A Novel Lytic Salmonella Phage Harboring an Unprecedented Tail-Protein Domain Combination Capable of Lysing Cross-Host-Transmitted Salmonella Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Bacterial Strains
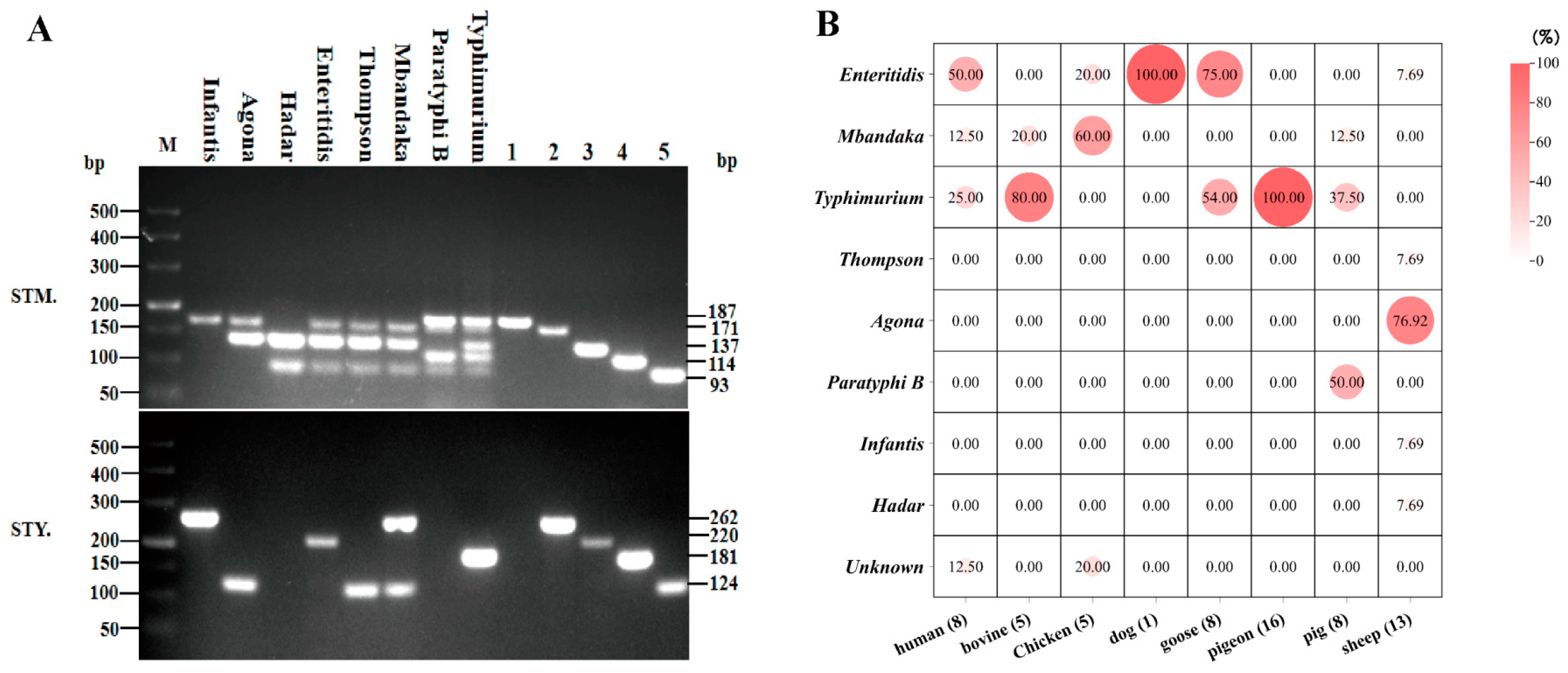
2.2. Identification of Salmonella Serotypes by Multiplex PCR
2.3. Phage Isolation and Purification
2.4. Transmission Electron Microscopy (TEM)
2.5. Host Rang of Phage
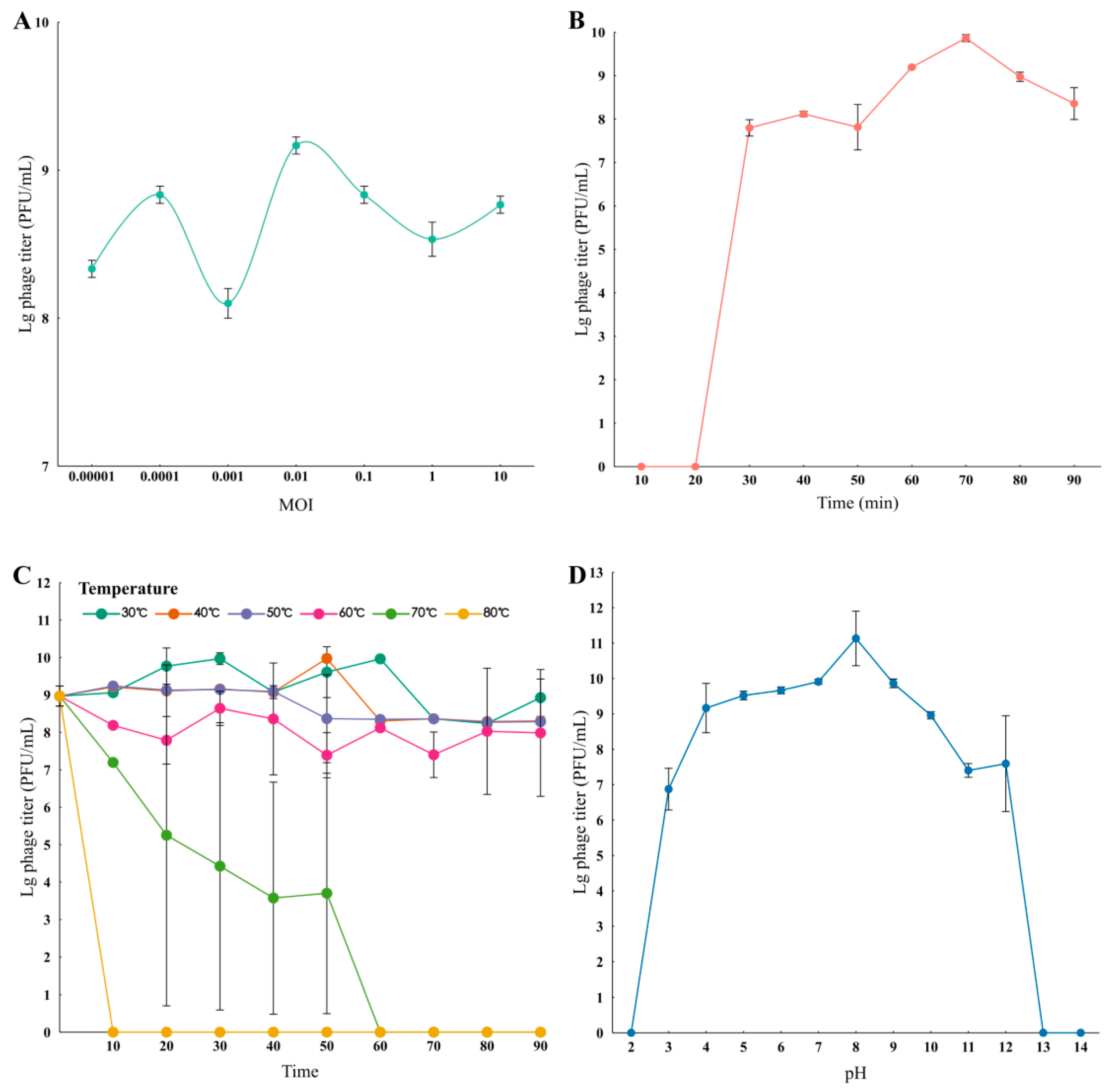
2.6. Growth Characteristics: The Optimal MOI and One-Step Experiments
2.7. Sensitivity of Temperature and pH
2.8. DNA Extraction and Complete Genome Sequencing
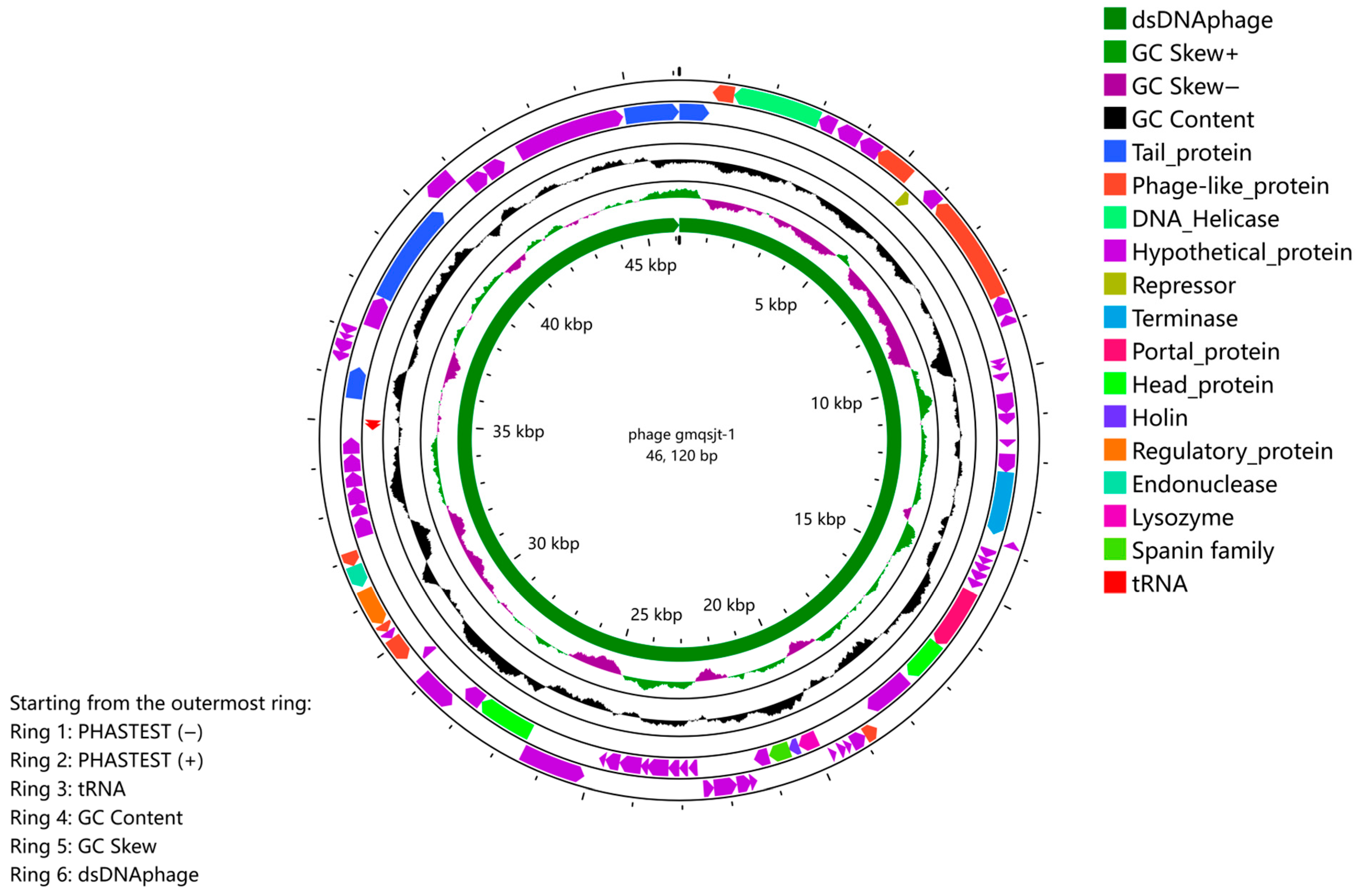
2.9. Annotation of the Genome
2.10. Phylogenetic and Taxonomic Analysis
2.11. Prediction of Tail Spike Protein Conserved Domain
2.12. Pulsed Field Gel Electrophoresis of Salmonella
3. Result
3.1. Serotype Identification
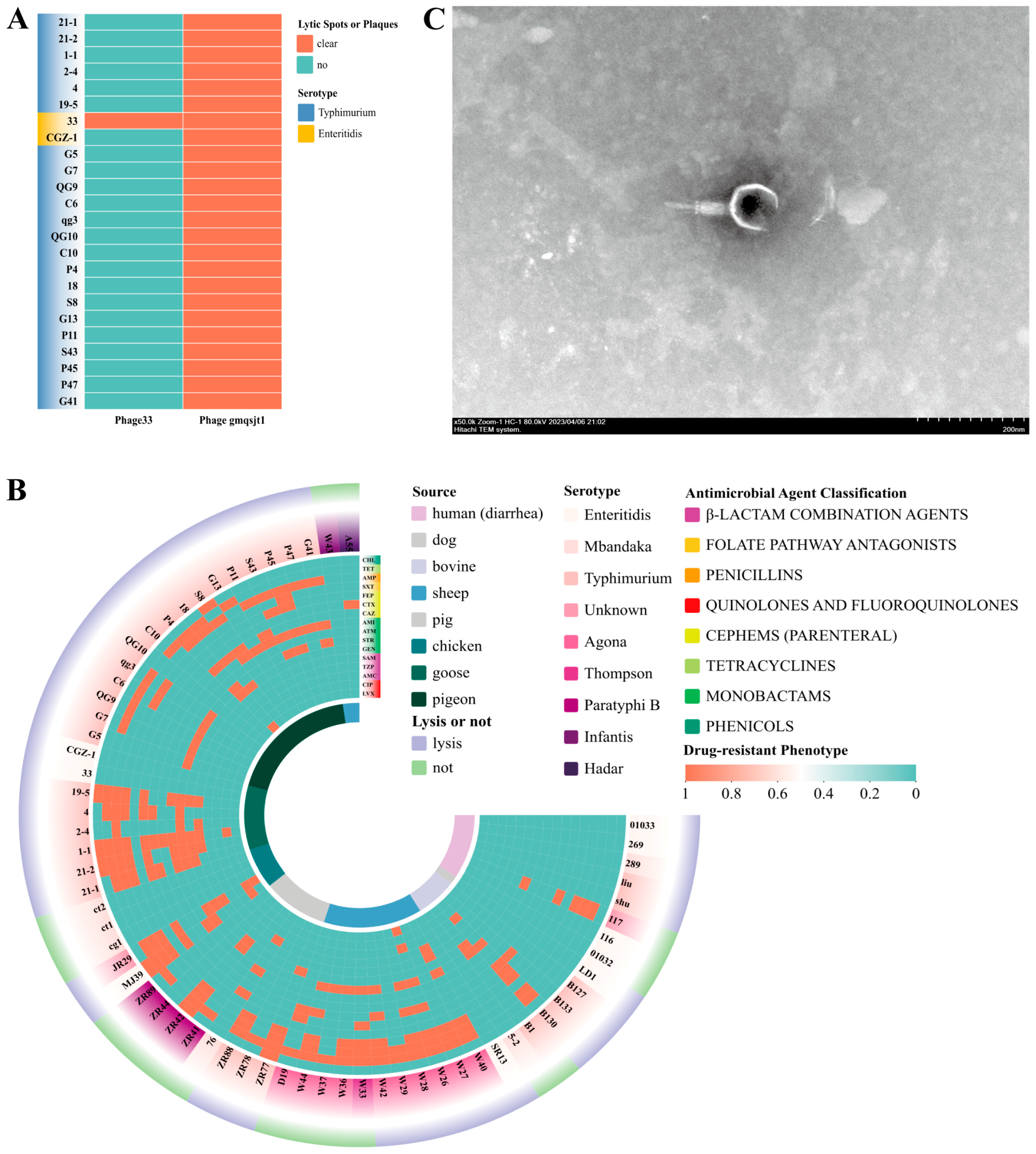
3.2. Isolation and Identification of Phages
3.3. Phages Host Range
3.4. Growth Characteristics of Phage
3.5. Genome Analysis
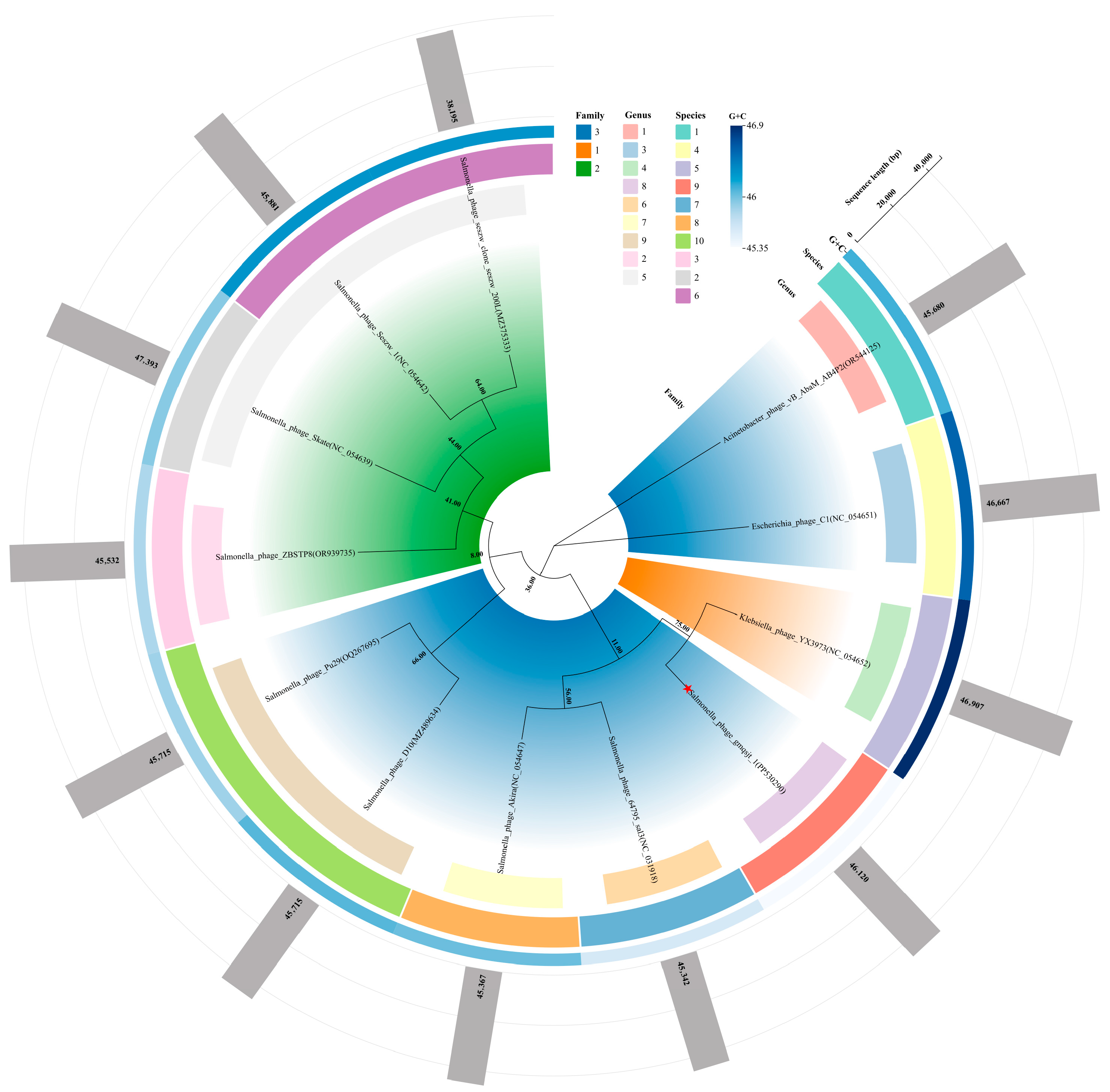
3.6. Phylogeny and Classification
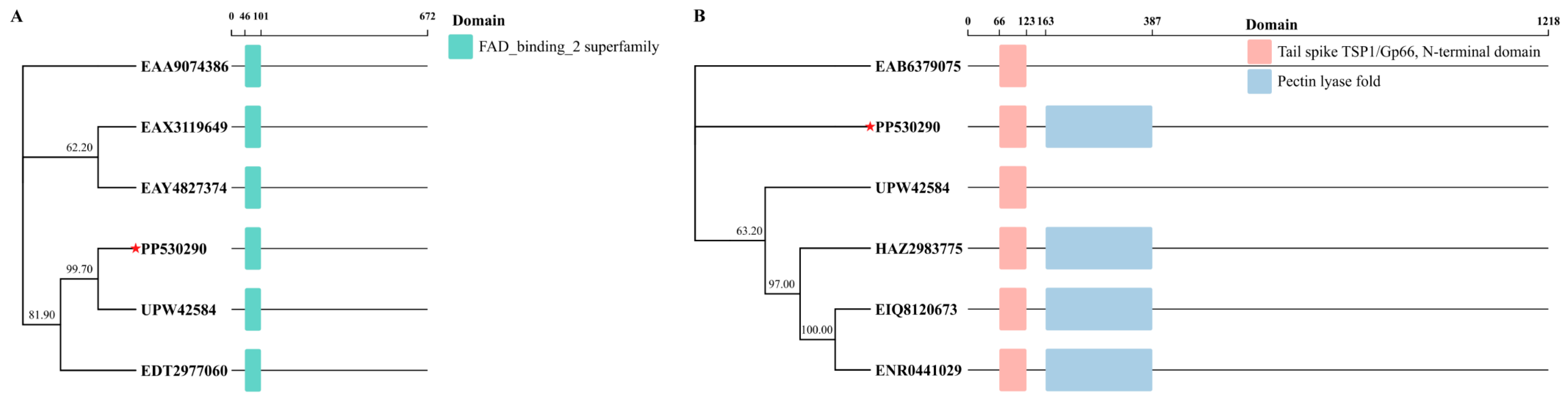
3.7. Tail Spike Protein Domain
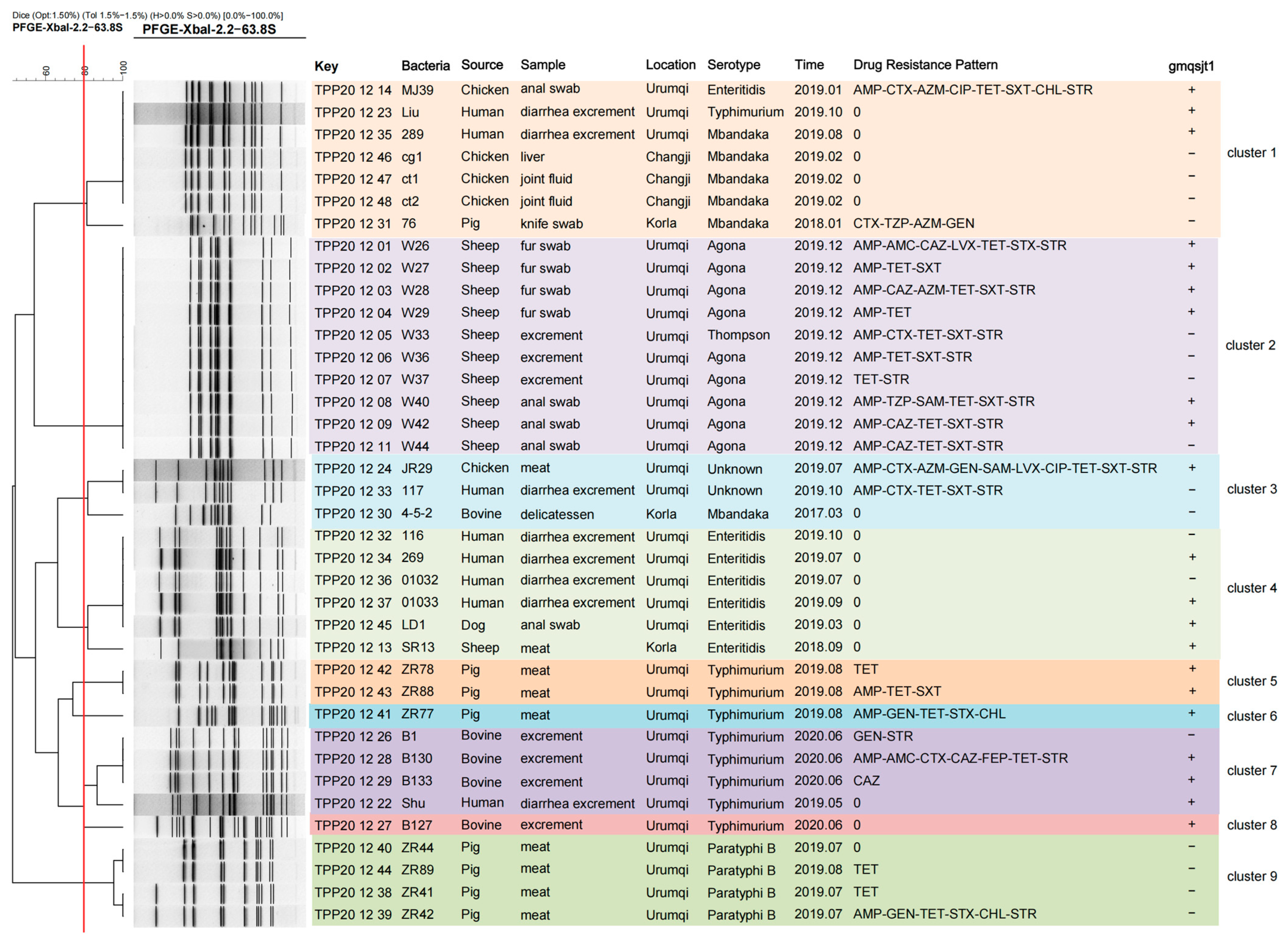
3.8. PFGE Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, J.; Wu, H.; Wu, S.; Wang, S.; Fan, H.; Ruan, H.; Qiao, J.; Caiyin, Q.; Wen, M. Salmonella: Infection Mechanism and Control Strategies. Microbiol. Res. 2025, 292, 128013. [Google Scholar] [CrossRef]
- Wang, J.; Sheng, H.; Xu, W.; Huang, J.; Meng, L.; Cao, C.; Zeng, J.; Meng, J.; Yang, B. Diversity of Serotype, Genotype, and Antibiotic Susceptibility of Salmonella Prevalent in Pickled Ready-to-Eat Meat. Front. Microbiol. 2019, 10, 2577. [Google Scholar] [CrossRef]
- Seelman Federman, S.; Jenkins, E.; Wilson, C.; DeLaGarza, A.; Schwensohn, C.; Schneider, B.; Nsubuga, J.; Literman, R.; Wellman, A.; Whitney, B.M.; et al. An Investigation of an Outbreak of Salmonella Typhimurium Infections Linked to Cantaloupe—United States, 2022. Food Control 2024, 166, 110733. [Google Scholar] [CrossRef]
- Soltan Dallal, M.M.; Nasser, A.; Karimaei, S. Characterization of Virulence Genotypes, Antimicrobial Resistance Patterns, and Biofilm Synthesis in Salmonella spp Isolated from Foodborne Outbreaks. Can. J. Infect. Dis. Med. Microbiol. 2024, 2024, 4805228. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Zhang, R.; Chen, L.; Zhang, H.; Qi, X.; Chen, J. Epidemiology of Foodborne Diseases Caused by Salmonella in Zhejiang Province, China, between 2010 and 2021. Front. Public Health 2023, 11, 1127925. [Google Scholar] [CrossRef] [PubMed]
- Konyali, D.; Guzel, M.; Soyer, Y. Genomic Characterization of Salmonella Enterica Resistant to Cephalosporin, Quinolones, and Macrolides. Curr. Microbiol. 2023, 80, 344. [Google Scholar] [CrossRef] [PubMed]
- Arsène, M.M.J.; Davares, A.K.L.; Viktorovna, P.I.; Andreevna, S.L.; Sarra, S.; Khelifi, I.; Sergueïevna, D.M. The Public Health Issue of Antibiotic Residues in Food and Feed: Causes, Consequences, and Potential Solutions. Vet. World 2022, 15, 662–671. [Google Scholar] [CrossRef]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and Bacterial Resistance—A Short Story of an Endless Arms Race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef]
- Vaughn, V.M.; Hersh, A.L.; Spivak, E.S. Antibiotic Overuse and Stewardship at Hospital Discharge: The Reducing Overuse of Antibiotics at Discharge Home Framework. Clin. Infect. Dis. 2022, 74, 1696–1702. [Google Scholar] [CrossRef]
- Perez-Bou, L.; Gonzalez-Martinez, A.; Gonzalez-Lopez, J.; Correa-Galeote, D. Promising Bioprocesses for the Efficient Removal of Antibiotics and Antibiotic-Resistance Genes from Urban and Hospital Wastewaters: Potentialities of Aerobic Granular Systems. Environ. Pollut. 2024, 342, 123115. [Google Scholar] [CrossRef]
- Yang, W.; Li, J.; Yao, Z.; Li, M. A Review on the Alternatives to Antibiotics and the Treatment of Antibiotic Pollution: Current Development and Future Prospects. Sci. Total Environ. 2024, 926, 171757. [Google Scholar] [CrossRef]
- Ajose, D.J.; Adekanmbi, A.O.; Kamaruzzaman, N.F.; Ateba, C.N.; Saeed, S.I. Combating Antibiotic Resistance in a One Health Context: A Plethora of Frontiers. One Health Outlook 2024, 6, 19. [Google Scholar] [CrossRef]
- Mehraj, I.; Hamid, A.; Gani, U.; Iralu, N.; Manzoor, T.; Saleem Bhat, S. Combating Antimicrobial Resistance by Employing Antimicrobial Peptides: Immunomodulators and Therapeutic Agents against Infectious Diseases. ACS Appl. Bio Mater. 2024, 7, 2023–2035. [Google Scholar] [CrossRef]
- Pacyga, K.; Pacyga, P.; Topola, E.; Viscardi, S.; Duda-Madej, A. Bioactive Compounds from Plant Origin as Natural Antimicrobial Agents for the Treatment of Wound Infections. Int. J. Mol. Sci. 2024, 25, 2100. [Google Scholar] [CrossRef]
- Yang, S.; Sadekuzzaman, M.; Ha, S.-D. Reduction of Listeria Monocytogenes on Chicken Breasts by Combined Treatment with UV-C Light and Bacteriophage ListShield. LWT 2017, 86, 193–200. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Zhan, W.; Li, Z.; Zou, L.; Zhao, Q. Challenges for the Application of Bacteriophages as Effective Antibacterial Agents in the Food Industry. J. Sci. Food Agric. 2022, 102, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Gildea, L.; Ayariga, J.A.; Robertson, B.K. Bacteriophages as Biocontrol Agents in Livestock Food Production. Microorganisms 2022, 10, 2126. [Google Scholar] [CrossRef] [PubMed]
- Hernández Villamizar, S.; Bonilla, J.A.; Jaramillo, Á.H.; Piñeros, R.; Ripoll, A.; Fonseca, L.; Riveros, K.; Vives, M.J.; Barato, P.; Clavijo, V. SalmoFree® Phage Additive Proves Its Safety for Laying Hens. PHAGE 2024, 5, 143–152. [Google Scholar] [CrossRef]
- Pino, M.; Mujica, K.; Mora-Uribe, P.; Garcias-Papayani, H.; Paillavil, B.; Avendaño, C.; Flores-Crisosto, D.; Norambuena, R.; Rojas-Martínez, V.; Aguilera, M.; et al. Research Note: Reduction of Salmonella Load in Brazilian Commercial Chicken Farms Using INSPEKTOR®: A Bacteriophage-Based Product. Poult. Sci. 2025, 104, 104544. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Villa, R.E.; Azimonti, G.; Bonos, E.; Christensen, H.; Durjava, M.; Dusemund, B.; Gehring, R.; Glandorf, B.; Kouba, M.; et al. Safety and Efficacy of a Feed Additive Consisting of the Bacteriophages PCM F/00069, PCM F/00070, PCM F/00071 and PCM F/00097 (Bafasal®) for All Poultry (Proteon Pharmaceuticals S.A.). EFSA J. 2024, 22, e9132. [Google Scholar] [CrossRef]
- Kim, S.; Frye, J.G.; Hu, J.; Fedorka-Cray, P.J.; Gautom, R.; Boyle, D.S. Multiplex PCR-Based Method for Identification of Common Clinical Serotypes of Salmonella Enterica subsp. Enterica. J. Clin. Microbiol. 2006, 44, 3608–3615. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, L.; Zhang, Y.; Tong, P.; Ma, W.; Wang, Y.; Liu, Y.; Su, Z. Isolation and Identification of Specific Enterococcus Faecalis Phage C-3 and G21-7 against Avian Pathogenic Escherichia Coli and Its Application to One-Day-Old Geese. Front. Microbiol. 2024, 15, 1385860. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; McEwen, G.K.; Margulies, E.H.; Birney, E. Pebble and Rock Band: Heuristic Resolution of Repeats and Scaffolding in the Velvet Short-Read de Novo Assembler. PLoS ONE 2009, 4, e8407. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding Pre-Assembled Contigs Using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward Almost Closed Genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Hunt, M.; Newbold, C.; Berriman, M.; Otto, T.D. A Comprehensive Evaluation of Assembly Scaffolding Tools. Genome Biol. 2014, 15, R42. [Google Scholar] [CrossRef]
- Wang, R.H.; Yang, S.; Liu, Z.; Zhang, Y.; Wang, X.; Xu, Z.; Wang, J.; Li, S.C. PhageScope: A Well-Annotated Bacteriophage Database with Automatic Analyses and Visualizations. Nucleic Acids Res. 2024, 52, D756–D761. [Google Scholar] [CrossRef]
- Wang, R.H.; Ng, Y.K.; Zhang, X.; Wang, J.; Li, S. Coding Genomes with Gapped Pattern Graph Convolutional Network. Bioinformatics 2023, 41, btaf336. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, Better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program: Table 1. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Göker, M.; García-Blázquez, G.; Voglmayr, H.; Tellería, M.T.; Martín, M.P. Molecular Taxonomy of Phytopathogenic Fungi: A Case Study in Peronospora. PLoS ONE 2009, 4, e6319. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete Genome Sequence of DSM 30083T, the Type Strain (U5/41T) of Escherichia Coli, and a Proposal for Delineating Subspecies in Microbial Taxonomy. Stand Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization by One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The Protein Sequence Classification Resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Su, Z.; Tong, P.; Zhang, L.; Zhang, M.; Wang, D.; Ma, K.; Zhang, Y.; Liu, Y.; Xia, L.; Xie, J. First Isolation and Molecular Characterization of blaCTX-M-121-Producing Escherichia Coli O157:H7 from Cattle in Xinjiang, China. Front. Vet. Sci. 2021, 8, 574801. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; Van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Chaudhari, R.; Singh, K.; Kodgire, P. Biochemical and Molecular Mechanisms of Antibiotic Resistance in Salmonella Spp. Res. Microbiol. 2023, 174, 103985. [Google Scholar] [CrossRef]
- Brashears, M.M.; Jimenez, R.L.; Portillo, R.M.; Bueno, R.; Montoya, B.D.; Echeverry, A.; Sanchez, M.X. Innovative Approaches to Controlling Salmonella in the Meat Industry. Meat Sci. 2025, 219, 109673. [Google Scholar] [CrossRef]
- Huang, S.; Rong, X.; Liu, M.; Liang, Z.; Geng, Y.; Wang, X.; Zhang, J.; Ji, C.; Zhao, L.; Ma, Q. Intestinal Mucosal Immunity-Mediated Modulation of the Gut Microbiome by Oral Delivery of Enterococcus Faecium Against Salmonella Enteritidis Pathogenesis in a Laying Hen Model. Front. Immunol. 2022, 13, 853954. [Google Scholar] [CrossRef]
- Nazir, J.; Manzoor, T.; Saleem, A.; Gani, U.; Bhat, S.S.; Khan, S.; Haq, Z.; Jha, P.; Ahmad, S.M. Combatting Salmonella: A Focus on Antimicrobial Resistance and the Need for Effective Vaccination. BMC Infect. Dis. 2025, 25, 84. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2023 Zoonoses Report. EFSA J. 2024, 22, e9106. [Google Scholar] [CrossRef]
- Marchello, C.S.; Birkhold, M.; Crump, J.A.; Martin, L.B.; Ansah, M.O.; Breghi, G.; Canals, R.; Fiorino, F.; Gordon, M.A.; Kim, J.-H.; et al. Complications and Mortality of Non-Typhoidal Salmonella Invasive Disease: A Global Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2022, 22, 692–705. [Google Scholar] [CrossRef]
- Plain Language Summary on The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, 211202. [CrossRef] [PubMed]
- Baskaran, V.; Karthik, L. Phages for Treatment of Salmonella Spp Infection. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2023; Volume 200, pp. 241–273. ISBN 978-0-323-95563-8. [Google Scholar]
- Abd-El Wahab, A.; Basiouni, S.; El-Seedi, H.R.; Ahmed, M.F.E.; Bielke, L.R.; Hargis, B.; Tellez-Isaias, G.; Eisenreich, W.; Lehnherr, H.; Kittler, S.; et al. An Overview of the Use of Bacteriophages in the Poultry Industry: Successes, Challenges, and Possibilities for Overcoming Breakdowns. Front. Microbiol. 2023, 14, 1136638. [Google Scholar] [CrossRef] [PubMed]
- Petrovic Fabijan, A.; Iredell, J.; Danis-Wlodarczyk, K.; Kebriaei, R.; Abedon, S.T. Translating Phage Therapy into the Clinic: Recent Accomplishments but Continuing Challenges. PLoS Biol. 2023, 21, e3002119. [Google Scholar] [CrossRef] [PubMed]
- Ranveer, S.A.; Dasriya, V.; Ahmad, M.F.; Dhillon, H.S.; Samtiya, M.; Shama, E.; Anand, T.; Dhewa, T.; Chaudhary, V.; Chaudhary, P.; et al. Positive and Negative Aspects of Bacteriophages and Their Immense Role in the Food Chain. npj Sci. Food 2024, 8, 1. [Google Scholar] [CrossRef]
- Yang, Y.; Dufault-Thompson, K.; Yan, W.; Cai, T.; Xie, L.; Jiang, X. Large-Scale Genomic Survey with Deep Learning-Based Method Reveals Strain-Level Phage Specificity Determinants. GigaScience 2024, 13, giae017. [Google Scholar] [CrossRef]
- Liu, S.; Lei, T.; Tan, Y.; Huang, X.; Zhao, W.; Zou, H.; Su, J.; Zeng, J.; Zeng, H. Discovery, Structural Characteristics and Evolutionary Analyses of Functional Domains in Acinetobacter Baumannii Phage Tail Fiber/Spike Proteins. BMC Microbiol. 2025, 25, 73. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Xia, Y.; Li, K.; Zhu, Q.; Ding, F.; Gu, H.; Zhang, Z.; Li, X.; Mi, X.; Chen, J.; et al. Dual Activation of Soybean Resistance against Phytophthora Sojae by Pectin Lyase and Degraded Pectin Oligosaccharides. Sci. China Life Sci. 2024, 67, 2746–2760. [Google Scholar] [CrossRef] [PubMed]
- Cahill, J.; Holt, A.; Theodore, M.; Moreland, R.; O’Leary, C.; Martin, C.; Bettridge, K.; Xiao, J.; Young, R. Spatial and Temporal Control of Lysis by the Lambda Holin. mBio 2024, 15, e01290-23. [Google Scholar] [CrossRef] [PubMed]
- Samir, S. Molecular Machinery of the Triad Holin, Endolysin, and Spanin: KeyPlayers Orchestrating Bacteriophage-Induced Cell Lysis and theirTherapeutic Applications. PPL 2024, 31, 85–96. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, X.; Zhou, Y.; Liu, M.; Li, P.; Gao, L.; Wang, H.; Ma, X.; Wang, L.; Huo, X.; et al. Isolation, Whole Genome Sequencing and Application of a Broad-Spectrum Salmonella Phage. Arch. Microbiol. 2024, 206, 335. [Google Scholar] [CrossRef]
- Cao, S.; Yang, W.; Zhu, X.; Liu, C.; Lu, J.; Si, Z.; Pei, L.; Zhang, L.; Hu, W.; Li, Y.; et al. Isolation and Identification of the Broad-Spectrum High-Efficiency Phage vB_SalP_LDW16 and Its Therapeutic Application in Chickens. BMC Vet. Res. 2022, 18, 386. [Google Scholar] [CrossRef]
- Shymialevich, D.; Błażejak, S.; Średnicka, P.; Cieślak, H.; Ostrowska, A.; Sokołowska, B.; Wójcicki, M. Biological Characterization and Genomic Analysis of Three Novel Serratia- and Enterobacter-Specific Virulent Phages. Int. J. Mol. Sci. 2024, 25, 5944. [Google Scholar] [CrossRef]
- Dendani Chadi, Z.; Arcangioli, M.-A. Pulsed-Field Gel Electrophoresis Analysis of Bovine Associated Staphylococcus Aureus: A Review. Pathogens 2023, 12, 966. [Google Scholar] [CrossRef]
- Napoleoni, M.; Villa, L.; Barco, L.; Busani, L.; Cibin, V.; Lucarelli, C.; Tiengo, A.; Dionisi, A.; Conti, F.; Da Silva Nunes, F.; et al. A Strong Evidence Outbreak of Salmonella Enteritidis in Central Italy Linked to the Consumption of Contaminated Raw Sheep Milk Cheese. Microorganisms 2021, 9, 2464. [Google Scholar] [CrossRef]
- Eun, J.S.; Han, J.; Lim, J.-H.; Shin, E.; Kim, J.; Ko, D.-J.; Yoo, J.; Kim, S.; Kim, J.S.; Park, J.S.; et al. Salmonellosis Outbreaks Linked to Eggs at 2 Gimbap Restaurants in Korea. Epidemiol. Health 2024, 46, e2024036. [Google Scholar] [CrossRef]
- Yuan, X.; Huang, Z.; Zhu, Z.; Zhang, J.; Wu, Q.; Xue, L.; Wang, J.; Ding, Y. Recent Advances in Phage Defense Systems and Potential Overcoming Strategies. Biotechnol. Adv. 2023, 65, 108152. [Google Scholar] [CrossRef]
- Kogay, R.; Wolf, Y.I.; Koonin, E.V. Defence Systems and Horizontal Gene Transfer in Bacteria. Environ. Microbiol. 2024, 26, e16630. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, Y.; Gu, C.; Luo, J.; Shen, Y.; Huang, X.; Xu, X.; Ahmed, T.; Alodaini, H.A.; Hatamleh, A.A.; et al. Strain-Specific Infection of Phage AP1 to Rice Bacterial Brown Stripe Pathogen Acidovorax Oryzae. Plants 2024, 13, 3182. [Google Scholar] [CrossRef]
- Shang, J.; Wang, K.; Zhou, Q.; Wei, Y. The Role of Quorum Sensing in Phage Lifecycle Decision: A Switch Between Lytic and Lysogenic Pathways. Viruses 2025, 17, 317. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Guo, M.; Ma, X.; Wang, W.; Ma, W.; Liu, Y.; Wei, J.; Su, Z. A Novel Lytic Salmonella Phage Harboring an Unprecedented Tail-Protein Domain Combination Capable of Lysing Cross-Host-Transmitted Salmonella Strains. Foods 2025, 14, 2850. https://doi.org/10.3390/foods14162850
Zhang L, Guo M, Ma X, Wang W, Ma W, Liu Y, Wei J, Su Z. A Novel Lytic Salmonella Phage Harboring an Unprecedented Tail-Protein Domain Combination Capable of Lysing Cross-Host-Transmitted Salmonella Strains. Foods. 2025; 14(16):2850. https://doi.org/10.3390/foods14162850
Chicago/Turabian StyleZhang, Ling, Mingqiang Guo, Xiaoyu Ma, Wei Wang, Wanpeng Ma, Yifan Liu, Junxiang Wei, and Zhanqiang Su. 2025. "A Novel Lytic Salmonella Phage Harboring an Unprecedented Tail-Protein Domain Combination Capable of Lysing Cross-Host-Transmitted Salmonella Strains" Foods 14, no. 16: 2850. https://doi.org/10.3390/foods14162850
APA StyleZhang, L., Guo, M., Ma, X., Wang, W., Ma, W., Liu, Y., Wei, J., & Su, Z. (2025). A Novel Lytic Salmonella Phage Harboring an Unprecedented Tail-Protein Domain Combination Capable of Lysing Cross-Host-Transmitted Salmonella Strains. Foods, 14(16), 2850. https://doi.org/10.3390/foods14162850




