; Kamikado, H.; Tanaka, T.; Honjoh, K.; Miyamoto, T. A new rapid real-time PCR method for detection of Listeria monocytogenes targeting the hlyA gene. Food Sci. Technol. Res. 2012, 18, 47–57. [Google Scholar] [CrossRef][Green Version]
- Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; M45. CLSI: Wayne, PA, USA, 2015. [Google Scholar][Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Moura, A.; Tourdjman, M.; Leclercq, A.; Hamelin, E.; Laurent, E.; Fredriksen, N.; Van Cauteren, D.; Bracq-Dieye, H.; Thouvenot, P.; Vales, G.; et al. Real-Time Whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017, 23, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Camargo, A.C.; Moura, A.; Avillan, J.; Herman, N.; McFarland, A.P.; Sreevatsan, S.; Call, D.R.; Woodward, J.J.; Lecuit, M.; Nero, L.A. Whole-genome sequencing reveals Listeria monocytogenes diversity and allows identification of long-term persistent strains in Brazil. Environ. Microbiol. 2019, 21, 4478–4487. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Van Stelten, A.; Simpson, J.M.; Ward, T.J.; Nightingale, K.K. Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 2010, 76, 2783–2790. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, J.; Osborne, J.; Kathariou, S. Listeria monocytogenes source distribution analysis indicates regional heterogeneity and ecological niche preference among serotype 4b clones. MBio 2018, 9, e00396-18. [Google Scholar] [CrossRef]
- Painset, A.; Björkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Félix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. LiSEQ–whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 2019, 5, e000257. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef]
- Marcus, R.; Hurd, S.; Mank, L.; Mshar, P.; Phan, Q.; Jackson, K.; Watarida, K.; Salfinger, Y.; Kim, S.; Ishida, M.L.; et al. Chicken salad as the source of a case of Listeria monocytogenes infection in Connecticut. J. Food Prot. 2009, 72, 2602–2606. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; APaganini, J.; Guo, R.; Coipan, C.E.; Friesema, I.H.M.; van Hoek, A.H.A.M.; van den Beld, M.; Kuiling, S.; Bergval, I.; Wullings, B.; et al. Source attribution of Listeria monocytogenes in the Netherlands. Int. J. Food Microbiol. 2025, 427, 110953. [Google Scholar] [CrossRef]
- Filipello, V.; Mughini-Gras, L.; Gallina, S.; Vitale, N.; Mannelli, A.; Pontello, M.; Decastelli, L.; Allard, M.W.; Brown, E.W.; Lomonaco, S. Attribution of Listeria monocytogenes human infections to food and animal sources in Northern Italy. Food Microbiol. 2020, 89, 103433. [Google Scholar] [CrossRef]
- Zhang, P.; Ji, L.; Wu, X.; Chen, L.; Yan, W.; Dong, F. Prevalence, genotypic characteristics, and antibiotic resistance of Listeria monocytogenes from retail foods in Huzhou, China. J. Food Prot. 2024, 87, 100307. [Google Scholar] [CrossRef]
- Soge, O.O.; Beck, N.K.; White, T.M.; No, D.B.; Roberts, M.C. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J. Antimicrob. Chemother. 2008, 62, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Gómez, D.; Azón, E.; Marco, N.; Carramiñana, J.J.; Rota, C.; Ariño, A.; Yangüela, J. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014, 42, 61–65. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, Q.; Li, S.; Liu, M. Prevalence and genetic diversity of Listeria monocytogenes isolated from retail pork in Wuhan, China. Front. Microbiol. 2021, 12, 620482. [Google Scholar] [CrossRef]
- Anwar, T.M.; Pan, H.; Chai, W.; Ed-Dra, A.; Fang, W.; Li, Y.; Yue, M. Genetic diversity, virulence factors, and antimicrobial resistance of Listeria monocytogenes from food, livestock, and clinical samples between 2002 and 2019 in China. Int. J. Food Microbiol. 2022, 366, 109572. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, B.; Li, Z.; Liang, Y.; Shi, T.; Dong, Q.; Chen, M.; Wu, H.; Zhang, H. Research on the genetic polymorphism and function of inlA with premature stop codons in Listeria monocytogenes. Foods 2025, 14, 2955. [Google Scholar] [CrossRef] [PubMed]
- Magagna, G.; Gori, M.; Russini, V.; De Angelis, V.; Spinelli, E.; Filipello, V.; Tranquillo, V.M.; De Marchis, M.L.; Bossù, T.; Fappani, C.; et al. Evaluation of the virulence potential of Listeria monocytogenes through the characterization of the truncated forms of Internalin, A. Int. J. Mol. Sci. 2023, 24, 10141. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Marquis, H.; Boor, K.J. σ B contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 2005, 151, 3215–3222. [Google Scholar] [CrossRef]
- Phelps, C.C.; Vadia, S.; Arnett, E.; Tan, Y.; Zhang, X.; Pathak-Sharma, S.; Gavrilin, M.A.; Seveau, S. Relative roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 2018, 86, e00555-18. [Google Scholar] [CrossRef]


| Antimicrobial Agent | Resistant (n = 38) | |
|---|---|---|
| n | % | |
| erythromycin | 8 | 21.1% |
| tetracycline | 8 | 21.1% |
| trimethoprim/sulfamethoxazole | 0 | 0.0% |
| ciprofloxacin | 0 | 0.0% |
| ampicillin | 0 | 0.0% |
| penicillin | 0 | 0.0% |
| vancomycin | 0 | 0.0% |
| meropenem | 0 | 0.0% |
| clindamycin | 0 | 0.0% |
| Phylogenetic Lineage/Serogroups | CC-ST | SL-CT | SSI | LGI | LIPI | No. of Isolates |
|---|---|---|---|---|---|---|
| lineage I/IIb | CC87-ST87 | SL87-CT58 | ND a | LGI-2 | LIPI-1/LIPI-4 | 3 |
| CC3-ST3 | SL3-CT13263 | SSI-1 | LGI-2/LGI-3 | LIPI-1/LIPI-3 | 1 | |
| lineage II/IIa | CC121-ST121 | SL121-CT13265 | SSI-2 | LGI-2/LGI-3 | LIPI-1 | 11 |
| CC121-ST121 | SL121-CT8211 | SSI-2 | LGI-2/LGI-3 | LIPI-1 | 4 | |
| CC14-ST91 | SL91-CT8302 | ND a | LGI-2 | LIPI-1 | 3 | |
| CC155-ST155 | SL155-CT5509 | SSI-1 | LGI-2 | LIPI-1 | 6 | |
| CC7-ST7 | SL7-CT13264 | SSI-1 | LGI-2 | LIPI-1 | 1 | |
| CC8-ST8 | SL8-CT1750 | SSI-1 | LGI-2/LGI-3 | LIPI-1 | 1 | |
| lineage II/IIc | CC9-ST9 | SL9-CT2502 | SSI-1 | LGI-2 | LIPI-1 | 6 |
| CC9-ST9 | SL9-CT2502 | SSI-1 | LGI-2/LGI-3 | LIPI-1 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Zhong, Y.; Jia, J.; Ma, L.; Lu, Y.; Wang, Q.; Gao, L.; Cao, J.; Dong, Y.; Zheng, Q.; et al. Genomic Landscape and Antimicrobial Resistance of Listeria monocytogenes in Retail Chicken in Qingdao, China. Foods 2025, 14, 3260. https://doi.org/10.3390/foods14183260
Wang W, Zhong Y, Jia J, Ma L, Lu Y, Wang Q, Gao L, Cao J, Dong Y, Zheng Q, et al. Genomic Landscape and Antimicrobial Resistance of Listeria monocytogenes in Retail Chicken in Qingdao, China. Foods. 2025; 14(18):3260. https://doi.org/10.3390/foods14183260
Chicago/Turabian StyleWang, Wei, Yao Zhong, Juntao Jia, Lidan Ma, Yan Lu, Qiushui Wang, Lijuan Gao, Jijuan Cao, Yinping Dong, Qiuyue Zheng, and et al. 2025. "Genomic Landscape and Antimicrobial Resistance of Listeria monocytogenes in Retail Chicken in Qingdao, China" Foods 14, no. 18: 3260. https://doi.org/10.3390/foods14183260
APA StyleWang, W., Zhong, Y., Jia, J., Ma, L., Lu, Y., Wang, Q., Gao, L., Cao, J., Dong, Y., Zheng, Q., & Xiao, J. (2025). Genomic Landscape and Antimicrobial Resistance of Listeria monocytogenes in Retail Chicken in Qingdao, China. Foods, 14(18), 3260. https://doi.org/10.3390/foods14183260








