Abstract
Ratoon rice offered higher yields and superior grain quality compared to main rice. This study compared differences in germination, chalkiness, and antioxidant enzyme activities between ratoon rice and main rice of cultivars 19X and NJXM, and analyzed the underlying quality improvement mechanisms from a transcriptome perspective. The results demonstrated that ratoon rice exhibited a significantly greater germination potential and germination rate, reduced chalky grain rate and chalkiness degree, and exhibited higher activities of superoxide dismutase (SOD) and catalase (CAT). Transcriptome analysis revealed that the enhanced germination in ratoon rice may result from significantly upregulated expression of genes qLTG3-1, OsLOX2, OsSAMDC2, and OsSAMDC4. The reduction in chalkiness in ratoon rice may involve the following three causes. First, the reduction of chalkiness may be due to the significantly upregulated GAD3 expression by enhancing high-temperature tolerance. Second, the significantly upregulated expression of peroxidase genes (prx86, POX8.1, Perox4) and significantly downregulated OsEBP89 expression potentially increased the oxidative stress tolerance to reduce the chalkiness of ratoon rice. Finally, the significantly upregulated OsNCED3 expression potentially modulated plant hormones to decrease the chalkiness of ratoon rice. These findings provided novel insights into revealing the mechanisms underlying the superior quality of ratoon rice.
1. Introduction
Rice (Oryza sativa L.) is one of the most important crops worldwide and currently serves as a staple food for more than half of the world’s population [1,2]. In these years, global population growth and industrialization have contributed to a progressive reduction in land available for rice cultivation [3]. Therefore, enhancing yields per unit area under conditions of finite arable land resources has emerged as a critical strategy to address this challenge. Ratoon rice cultivation serves as a viable agricultural intervention, with the literature demonstrating its capacity to enhance total grain production by 58.3% through optimized resource recycling from the first crop, achieving grain yields of up to 3.5 t/ha [4].
Ratooning rice refers to an agricultural practice where new panicles regenerate from remaining axillary buds on the stubble nodes following the harvest of the main crop, thereby enabling an additional harvest from the same cultivation system [5,6]. This dual-cropping mode has demonstrated significant potential for yield augmentation, minimizing requirements for both labor and agricultural land [7,8]. Additionally, the available data indicates that ratooning rice cultivation can reduce global greenhouse gas emissions and enhance the net ecosystem economic benefit of the crop [9]. It was reported that major rice-producing countries, including India, Indonesia, Iran, Japan, Nigeria, and the United States, were accelerating the promotion of ratoon rice cultivation systems [10]. As the northernmost critical zone for the distribution of this system, Japan’s Kanto region (35–36° N) was constrained by climatic conditions and the length of the growing season, resulting in the adoption of short-duration cold-tolerant japonica rice varieties. Notably, current ratoon rice practices in other parts of Asia predominantly rely on high-yielding indica or indica hybrid rice adapted to warm environments [11].
Currently, several studies have been conducted on the quality differences between ratoon rice and main rice. It was reported that the main harvest rice had a significantly higher length-to-width ratio, while its grain yield and chalkiness were significantly lower than those of ratoon rice. Additionally, the amylase content and alkali spreading value of the main rice were significantly lower than those of ratoon rice [12]. Lin et al. demonstrated that the seed setting rate, chalky grain rate, and chalkiness of ratoon rice were significantly reduced compared to the main rice. Such improvements correlated with a shift in metabolomic composition, especially regarding variations in sugar components in carbohydrate metabolism. Furthermore, rice ratooning elevated ascorbate levels and oxidative stress resilience in rice grains [2]. A previous study indicated that ratoon rice exhibited increased amylose content, decreased protein content, reduced relative crystallinity of starch, and higher hardness, cohesiveness, adhesiveness, and resilience compared to main-season rice, along with higher sensory scores [4].
Transcriptome analysis can profile all transcripts produced in plant cells and tissues, including the number of transcripts, expression dynamics at specific developmental stages, post-transcriptional modifications, and regulation of non-coding RNAs [13,14]. Transcriptome analysis has been widely applied in rice research. It primarily revealed important biological processes in rice, such as stress resistance, growth and development mechanisms, and metabolic regulation, by analyzing gene expression patterns and regulatory networks under different conditions [15]. Recent research integrating physiological, molecular, and transcriptome analyses revealed the molecular mechanisms underlying yield variation and chalkiness improvement in ratoon rice. These changes were found to be associated with the gene GF14f, which exerted negative impacts on antioxidant capacity and environmental stress resistance in ratoon rice. Furthermore, the detrimental effects of GF14f exhibited constitutive expression, being independent of seasonal or environmental conditions [16]. To elucidate the molecular mechanisms underlying antioxidant properties in rice during the post-grain filling stage, transcriptome analysis was employed to study the changes of gene expression at five distinct time points following grain filling. Through the construction of a weighted gene co-expression network (WGCNA) and modular clustering analysis of genes, specific modules exhibiting significant associations with antioxidant enzyme activities were successfully identified. Within these modules, hub genes that were significantly correlated with antioxidant traits were further characterized [17].
This study aimed to elucidate the molecular basis of quality trait differences between main rice and ratoon rice. We investigated key physiological quality parameters—including germination potential, germination rate, chalky grain incidence, and chalkiness degree—alongside antioxidative enzyme profiles (SOD, CAT activities). Utilizing transcriptome analysis, we identified differentially expressed genes (DEGs). This work could provide a theoretical foundation to support the broader promotion and application of ratoon rice systems.
2. Materials and Methods
2.1. Experimental Materials
Two indica rice cultivars, 19 Xiang and Nanjing Xiangmi, were used as experimental materials. They were cultivated in Jiangxi Province, China, with both main rice (MR) and ratoon rice (RR) harvested in August and October 2023, respectively. The two varieties of the main rice were designated as 19X-MR and NJXM-MR, and the two varieties of the ratoon rice were denoted as 19X-RR and NJXM-RR.
The intact paddy grains were selected for determining germination potential and germination rate. Subsequently, the paddy grains were dehulled using a hulling machine (Satake Corporation, Hiroshima, Japan), and brown rice samples were obtained after removing husks and broken particles. The brown rice samples can be used for antioxidant enzyme activity analysis. Some of the brown rice samples were placed in a rice milling machine (Suzhou Jiangdong Precision Instrument Co., Ltd., Suzhou, China) and the milling time was adjusted to 40 s to remove the cortex and embryo. After sieving, intact milled rice grains were collected for the determination of chalky grain rate and chalkiness degree.
2.2. Experimental Design
The MR and the RR were harvested in August 2023 and October 2023, respectively. After harvest, they were naturally air-dried, followed by manual removal of impurities. Then, the germination potential, germination rate, chalkiness, activities of superoxide dismutase (SOD) and catalase (CAT), as well as transcriptomics, were determined.
2.3. Determination of Germination Potential and Germination Rate
The determination of germination potential and germination rates was conducted with reference to the Chinese National Standard GB/T 5520-2011. Paddy grains were first soaked in water, and 100 paddy grains were then placed on each germination bed. The germination bed was cultured at 30 °C for a total of 10 days with daily watering. Germination potential referred to the percentage of normally germinated grains on the 3rd day, accounting for the number of test grains, while germination rate was the percentage of all normally germinated grains on the 10th day, accounting for the number of test grains. Four biological replicates were set for all these experiments.
2.4. Determination of Chalky Grain Rate and Chalkiness Degree
The chalky grain rate and chalkiness degree of rice grains were measured using a Rice Appearance Quality Tester (CanoScan 9000F MarkII, Canon, Beijing, China). Rice samples were placed on the instrument scanning platform, and then the sample images were captured.
The calculation formula for chalky grain rate was
The calculation formula for chalkiness degree was
2.5. Determination of SOD Activity
The activity of superoxide dismutase (SOD) was measured using a commercial kit (Solarbio Life Science, BC 0170, Beijing, China). A total of 0.1 g of brown rice flour was homogenized with 1 mL of phosphate-buffered saline (PBS) on ice. The homogenate was centrifuged at 8000× g for 10 min at 4 °C. The resulting supernatant was collected and maintained on ice for subsequent analysis. For superoxide dismutase (SOD) activity determination, the supernatant was incubated with nitroblue tetrazolium (from the assay kit) at 37 °C for 30 min. The absorbance was immediately measured at 560 nm using a spectrophotometer (T700A UV-Vis Spectrophotometer, Beijing Purkinje General Instrument Co., Ltd., Beijing, China). SOD activity was calculated according to the manufacturer’s protocol based on the recorded absorbance values.
2.6. Determination of CAT Activity
The activity of catalase (CAT) was measured using a commercial kit (Solarbio Life Science, BC 0200, Beijing, China). Specifically, 0.1 g of brown rice flour was weighed and homogenized with 1 mL of extraction buffer in an ice bath. The homogenate was centrifuged at 8000× g for 10 min at 4 °C, and the supernatant was collected and stored on ice for subsequent analysis. For catalase (CAT) activity determination, a CAT assay working solution was prepared according to the manufacturer’s specifications (volume-adjusted as needed). The working solution was equilibrated at 25 °C for 10 min. Subsequently, 1 mL of the working solution was transferred to a 1 mL quartz cuvette, followed by the addition of a 35 μL sample. The mixture was immediately mixed by vortexing for 5 s. Absorbance measurements were recorded at 240 nm immediately (t = 0) and after 1 min (t = 1 min) using a spectrophotometer (T700A UV-Vis Spectrophotometer, Beijing Purkinje General Instrument Co., Ltd., Beijing, China) at ambient temperature. CAT activity was calculated from the absorbance difference according to the kit manufacturer’s protocol.
2.7. Determination of Transcriptomics
Total RNA was isolated from fresh rice (0.1 g per sample, immediately frozen in liquid nitrogen after sampling and stored at −80 °C) using the TRIeasy Plus RNA Kit (YeaSen, Shanghai, China) [18]. To ensure RNA quality, RNA integrity was assessed by 1.2% agarose gel electrophoresis (120 V for 30 min), purity was determined using a NanoPhotometer spectrophotometer (Implen, Munich, Germany) with an acceptable OD260/280 ratio of 1.8–2.0, concentration was measured via a Qubit 4.0 fluorometer (Invitrogen, Carlsbad, CA, USA), and the RNA integrity number (RIN) was evaluated using a Qsep400 bioanalyzer (BiOptic, New Taipei City, Taiwan), with a passing threshold of RIN ≥ 7.0. mRNA with polyA tails was enriched using Oligo (dT) magnetic beads (Thermo Fisher, Waltham, MA, USA). Subsequently, RNA was fragmented into short fragments by adding fragmentation buffer (Illumina, San Diego, CA, USA, catalog number RS-122-2103).
Using the short fragment RNA as the template, first-strand cDNA was synthesized with hexamer random primers (Thermo Fisher, Waltham, MA, USA). Then, second-strand cDNA was generated by adding buffer, dNTPs (including dTTP, dATP, dGTP, and dCTP; Takara, Beijing, China), and DNA polymerase (New England Biolabs, Ipswich, MA, USA) I. Double-stranded cDNA was purified using DNA purification magnetic beads (Beckman, Brea, CA, USA). The purified double-stranded cDNA was subjected to end repair, adenylation, and ligation of sequencing adaptors (Illumina). Size selection was performed using DNA purification magnetic beads. Finally, the final cDNA library was obtained via PCR amplification. After passing quality inspection (concentration ≥ 10 ng/μL and expected fragment distribution), sequencing was performed on the Illumina NovaSeq 6000 platform, with three independent biological replicates set for each treatment.
2.8. Statistical Analysis
Four biological replicates were set for the determination of germination potential and germination rate, while three replicates were used for the detection of chalky grain rate, chalkiness degree, superoxide dismutase (SOD) and catalase (CAT) activities, as well as transcriptome analysis.
Statistical analyses were performed using SPSS 27.0 software (IBM Corp., USA) through analysis of variance (ANOVA) followed by Duncan’s multiple range test, with differences considered statistically significant at the 95% confidence level (p < 0.05). Germination potential and germination rate, chalky grain rate and chalkiness degree, superoxide dismutase (SOD), and catalase (CAT) activity graphs were generated using Origin 2021, while transcriptome analysis graphs were created using the online platform of Weishengxin (https://www.bioinformatics.com.cn/, accessed on 10 May 2025).
Gene function analysis was carried out through the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/, accessed on 20 May 2025), the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/index.html, accessed on 25 May 2025), and the National Rice Database of the National Rice Data Center (https://www.ricedata.cn/reference/, accessed on 1 June 2025); the data access dates were from 10 May 2025 to 10 June 2025.
3. Results
3.1. Comparison of Germination Potential (GP) and Germination Rate (GR) Between Main Rice and Ratoon Rice
Seed GP and GR were key indicators for assessing seed quality and germination capacity while serving as a critical metric for evaluating growth potential and plant growing performance [19]. As illustrated in Figure 1, both 19X-RR and NJXM-RR demonstrated significantly higher GP and GR compared to their main rice (p < 0.05) counterparts. Collectively, these results confirmed ratoon rice’s superior seed germination traits.
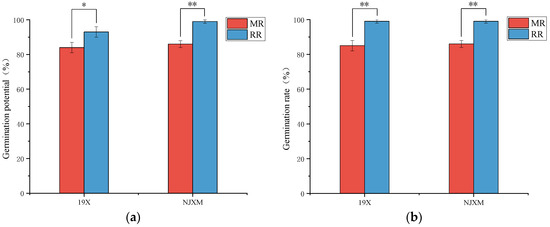
Figure 1.
Comparison of (a) germination potential and (b) germination rate between main rice (MR, red bars) and ratoon rice (RR, blue bars) for two varieties: 19X and NJXM. “*” indicated p < 0.05, “**” indicated p < 0.01.
3.2. Comparison of Chalky Grain Rate and Chalkiness Degree Between Main Rice and Ratoon Rice
Chalkiness referred to the opaque regions in rice grains. Additionally, chalkiness increased the brittleness of rice grains and reduced their market value [20]. As shown in Figure 2, both 19X and NJXM ratoon rice exhibited significantly lower chalky grain rate and chalkiness degree than the main rice (p < 0.05). These results demonstrated significantly superior grain appearance quality in ratoon rice. Alizadeh [12] and Huang [21] et al. had also reported that the chalkiness of ratoon rice was significantly lower than that of main rice, which was consistent with the findings of our study.
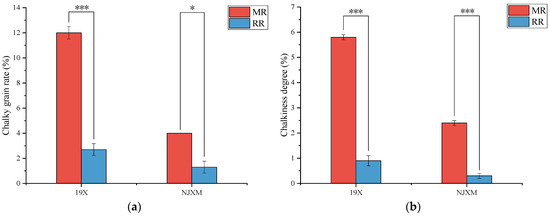
Figure 2.
Comparison of (a) chalky grain rate and (b) chalkiness degree between main rice (MR, red bars) and ratoon rice (RR, blue bars) for two varieties: 19X and NJXM. “*” indicated p < 0.05, “***” indicated p < 0.001.
3.3. Comparison of Superoxide Dismutase (SOD) and Catalase (CAT) Activities Between Main Rice and Ratoon Rice
Superoxide dismutase (SOD) and catalase (CAT) are key antioxidant enzymes in plants, playing essential roles in their metabolic activities. Their primary functions include scavenging reactive oxygen species (ROS), maintaining redox balance, and protecting cells from oxidative damage [22]. SOD serves as the first line of defense in the plant antioxidant system, functioning to catalyze the conversion of superoxide anion radicals (O2−) into hydrogen peroxide (H2O2) and oxygen (O2), thereby reducing the accumulation of reactive oxygen species (ROS) [23]. CAT is responsible for decomposing hydrogen peroxide (H2O2) into water and oxygen, thereby preventing the excessive accumulation of H2O2 in cells [24]. As shown in Figure 3, both 19X-RR and NJXM-RR exhibited significantly higher superoxide dismutase (SOD) and catalase (CAT) activities than the main rice (p < 0.05).
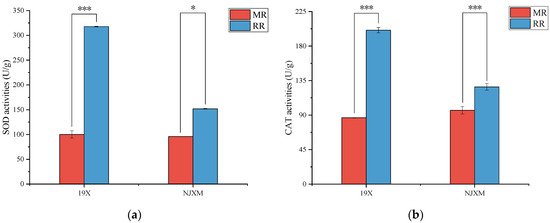
Figure 3.
Comparison of (a) SOD activity and (b) CAT activity between main rice (MR, red bars) and ratoon rice (RR, blue bars) for two varieties: 19X and NJXM. “*” indicated p < 0.05, “***” indicated p < 0.001.
3.4. Transcriptome Analysis
To investigate the differences in quality traits between main rice and ratoon rice, this study employed a transcriptomic approach using high-throughput RNA sequencing to identify differentially expressed genes (DEGs) in the main and ratoon rice of the two paddy varieties.
3.4.1. Quality Analysis of Transcriptome Sequencing
In this experiment, transcriptome sequencing analysis was performed on 12 test samples, including four groups of samples (ratoon rice and main rice of two varieties) with three biological replicates each. A total of 648,859,190 raw reads (raw data read counts) were obtained, and 614,027,496 clean reads (high-quality read counts after raw data filtering) were generated after filtering. The average values of Q20 (the percentage of bases with a Qphred score ≥ 20 in the total number of bases) and Q30 (the percentage of bases with a Qphred score ≥ 30 in the total number of bases) for the 12 samples were 97.75% and 94.13%, respectively, and the mean GC content (the percentage of the sum of guanine G and cytosine C counts in high-quality reads to the total number of bases) was 54.13% (Table 1). This indicated that the quality of transcriptome sequencing data met the standards for differentially expressed gene analysis.

Table 1.
Sequencing data quality statistics.
3.4.2. Analysis of Differentially Expressed Genes (DEGs)
Principal Component Analysis (PCA), a statistical method for analyzing multi-dimensional data in unsupervised pattern recognition, was employed to assess the overall differences in gene expression between main rice and ratoon rice. The PCA results for the two groups of samples are shown in Figure 4a,b, where PC1 and PC2 denote the first and second principal components, respectively. Each scatter point represents an individual sample, revealing substantial differences in gene expression between the main rice and ratoon rice of both 19X and NJXM varieties. To screen for genes with significantly differential expression, thresholds of fold change (FC) ≥ 2.0 or ≤0.5 and false discovery rate (FDR) < 0.05 were applied [25] (FDR refers to the false discovery rate after multiple hypothesis testing correction for p-values.).
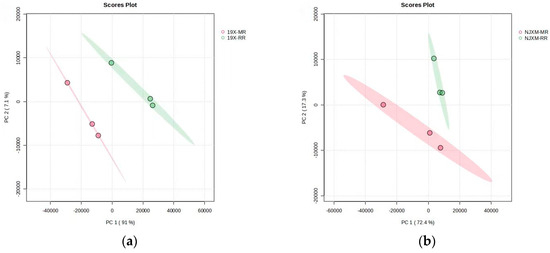
Figure 4.
Comparison of differential gene expression between main rice and ratoon rice. (a) PCA plot of 19X main rice and ratoon rice; (b) PCA plot of NJXM main rice and ratoon rice. PC1 and PC2 represent the first and second principal components, respectively. Each scatter point stands for an individual sample. The plots revealed substantial differences in gene expression between the main rice and ratoon rice of both 19X (left plot) and NJXM (right plot) varieties, as samples of different rice types (distinguished by colors) showed clear separation along the PC1–PC2 axes.
The results of the screening are presented in Figure 5a,b, where each point represents a gene. In the comparison of the main and ratoon rice of 19X, a total of 703 genes showed significantly upregulated expression levels, while 427 genes showed significantly downregulated expression. For NJXM, the comparison between main and ratoon rice identified 660 significantly upregulated expression genes and 705 significantly downregulated expression genes. Subsequently, we compared the DEGs from 19X and NJXM to find genes consistently differentially expressed in both varieties. The results are presented in Figure 6a,b. Similarly, 127 common upregulated DEGs and 92 common downregulated DEGs were identified, which showed a total of 219 common DEGs for 19X-RR vs. 19X-MR and NJXM-RR vs. NJXM-MR.
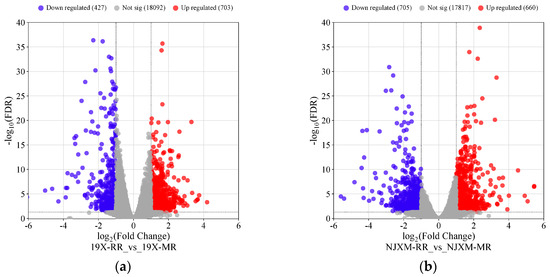
Figure 5.
Comparison of differential gene expression between main rice and ratoon rice (a) Volcano plot of 19X main rice and ratoon rice (b) Volcano plot of NJXM main rice and ratoon rice. Red points denote genes with significantly upregulated expression levels, blue points indicate genes with significantly downregulated expression levels, and gray points represent genes with non-significant expression.
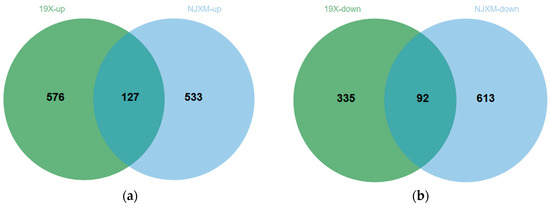
Figure 6.
Intersection of differentially expressed genes (DEGs) in ratoon rice between varieties 19X and NJXM. (a) Upregulated DEGs. The Venn diagram shows 576 upregulated gene expressions for 19X, 533 for NJXM, and 127 shared by both varieties. (b) Downregulated DEGs. The Venn diagram shows 335 downregulated gene expressions for 19X, 613 for NJXM, and 92 shared by both varieties.
3.4.3. Effects of Related Genes on Germination of Main Rice and Ratoon Rice
As indicated in Results 3.1, both the germination potential and germination rate of ratoon rice were significantly improved compared with those of main rice. As presented in Section 3.4.2, comparative transcriptome analysis identified 219 DEGs in both 19X and NJXM. Through systematic integration of the NCBI database, Rice Annotation Project Database (RAP-DB), National Rice Data Center, and the literature, four seed germination-associated genes were screened out. All these four genes demonstrated upregulated expression.
As shown in RAP-DB, the gene qLTG3-1 (Os03g0103300) was related to low-temperature germinability and pre-harvest sprouting resistance. It functions as a major regulator of germination potential in rice, participating in seed germination processes [26]. The expression of gene qLTG3-1 exhibited significant upregulation in both cultivars, with 2.24-fold and 2.04-fold higher expression in 19X-RR vs. 19X-MR and NJXM-RR vs. NJXM-MR, respectively. As shown in RAP-DB, the expression of gene OsLOX2 (Os03g0738600) was also critically involved in seed germination and longevity. Huang et al. [27] employed overexpression and knockout approaches to elucidate the role of the rice OsLOX2 gene in seed germination. Their results demonstrated that enhanced OsLOX2 expression in transgenic lines led to significantly higher germination rates. Conversely, reduced OsLOX2 expression resulted in slower germination. As shown in Table 2, the expression of gene OsLOX2 showed significant upregulation in both 19X-RR vs. 19X-MR and NJXM-RR vs. NJXM-MR, with fold change values of 5.92 and 9.26, respectively. Therefore, upregulation of this gene expression would accelerate germination and enhance the germination rate of ratoon rice seeds.
Genes OsSAMDC2 (OS02G0611200) and OsSAMDC4 (OS09G0424300) were also implicated in seed germination. As indicated in RAP-DB, the genes OsSAMDC2 (Os02g0611200) and OsSAMDC4 (Os09g0424300) both encoded S-adenosylmethionine decarboxylase; additionally, OsSAMDC2 was involved in polyamine biosynthesis and played a role in plant resistance to abiotic stresses. Chen [28] et al. generated transgenic rice plants with OsSAMDC2 and OsSAMDC4 knockdown via an antisense RNA interference (RNAi) strategy. Their study revealed that the transgenic lines exhibited reduced plant height, significantly decreased seed germination rates and final germination percentages, diminished pollen viability, and reduced tolerance to abiotic stresses. As shown in Table 2, the expression of genes OsSAMDC2 and OsSAMDC4 both exhibited significant upregulation in both cultivars, with a 2.68-fold and 3.87-fold higher expression in 19X-RR vs. 19X-MR, and 2.57-fold and 3.33-fold higher expression in NJXM-RR vs. NJXM-MR, respectively. Therefore, upregulation of OsSAMDC2 and OsSAMDC4 in ratoon rice may enhance the germination rate of its seeds.
3.4.4. Effects of Related Genes on Chalkiness of Main Rice and Ratoon Rice
As indicated in Results 3.2, compared with the main rice varieties, the chalky grain rate and the chalkiness degree of both 19X-RR and NJXM-RR were significantly reduced. Section 3.4.2 reported 219 DEGs of 19X and NJXM cultivars. An integrated screening framework incorporating the NCBI database, Rice Annotation Project Database (RAP-DB), National Rice Data Center, and the literature identified six seed chalkiness-associated genes, with five genes exhibiting upregulated expression and one showing downregulated expression. Chalkiness degree was a complex trait controlled by multiple genes and was highly susceptible to environmental influences [29].
Research has shown that high temperature/heat stress was a critical factor inducing chalkiness in rice grains by affecting the starch content and composition, changing the fine structure of amylopectin, and increasing the ratio of long chains to short chains of amylopectin [30]. As revealed by RAP-DB, the gene GAD3 (Os03g0236200) encoded glutamate decarboxylase. As reported in the relevant literature, its expression was regulated by the transcription factor OsMYB55, closely related to rice thermotolerance [31]. As shown in Table 2, the expression of gene GAD3 showed significant upregulation in both 19X-RR vs. 19X-MR and NJXM-RR vs. NJXM-MR, with fold change values of 2.21 and 2.38, respectively. Therefore, the upregulation expression of this gene in ratoon rice may enhance its tolerance to high-temperature stress, thereby reducing chalkiness.
Moreover, reactive oxygen species ROS produced under heat stress were involved in α-amylase induction in maturating rice grains through the regulating of the levels of gibberellin (GA) and abscisic acids (ABA), ultimately contributing to grain chalkiness [32]. ROS can also be produced under other stresses, such as wounding, pathogen attack, and oxidative stress [33]. In RAP-DB, gene Prx86 (Os06g0547400) was described as similar to Peroxidase P7, while gene POX8.1 (Os07g0677100) was described as a peroxidase, and gene Perox4 (Os07g0677200) was described as a peroxidase involved in the negative regulation of rice resistance to blast disease. These genes play a role in responding to environmental stresses by participating in the process of hydrogen peroxide (H2O2) scavenging [34]. As shown in Table 2, the expressions of genes Prx86, POX8.1, and Perox4 all exhibited significant upregulation in both cultivars, with 2.06-fold, 5.93-fold, and 9.88-fold higher expression in 19X-RR vs. 19X-MR, and 2.55-fold, 5.69-fold, and 3.59-fold higher expression in NJXM-RR vs. NJXM-MR, respectively. The upregulation expression of these genes in ratoon rice suggested that ratooning may enhance its tolerance to oxidative stress.
In RAP-DB, the gene OsEBP89 (Os03g0182800) was described as an APETALA2/ethylene responsive factor belonging to the ERF transcription factor family, which was associated with tolerance to drought and submergence stress. Studies have demonstrated that OsEBP89 (Os03g0182800) knockout lines exhibit enhanced reactive oxygen species (ROS) scavenging capacity [35]. As shown in Table 2, the expression of gene OsEBP89 showed significant downregulation in both 19X-RR vs. 19X-MR and NJXM-RR vs. NJXM-MR, with fold change values of 0.24 and 0.27, respectively. Consequently, the downregulation of OsEBP89 in ratoon rice may reduce its chalkiness by improving tolerance to oxidative stress. The activities of SOD and CAT were significantly higher for ratoon rice (showed in Section 3.3). This indicates that the increased antioxidant enzyme activity may mitigate oxidative damage to the endosperm structure, thereby reducing chalkiness.
Furthermore, it was reported that phytohormones, particularly ABA and auxin, were involved in the regulation of rice chalkiness formation through the interaction of multiple transcription factors and their downstream regulatory factors [36]. As indicated in RAP-DB, OsNCED3 (Os03g0645900), encoded 9-cis-epoxycarotenoid dioxygenase serves as a key enzyme in ABA biosynthesis. Overexpression of OsNCED3 enhanced ABA content, reduced gibberellin (GA) levels, elevated the ABA/GA ratio, and improved tolerance to water stress [37], whereas knockout of OsNCED3 led to the opposite outcomes. As shown in Table 2, the expression of gene OsNCED3 exhibited significant upregulation in both cultivars, with 3.91-fold and 3.96-fold higher expression in 19X-RR vs. 19X-MR and NJXM-RR vs. NJXM-MR, respectively. In ratoon rice, upregulation expression of the OsNCED3 gene can also lead to chalkiness reduction.

Table 2.
Summary of gene expression p-values and fold change (FC) values.
Table 2.
Summary of gene expression p-values and fold change (FC) values.
| Indicator | Gene Name | 1 p-Value (19X) | 1 p-Value (NJXM) | 2 Fold Change (19X) | 2 Fold Change (NJXM) |
|---|---|---|---|---|---|
| Germination | qLTG3-1 (Os03g0103300) | 0.000000527 | 0.000000841 | 2.24 | 2.04 |
| OsLOX2 (Os03g0738600) | 0.000000000 | 0.000000000 | 5.92 | 9.26 | |
| OsSAMDC2 (OS02g0611200) | 0.000000000 | 0.000010445 | 2.68 | 2.57 | |
| OsSAMDC4 (OS09g0424300) | 0.000000000 | 0.000035246 | 3.87 | 3.33 | |
| Chalkiness | GAD3 (Os03g0236200) | 0.000000003 | 0.000000175 | 2.21 | 2.38 |
| prx86 (Os06g0547400) | 0.000000061 | 0.000000000 | 2.06 | 2.55 | |
| POX8.1 (OS07G0677100) | 0.000002788 | 0.000000667 | 5.93 | 5.69 | |
| Perox4 (OS07G0677200) | 0.000000000 | 0.000000046 | 9.88 | 3.59 | |
| OsEBP89 (Os03g0182800) | 0.000000000 | 0.000000001 | 0.24 | 0.27 | |
| OsNCED3 (Os03g0645900) | 0.000000000 | 0.000000001 | 3.91 | 3.96 |
1 p-value: Statistical significance of differential expression (adjusted for multiple testing). 2 Fold change (FC): Magnitude and direction of change (FC > 1 = upregulated; FC < 1 = downregulated).
4. Conclusions
The results indicated that the ratoon rice of cultivars 19X and NJXM exhibited significant improvement in physiological activity and appearance quality over their main rice varieties. The ratoon rice displayed enhanced germination ability, reduced chalkiness, and increased antioxidant enzyme activity. Transcriptome analysis identified key gene candidates and global expression trends correlating with quality improvements in ratoon rice. Enhanced germination was associated with the upregulated expression of genes qLTG3-1, OsLOX2, OsSAMDC2, and OsSAMDC4. The reduction in chalkiness involved the coordinated effects of the upregulated expression of genes GAD3, prx86, POX8.1, Perox4, and OsNCED3, coupled with the downregulated expression of OsEBP89. The findings of this study established a theoretical foundation for the improvement of ratoon rice quality and the advancement of high-efficiency cultivation practices.
Author Contributions
Conceptualization, F.X., and C.Q.; methodology, F.X., X.F., and C.Q.; software, F.X.; formal analysis, F.X., L.N., Z.Q., and X.F.; writing—original draft preparation, F.X., and C.Q.; writing—review and editing, F.X., and C.Q.; supervision, C.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 32372402).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang, W.; Luo, X.; Zhan, Z.; Shu, Z.; Wang, P.; Ding, W.; Zeng, X.; Shi, Y.-C. Comparison of the Structural and Functional Properties of Starches in Rice from Main and Ratoon Crops. J. Cereal Sci. 2021, 99, 103233. [Google Scholar] [CrossRef]
- Lin, F.; Rensing, C.; Pang, Z.; Zou, J.; Lin, S.; Letuma, P.; Zhang, Z.; Lin, W. Metabolomic Analysis Reveals Differential Metabolites and Pathways Involved in Grain Chalkiness Improvement under Rice Ratooning. Field Crops Res. 2022, 283, 108521. [Google Scholar] [CrossRef]
- Huang, X.; Yang, J.; Zhou, W.; Zhang, G.; Liao, B.; Wahab, A.; Yi, Z.; Tu, N. Comparison of the Source–Sink Characteristics between Main Season and Ratooning in Rice (Oryza sativa L.). Agronomy 2023, 13, 1731. [Google Scholar] [CrossRef]
- Zhang, W.; Zhan, Z.; Wang, H.; Shu, Z.; Wang, P.; Zeng, X. Structural, Pasting and Sensory Properties of Rice from Main and Ratoon Crops. Int. J. Food Prop. 2021, 24, 965–975. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Su, Y.; Shen, H. Rice Responses to Abiotic Stress: Key Proteins and Molecular Mechanisms. Int. J. Mol. Sci. 2025, 26, 896. [Google Scholar] [CrossRef]
- Yang, W.; Mo, X.; Zhang, Y.; Liu, Z.; Tang, Q.; Xu, J.; Pan, S.; Wang, Y.; Chen, G.; Hu, Y. Appropriate Stubble Height Can Effectively Improve the Rice Quality of Ratoon Rice. Foods 2024, 13, 1392. [Google Scholar] [CrossRef]
- Yue, W.; Li, Y. Mapping the Ratoon Rice Suitability Region in China Using Random Forest and Recursive Feature Elimination Modeling. Field Crops Res. 2023, 301, 109016. [Google Scholar] [CrossRef]
- Kuang, N.; Zheng, H.; Tang, Q.; Chen, Y.; Wang, X.; Luo, Y. Amylose Content, Morphology, Crystal Structure, and Thermal Properties of Starch Grains in Main and Ratoon Rice Crops. Phyton 2021, 90, 1119–1230. [Google Scholar] [CrossRef]
- Ye, M.; Song, Y.; Baerson, S.R.; Long, J.; Wang, J.; Pan, Z.; Lin, W.; Zeng, R. Ratoon Rice Generated from Primed Parent Plants Exhibit Enhanced Herbivore Resistance. Plant Cell Environ. 2017, 40, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Yang, F.; Li, Q.; Zeng, Y.; Li, B.; Zhong, X.; Lu, H.; Wang, L.; Chen, H.; Chen, Y.; et al. Differences in Starch Structural and Physicochemical Properties and Texture Characteristics of Cooked Rice between the Main Crop and Ratoon Rice. Food Hydrocoll. 2021, 116, 106643. [Google Scholar] [CrossRef]
- Xie, W.; Ata-Ul-Karim, S.T.; Shiotsu, F.; Kato, Y. Crop Productivity in a Rice–Ratoon-Rice System vs. a Single-Crop System in Central Japan. Field Crops Res. 2023, 303, 109140. [Google Scholar] [CrossRef]
- Alizadeh, M.R.; Habibi, F. A Comparative Study on the Quality of the Main and Ratoon Rice Crops. J. Food Qual. 2016, 39, 669–674. [Google Scholar] [CrossRef]
- Liang, R.; Su, Y.; Qin, X.; Gao, Z.; Fu, Z.; Qiu, H.; Lin, X.; Zhu, J. Comparative Transcriptomic Analysis of Two Cucumis melo var. saccharinus Germplasms Differing in Fruit Physical and Chemical Characteristics. BMC Plant Biol. 2022, 22, 193. [Google Scholar] [CrossRef]
- Lin, F.; Huang, J.; Lin, S.; Letuma, P.; Xie, D.; Rensing, C.; Lin, W. Physiological and transcriptomic analysis reveal the regulatory mechanism underlying grain quality improvement induced by rice ratooning. J. Sci. Food Agric. 2023, 103, 3569–3578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Xiao, W.; Chen, D.; Hu, J.; Gao, N.; Huang, M.; Ye, X. Comparative Transcriptomic Analysis Reveals the Important Process in Two Rice Cultivars with Differences in Cadmium Accumulation. Ecotoxicol. Environ. Saf. 2023, 252, 114629. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Lin, S.; Zhang, Z.; Lin, W.; Rensing, C.; Xie, D. GF14f Gene Is Negatively Associated with Yield and Grain Chalkiness under Rice Ratooning. Front. Plant Sci. 2023, 14, 1112146. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Bai, D.; Wang, X.; Dou, G.; Lv, J.; Bao, Y.; Wang, N.; Yu, L.; Zhou, Y.; Zhang, J.; et al. Identification of Candidate Genes for Hypoxia Tolerance in Rice by Genome-Wide Association Analysis and Transcriptome Sequencing. Rice 2025, 18, 10. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Zhu, L. Amino Acid Transporter as a Potential Carrier Protein for the Root-to-Shoot Translocation of Polybrominated Diphenyl Ethers in Rice. Environ. Sci. Technol. 2023, 57, 9722–9731. [Google Scholar] [CrossRef]
- Yao, Q.; Zheng, X.; Zhou, G.; Zhang, J. SGR-YOLO: A Method for Detecting Seed Germination Rate in Wild Rice. Front. Plant Sci. 2024, 14, 1305081. [Google Scholar] [CrossRef]
- Qian, J.; Mo, X.; Wang, Y.; Li, Q. Seed Priming with 2,4-Epibrassionolide Enhances Seed Germination and Heat Tolerance in Rice by Regulating the Antioxidant System and Plant Hormone Signaling Pathways. Antioxidants 2025, 14, 242. [Google Scholar] [CrossRef]
- Huang, J.; Pan, Y.; Chen, H.; Zhang, Z.; Fang, C.; Shao, C.; Amjad, H.; Lin, W.; Lin, W. Physiochemical mechanisms involved in the improvement of grain-filling, rice quality mediated by related enzyme activities in the ratoon cultivation system. Field Crops Res. 2020, 258, 107962. [Google Scholar] [CrossRef]
- Thongnok, S.; Siripornadulsil, W.; Siripornadulsil, S. Responses to Arsenic Stress of Rice Varieties Coinoculated with the Heavy Metal-Resistant and Rice Growth-Promoting Bacteria Pseudomonas Stutzeri and Cupriavidus Taiwanensis. Plant Physiol. Biochem. 2022, 191, 42–54. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, R.; Yan, X.; Fan, K. Superoxide Dismutase Nanozymes: An Emerging Star for Anti-Oxidation. J. Mater. Chem. B 2021, 9, 6939–6957. [Google Scholar] [CrossRef] [PubMed]
- Chioti, V.; Zervoudakis, G. Is Root Catalase a Bifunctional Catalase-Peroxidase? Antioxidants 2017, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-X.; Zhu, F.-Y.; Gao, B.; Ma, K.-L.; Zhang, Y.; Fernie, A.R.; Luo, J.; Yi, S.-Y.; Liu, Y.-X.; Xu, Z.-H.; et al. Full-Length Transcript-Based Proteogenomics of Rice Improves its Genome and Proteome Annotation. Plant Physiol. 2020, 183, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular Identification of a Major Quantitative Trait Locus, qLTG3–1, Controlling Low-Temperature Germinability in Rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef]
- Huang, J.; Cai, M.; Long, Q.; Liu, L.; Lin, Q.; Jiang, L.; Chen, S.; Wan, J. OsLOX2, a Rice Type I Lipoxygenase, Confers Opposite Effects on Seed Germination and Longevity. Transgenic Res. 2014, 23, 643–655. [Google Scholar] [CrossRef]
- Chen, M.; Chen, J.; Fang, J.; Guo, Z.; Lu, S. Down-Regulation of S-Adenosylmethionine Decarboxylase Genes Results in Reduced Plant Length, Pollen Viability, and Abiotic Stress Tolerance. Plant Cell Tissue Organ Cult. 2014, 116, 311–322. [Google Scholar] [CrossRef]
- Qin, Y.; Kim, S.-M.; Sohn, J.-K. Genetic Analysis and QTL Mapping for Grain Chalkiness Characteristics of Brown Rice (Oryza sativa L.). Genes Genom. 2009, 31, 155–164. [Google Scholar] [CrossRef]
- Du, Y.; Long, C.; Deng, X.; Zhang, Z.; Liu, J.; Xu, Y.; Liu, D.; Zeng, Y. Physiological Basis of High Nighttime Temperature-Induced Chalkiness Formation during Early Grain-Filling Stage in Rice (Oryza sativa L.). Agronomy 2023, 13, 1475. [Google Scholar] [CrossRef]
- El-kereamy, A.; Bi, Y.-M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The Rice R2R3-MYB Transcription Factor OsMYB55 Is Involved in the Tolerance to High Temperature and Modulates Amino Acid Metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [PubMed]
- Suriyasak, C.; Harano, K.; Tanamachi, K.; Matsuo, K.; Tamada, A.; Iwaya-Inoue, M.; Ishibashi, Y. Reactive Oxygen Species Induced by Heat Stress during Grain Filling of Rice (Oryza sativa L.) Are Involved in Occurrence of Grain Chalkiness. J. Plant Physiol. 2017, 216, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D.; et al. ROS Generated from Biotic Stress: Effects on Plants and Alleviation by Endophytic Microbes. Front. Plant Sci. 2022, 13, 1042936. [Google Scholar] [CrossRef]
- The UniProt, C.o.n.s.o.r.t.i.u.m.; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Chen, S.; Ma, X.; Wei, H.; Chen, C.; Gao, N.; Zou, Y.; Kong, D.; Li, T.; et al. An APETALA2/Ethylene Responsive Factor, OsEBP89 Knockout Enhances Adaptation to Direct-Seeding on Wet Land and Tolerance to Drought Stress in Rice. Mol. Genet. Genom. 2020, 295, 941–956. [Google Scholar] [CrossRef]
- Xie, Q.; Xu, J.; Huang, K.; Su, Y.; Tong, J.; Huang, Z.; Huang, C.; Wei, M.; Lin, W.; Xiao, L. Dynamic Formation and Transcriptional Regulation Mediated by Phytohormones during Chalkiness Formation in Rice. BMC Plant Biol. 2021, 21, 308. [Google Scholar] [CrossRef]
- Chen, Y.; Xiang, Z.; Liu, M.; Wang, S.; Zhang, L.; Cai, D.; Huang, Y.; Mao, D.; Fu, J.; Chen, L. ABA Biosynthesis Gene OsNCED3 Contributes to Preharvest Sprouting Resistance and Grain Development in Rice. Plant Cell Environ. 2023, 46, 1384–1401. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).