Identification of Candidate Gene Networks Controlling Soluble Sugar Metabolism During Brassica napus L. Development by Integrated Analysis of Metabolic and Transcriptomic Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Determination of Soluble Sugar and Starch Content
2.3. RNA Extraction, Sequencing, and Transcriptome Data Analysis
2.4. Metabolite Detection and Data Analysis
2.5. Quantitative Real-Time PCR Validation
2.6. Statistical Analysis
3. Results
3.1. Soluble Sugar and Starch Concentration During Development
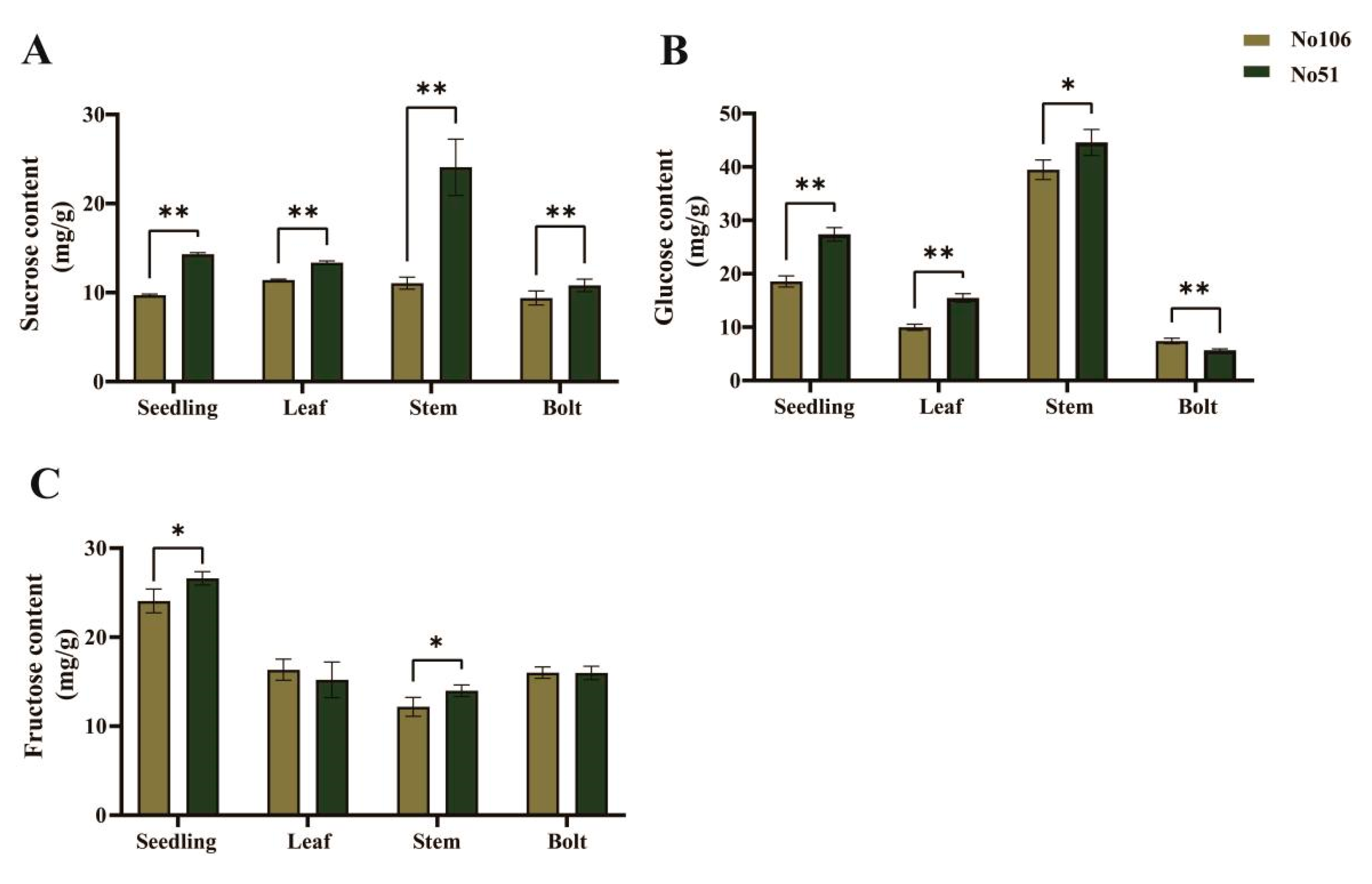
3.2. Analysis of Soluble Sugar Content in Different Rapeseeds
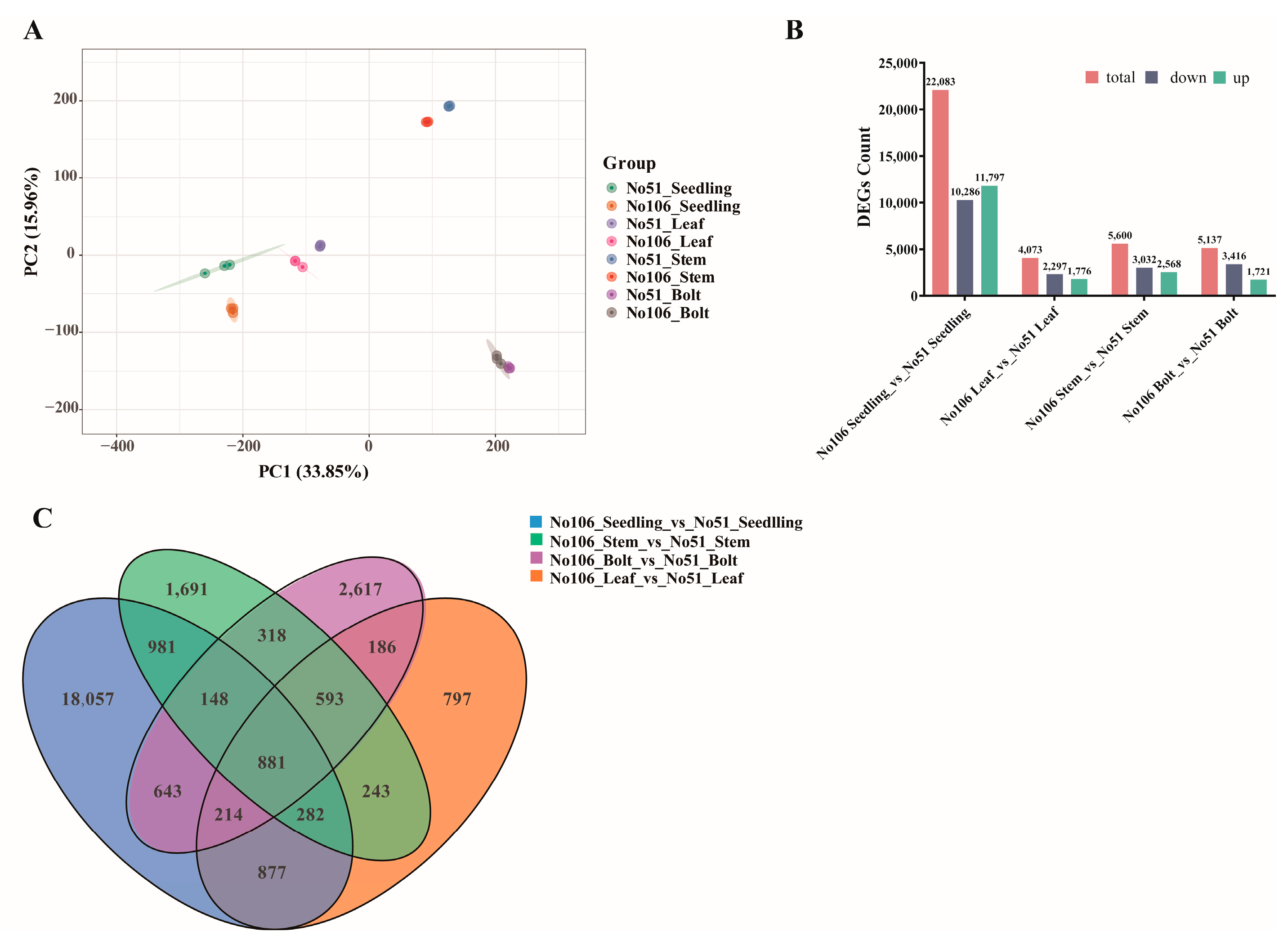
3.3. Transcriptome Analysis and DEGs
3.4. Differentially Expressed Metabolites (DEMs) of Two Different Rapeseeds
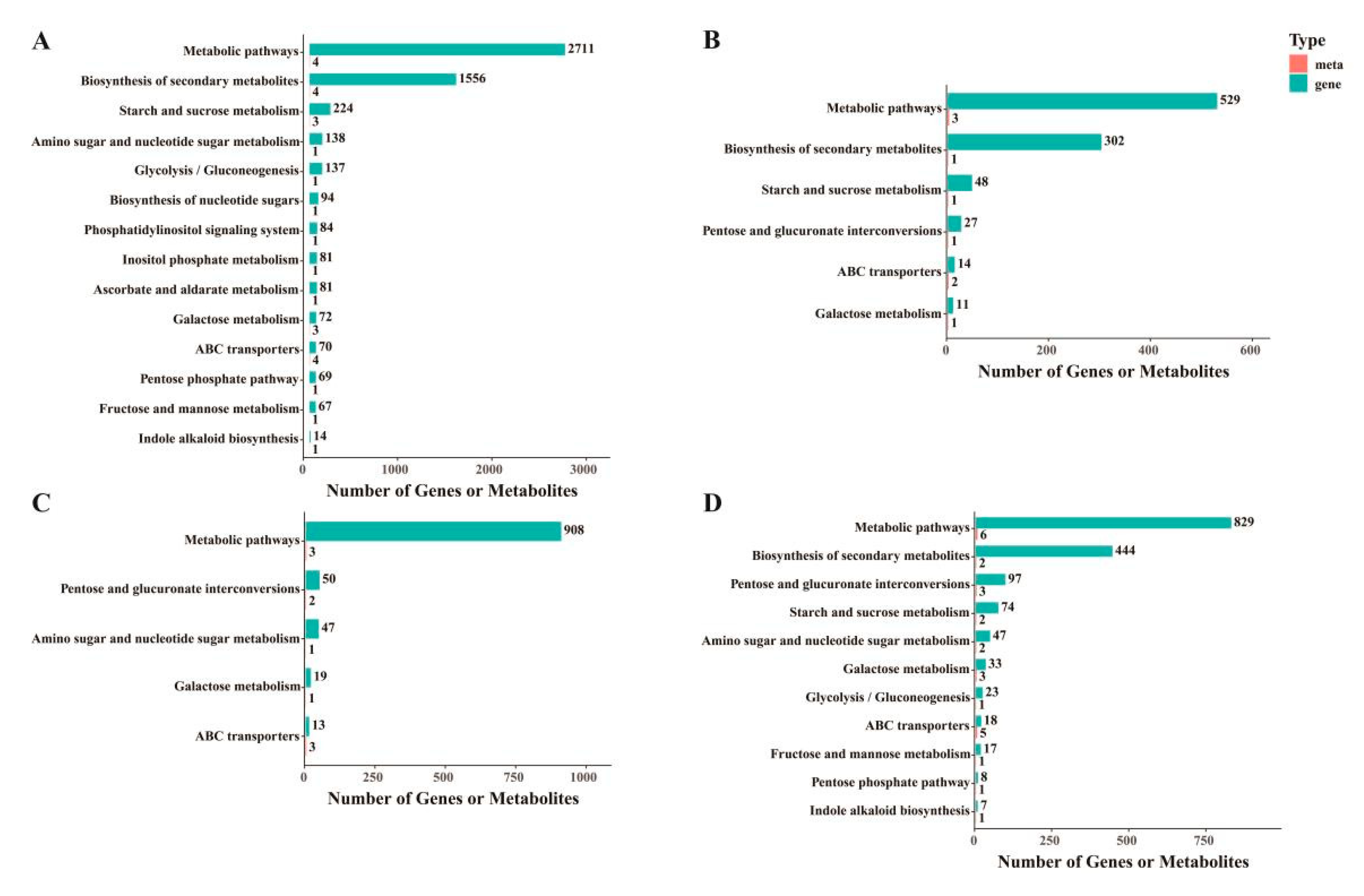
3.5. Combined Transcriptome and Metabolome Analysis
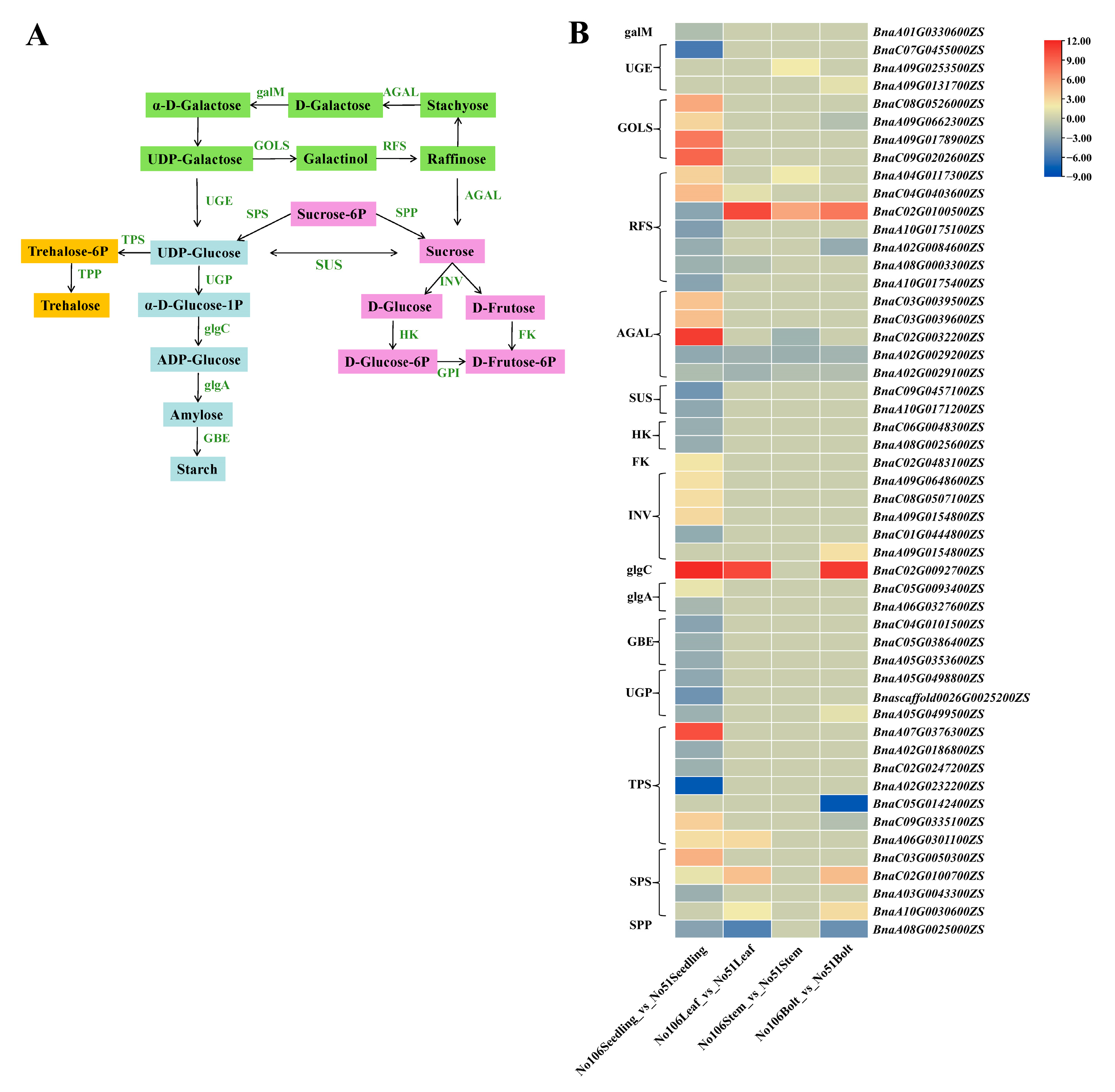
3.6. Gene Networks Regulating Soluble Sugar and Organic Acid Metabolism
3.7. qRT-PCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiao, Z.; Pan, Y.; Wang, C.; Li, X.; Lu, Y.; Tian, Z.; Kuang, L.; Wang, X.; Dun, X.; Wang, H. Multi-Functional Development and Utilization of Rapeseed: Comprehensive Analysis of the Nutritional Value of Rapeseed Sprouts. Foods 2022, 11, 778. [Google Scholar] [CrossRef]
- Li, X.; Huang, W.; Yang, Z.; Hu, W.; Zhou, Z.; Chen, B. Leaf and pod growth affect seed yield after shoot removal and different nitrogen rates of dual-purpose rapeseed (Brassica napus L.). J. Integr. Agric. 2025; in press. [Google Scholar] [CrossRef]
- Wu, X.; Chen, F.; Zhao, X.; Pang, C.; Shi, R.; Liu, C.; Sun, C.; Zhang, W.; Wang, X.; Zhang, J. QTL Mapping and GWAS Reveal the Genetic Mechanism Controlling Soluble Solids Content in Brassica napus Shoots. Foods 2021, 10, 2400. [Google Scholar] [CrossRef]
- Dagar, H.; Hooda, V.; Raj, D.; Dagar, C.; Rathore, A.; Dhanda, A.; Dhanker, P. Effect of seed rate and fertilizer levels on fodder quality, yield and economics of dual purpose wheat in western zone of Haryana. J. Plant Nutr. 2023, 7, 1186–1196. [Google Scholar] [CrossRef]
- Shan, Y.; Liao, Q.; Yuan, J.; Wan, X.; Liao, Y. Effects of Cutting Parameters on the Shearing Properties of Oilseed Rape (Brassica napus L.) Shoots in Harvesting. Appl. Eng. Agric. 2024, 40, 339–349. [Google Scholar] [CrossRef]
- Tao, H.; Zhao, Y.; Li, L.; He, Y.; Zhang, X.; Zhu, Y.; Hong, G. Comparative metabolomics of flavonoids in twenty vegetables reveal their nutritional diversity and potential health benefits. Food Res. Int. 2023, 164, 112384. [Google Scholar] [CrossRef] [PubMed]
- Kristal, A.; Lampe, J. Brassica vegetables and prostate cancer risk: A review of the epidemiological evidence. Nutr. Cancer 2002, 42, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Giovannucci, E.; Hunter, D.; Neuberg, D.; Su, L.; Christiani, D. Dietary intake of Cruciferous vegetables, Glutathione S-transferase (GST) polymorphisms and lung cancer risk in a Caucasian population. Cancer Causes Control. 2004, 15, 977–985. [Google Scholar] [CrossRef]
- Fenwick, G.; Griffiths, N.; Heaney, R. Bitterness in brussels sprouts (Brassica oleracea L. vargemmifera): The role of glucosinolates and their breakdown products. J. Sci. Food Agric. 1983, 34, 73–80. [Google Scholar] [CrossRef]
- Padilla, G.; Cartea, M.; Velasco, P.; de Haro, A.; Ordás, A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry 2007, 68, 536–545. [Google Scholar] [CrossRef]
- Dinehart, M.; Hayes, J.; Bartoshuk, L.; Lanier, S.; Duffy, V. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake. Physiol. Behav. 2006, 87, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Engel, E.; Baty, C.; Le Corre, D.; Souchon, I.; Martin, N. Flavor-active compounds potentially implicated in cooked cauliflower acceptance. J. Agric. Food Chem. 2002, 50, 6459–6467. [Google Scholar] [CrossRef]
- Hussain, S.; Guo, L.; Shi, C.; Khan, M.; Bai, Y.; Du, W.; Liu, Y. Assessment of sugar and sugar accumulation-related gene expression profiles reveal new insight into the formation of low sugar accumulation trait in a sweet orange (Citrus sinensis) bud mutant. Mol. Biol. Rep. 2020, 47, 2781–2791. [Google Scholar] [CrossRef]
- Auerswald, H.; Schwarz, D.; Kornelson, C.; Brückner, B. Sensory analysis, sugar and acid content of tomato at different EC values of the nutrient solution. Sci. Hortic. 1999, 82, 227–242. [Google Scholar] [CrossRef]
- Jahangir, M.; Kim, H.; Choi, Y.; Verpoorte, R. Health-Affecting Compounds in Brassicaceae. Compr. Rev. Food Sci. Food Saf. 2010, 8, 31–43. [Google Scholar] [CrossRef]
- Ren, R.; Yue, X.; Li, J.; Xie, S.; Guo, S.; Zhang, Z. Coexpression of Sucrose Synthase and the SWEET Transporter, Which Are Associated with Sugar Hydrolysis and Transport, Respectively, Increases the Hexose Content in Vitis vinifera L. Grape Berries Front. Plant Sci. 2020, 11, 321. [Google Scholar] [CrossRef] [PubMed]
- Miron, D.; Schaffer, A. Sucrose Phosphate Synthase, Sucrose Synthase, and Invertase Activities in Developing Fruit of Lycopersicon esculentum Mill and the Sucrose Accumulating Lycopersicon hirsutum Humb and Bonpl. Plant Physiol. 1991, 95, 623–627. [Google Scholar] [CrossRef]
- Granot, D.; David-Schwartz, R.; Kelly, G. Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci. 2013, 4, 44. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Yu, O.; Tang, J.; Gu, X.; Wan, X.; Fang, C. Metabolic profiling of strawberry (Fragaria × ananassa Duch) during fruit development and maturation. J. Exp. Bot. 2011, 62, 1103–1118. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Tian, C.; Ji, S.; Ni, F.; Fan, X.; Yang, Y.; Sun, C.; Gong, H.; Zhang, A. Integrative analyses of metabolome and transcriptome reveals metabolomic variations and candidate genes involved in sweet cherry (Prunus avium L.) fruit quality during development and ripening. PLoS ONE 2021, 16, e0260004. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.; Li, J.; Zhang, Y.; Zhou, D.; Li, C.; He, L.; Li, H.; Wang, F.; Gao, J. Identification of key genes controlling soluble sugar and glucosinolate biosynthesis in Chinese cabbage by integrating metabolome and genome-wide transcriptome analysis. Front. Plant Sci. 2022, 13, 1043–1489. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, W.; Feng, Y.; Tu, M.; Wittich, P.; Bate, N.; Messing, J. Transcriptome and metabolome reveal distinct carbon allocation patterns during internode sugar accumulation in different sorghum genotypes. Plant Biotechnol. J. 2019, 17, 472–487. [Google Scholar] [CrossRef]
- Sanyal, R.; Pradhan, B.; Jawed, D.; Tribhuvan, K.; Dahuja, A.; Kumar, M.; Kumar, N.; Mishra, G.; Ram, C.; Mahatma, M.; et al. Spatio-temporal expression pattern of Raffinose Synthase genes determine the levels of Raffinose Family Oligosaccharides in peanut (Arachis hypogaea L) seed. Sci. Rep. 2023, 13, 795. [Google Scholar] [CrossRef] [PubMed]
- Roe, J. The determination of sugar in blood and spinal fluid with anthrone reagent. J. Biol. Chem. 1955, 212, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Zhang, H.; Fan, Y.; Yan, L. Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum). BMC Plant Biol. 2022, 22, 358. [Google Scholar] [CrossRef]
- Gómez-González, S.; Ruiz-Jiménez, J.; Priego-Capote, F.; Luque de Castro, M.D. Qualitative and quantitative sugar profiling in olive fruits, leaves, and stems by gas chromatography-tandem mass spectrometry (GC-MS/MS) after ultrasound-assisted leaching. J. Agric. Food Chem. 2010, 58, 12292–12299. [Google Scholar] [CrossRef]
- Pfaffl, M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2003–2007. [Google Scholar] [CrossRef]
- Cheng, J.; Wen, S.; Li, K.; Zhou, Y.; Zhu, M.; Neuhaus, H.; Bie, Z. The hexose transporters CsHT3 and CsHT16 regulate postphloem transport and fruit development in cucumber. Plant Physiol. 2025, 16, kiae597. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, J.; Li, G. Dynamic epigenetic modifications in plant sugar signal transduction. Trends Plant Sci. 2022, 27, 379–390. [Google Scholar] [CrossRef]
- Wang, R.; Shu, P.; Zhang, C.; Zhang, J.; Chen, Y.; Zhang, Y.; Du, K.; Xie, Y.; Li, M.; Ma, T.; et al. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol. 2022, 233, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Pangborn, R. Relative Taste Intensities of Selected Sugars and Organic. Acidsa J. Food Sci. 1963, 28, 726–733. [Google Scholar] [CrossRef]
- Lee, J.; Dong, X.; Choi, K.; Song, H.; Yi, H.; Hur, Y. Identification of source-sink tissues in the leaf of Chinese cabbage (Brassica rapa ssp. pekinensis) by carbohydrate content and transcriptomic analysis. Genes Genom. 2020, 42, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yue, Z.; Zhong, X.; Lei, J.; Tao, P.; Li, B. Distribution of primary and secondary metabolites among the leaf layers of headed cabbage (Brassica oleracea var capitata). Food Chem. 2020, 312, 126028. [Google Scholar] [CrossRef]
- Xie, F.; Xuan, Z.; Chen, W.; Du, J.; Wang, Y.; Huo, E.; Guo, L.; Li, M. Metabolomics Analysis Reveals Dynamic Accumulation of Sugar and Acid During Stem Development of Brassica juncea. Agronomy 2022, 12, 3227. [Google Scholar] [CrossRef]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source-sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef]
- Liu, Q.; Li, J.; Liu, W. Sugar accumulation and characterization of metabolizing enzyme genes in leafy head of Chinese cabbage (Brassica campestris L. ssp. pekinensis). Hortic. Environ. Biotechnol. 2021, 62, 17–29. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, M.; Yin, X.; Liu, Z.; Zhou, L.; Hu, X.; Yi, J. Metabolite accumulation pathway of watercore in apple fruit: An integrative transcriptome and metabolome analysis. Sci. Hortic. 2025, 340, 113954. [Google Scholar] [CrossRef]
- Tieman, D.; Zhu, G.; Resende, M.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.; Beltran, K.; Taylor, M.; Zhang, B.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef]
- Cheng, H.; Kong, W.; Tang, T.; Ren, K.; Zhang, K.; Wei, H.; Lin, T. Identification of Key Gene Networks Controlling Soluble Sugar and Organic Acid Metabolism During Oriental Melon Fruit Development by Integrated Analysis of Metabolic and Transcriptomic Analyses. Front. Plant Sci. 2022, 13, 830517. [Google Scholar] [CrossRef]
- Umer, M.; Bin, S.; Gebremeskel, H.; Zhao, S.; Yuan, P.; Zhu, H.; Kaseb, M.; Anees, M.; Lu, X.; He, N.; et al. Identification of key gene networks controlling organic acid and sugar metabolism during watermelon fruit development by integrating metabolic phenotypes and gene expression profiles. Hortic. Res. 2020, 7, 193. [Google Scholar] [CrossRef]
- Liao, G.; Li, Y.; Wang, H.; Liu, Q.; Zhong, M.; Jia, D.; Huang, C.; Xu, X. Genome-wide identification and expression profiling analysis of sucrose synthase (SUS) and sucrose phosphate synthase (SPS) genes family in Actinidia chinensis and A. eriantha. BMC Plant Biol. 2022, 22, 215. [Google Scholar] [CrossRef] [PubMed]
- Coculo, D.; Lionetti, V. The Plant Invertase/Pectin Methylesterase Inhibitor Superfamily. Front. Plant Sci. 2022, 13, 863–892. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Guo, X.; Du, J.; Yu, M. ALA Promotes Sucrose Accumulation in Early Peach Fruit by Regulating SPS Activity Current. Issues Mol. Biol. 2024, 46, 7944–7954. [Google Scholar] [CrossRef]
- Albi, T.; Ruiz, M.; de Los Reyes, P.; Valverde, F.; Romero, J. Characterization of the Sucrose Phosphate Phosphatase (SPP) Isoforms from Arabidopsis thaliana and Role of the S6PPc Domain in Dimerization. PLoS ONE 2016, 11, e0166308. [Google Scholar] [CrossRef]
- De Jesus, A.; Keyhani-Nejad, F.; Pusec, C.; Goodman, L.; Geier, J.; Stoolman, J.; Stanczyk, P.; Nguyen, T.; Xu, K.; Suresh, K.; et al. Hexokinase 1 cellular localization regulates the metabolic fate of glucose. Mol. Cell 2022, 82, 1261–1277. [Google Scholar] [CrossRef]
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2014, 65, 809–819. [Google Scholar] [CrossRef]
- Bahaji, A.; Baroja-Fernández, E.; Ricarte-Bermejo, A.; Sánchez-López, Á.; José Muñoz, F.; Jose, M.; Ruiz, M.; Baslam, M.; Almagro, G.; Sesma, M.; et al. Characterization of multiple SPS knockout mutants reveals redundant functions of the four Arabidopsis sucrose phosphate synthase isoforms in plant viability, and strongly indicates that enhanced respiration and accelerated starch turnover can alleviate the blockage of sucrose biosynthesis. Plant Sci. 2015, 238, 135–147. [Google Scholar] [CrossRef]
- Anur, R.; Mufithah, N.; Sawitri, W.; Sakakibara, H.; Sugiharto, B. Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane. Plants 2020, 9, 200. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Canam, T.; Kang, K.; Ellis, D.; Mansfield, S. Over-expression of an arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 2008, 17, 181–192. [Google Scholar] [CrossRef]
- Bagnato, L.; Tosato, E.; Gurrieri, L.; Trost, P.; Forlani, G.; Sparla, F. Arabidopsis thaliana Sucrose Phosphate Synthase A2 Affects Carbon Partitioning and Drought. Response Biol. 2023, 12, 685. [Google Scholar] [CrossRef]
- Sengupta, S.; Mukherjee, S.; Basak, P.; Majumder, A. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 2015, 6, 656. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, Y.; Liu, Y.; Li, X.; Hao, G.; Han, Q.; Dirk, L.; Downie, A.; Ruan, Y.; Wang, J.; et al. Raffinose synthase enhances drought tolerance through raffinose synthesis or galactinol hydrolysis in maize and Arabidopsis plants. J. Biol. Chem. 2020, 295, 8064–8077. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, B.; Guan, C.; Guan, M. Identification of Candidate Gene Networks Controlling Soluble Sugar Metabolism During Brassica napus L. Development by Integrated Analysis of Metabolic and Transcriptomic Analyses. Foods 2025, 14, 2874. https://doi.org/10.3390/foods14162874
Zhou B, Guan C, Guan M. Identification of Candidate Gene Networks Controlling Soluble Sugar Metabolism During Brassica napus L. Development by Integrated Analysis of Metabolic and Transcriptomic Analyses. Foods. 2025; 14(16):2874. https://doi.org/10.3390/foods14162874
Chicago/Turabian StyleZhou, Bingqian, Chunyun Guan, and Mei Guan. 2025. "Identification of Candidate Gene Networks Controlling Soluble Sugar Metabolism During Brassica napus L. Development by Integrated Analysis of Metabolic and Transcriptomic Analyses" Foods 14, no. 16: 2874. https://doi.org/10.3390/foods14162874
APA StyleZhou, B., Guan, C., & Guan, M. (2025). Identification of Candidate Gene Networks Controlling Soluble Sugar Metabolism During Brassica napus L. Development by Integrated Analysis of Metabolic and Transcriptomic Analyses. Foods, 14(16), 2874. https://doi.org/10.3390/foods14162874




