Abstract
Probiotics have been widely explored for their potential in managing hyperuricemia. However, their isolation and identification are fundamental prerequisites for practical application. In this study, 254 lactic acid bacteria (LAB) strains were isolated from Chinese sauerkraut and screened for probiotic potential based on genomic and phenotypic characteristics, as well as nucleoside-degrading activity relevant to decrease serum urate. Among them, Lactiplantibacillus plantarum (L. plantarum) F42 exhibited the highest bile salt tolerance (survivor rate: 19.46 ± 4.33%), strong adhesion to Caco-2 cells (1.89 ± 0.12%), effective nucleoside degradation (inosine: 5.46 ± 0.67 mg∙L−1∙min−1; guanosine: 3.84 ± 0.11 mg∙L−1∙min−1), and notable anti-listeria activity (inhibition zone: 6.9 ± 0.3 mm). Based on its functional profile, L. plantarum F42 was selected as a promising probiotic candidate for further investigation of its urate-lowering effects. This work provides a new insight into anti-hyperuricemia probiotic selection based on in vitro nucleoside-degrading activity.
1. Introduction
Hyperuricemia (HUA) has become a major public health concern worldwide due to its increasing prevalence and incidence [1]. Notably, this increasing prevalence contributes to the growing cost of urate-lowering and associated analgesic therapies [2]. Another major concern is the occurrence of serious adverse effects due to drug treatments, such as subclinical hypothyroidism [3] and kidney function impairment [4] induced by the administration of the urate-lowering medicine allopurinol, as well as an increased risk of cardiovascular death caused by supplementation with another urate-lowering medicine, febuxostat [5]. In contrast, probiotic-based interventions have recently attracted attention as a potential alternative strategy for managing HUA, owing to their promising safety and efficacy profiles.
Increasing evidence highlights the beneficial effects of probiotics, especially LAB in HUA management. For example, Lactobacillus gasseri PA-3, isolated from human feces, has been found to reduce serum urate levels in rats by inhibiting the absorption of purine-based compounds derived from dietary sources, as demonstrated in both in vitro [6] and in vivo studies [7]. Similarly, the Lacticaseibacillus rhamnosus Fmb14 strain has been shown to utilize dietary inosine as an energy source and engage in the production of folic acid as well as riboflavin, which probably results in a decrease in inosine uptake in HUA mice, thus demonstrating a suppressive effect on urate synthesis [8]. In addition, numerous in vitro studies have shown that various Lactobacillus strains have the potential to act as functional probiotics against HUA [9,10].
In accordance with the definition of ‘probiotics’ by the WHO, the main criteria for identifying a probiotic candidate include its tolerance to acid and bile salts, adhesive ability to gut epithelial cells, antagonistic activity against pathogens, and overall safety profile [11]. Moreover, ability to degrade nucleosides, such as inosine and guanosine, has been widely regarded as a supplemental criterion for selecting probiotic candidates with the potential function of attenuating HUA, supported by the promising correlation between its ability in vitro and decrease in serum urate levels in vivo [9,10,12]. Although the majority of probiotic strains are known to colonize the colon, studies have identified viable Lactobacillus species in the jejunum where nucleoside absorption predominantly occurs. This suggests that oral Lactobacillus strains with acid and bile tolerance may reach and remain active in the small intestine long enough to exert nucleoside-degrading activity before dietary nucleosides are absorbed into the bloodstream, thereby reducing the systemic purine load and subsequent uric acid production.
Despite growing interest in probiotic supplementation for HUA, few studies have systematically screened probiotics from traditional fermented foods based on their nucleoside-degrading activity. Moreover, comprehensive genomic and phenotypic evaluations of such strains remain limited. This study aimed to isolate a specific probiotic candidate with a high potential to attenuate HUA. Nucleoside degradation was used as an index to isolate LAB from Chinese sauerkraut. Subsequently, their genomic feature and probiotic properties—such as resistance to bile salts and low pH, potential colonization in the intestine, bacteriostatic ability, and safety evaluation—were compared to select one strain of Lactobacillus with great potential as a probiotic for further investigation of its function and mechanism in attenuating HUA.
2. Materials and Methods
2.1. Materials
Chinese sauerkraut, a traditional fermented food using Chinese cabbage as raw material, was provided by local market (Guangzhou, China). De Man–Rogosa–Sharpe (MRS), Luria–Bertani (LB) broth, and Columbia blood agar plate were purchased from Huankai Microbial Sci & Tech Co., Ltd. (Guangzhou, China). Escherichia coli (E. coli) ATCC 25922, Staphylococcus aureus (S. aureus) ATCC 51650, Listeria monocytogenes (L. monocytogenes) CMCC 54002, Salmonella enterica (S. enterica) GIM 1.237 and Caco-2 cells were purchased from the Guangdong Microbial Culture Center (Guangzhou, China). Basic media for human cell culture, such as DMEM, penicillin–streptomycin, trypsin, and fetal bovine serum (FBS), were purchased from Gibo Co., Ltd. (New York, NY, USA). Inosine and guanosine (purity > 99%) were provided by Macklin Co., Ltd. (Shanghai, China). All other chemicals used were of analytical grade or higher quality.
2.2. Isolation of LAB from Chinese Sauerkraut
One milliliter of Chinese sauerkraut juice was mixed with 9 mL of saline (0.85% w/v NaCl) for dilution ten times. Similarly, the appropriate decimal dilutions were performed. MRS agar containing 0.1% w/v bromocresol purple powder was employed as growth medium of LAB. Both pour and spread plate methods were used for LAB isolation; however, the spread plate method was selected for LAB enumeration. After 48 h of incubation at 37 °C, each colony surrounded by a yellow circle in the agar was streaked on the surface of a new MRS agar for second isolation. A single colony was then inoculated in a new MRS broth for microbial proliferation. Isolated LAB was collected in sterile tubes with a final concentration of glycerol of 25% v/v and stored at −80 °C until use.
2.3. Degradation of Nucleosides
Nucleoside degradation was measured using the method described by Yamada et al. [6] with minor modifications. Each isolated LAB culture was incubated in MRS broth at 37 °C for 18 h. Then, 1 mL of cell culture was collected, and MRS was replaced with the same volume of sodium phosphate buffer (0.1 M, pH 7.4) containing 1.3 mM inosine and 1.3 mM guanosine for a 1 h reaction at 37 °C. Subsequently, 0.25 mL of 5% v/v TFA was added to terminate the degradation reaction. Centrifugation was performed to collect the supernatant, which was then filtered through a 0.22 μm membrane filter for further quantification by high-performance liquid chromatography (HPLC) equipped with a detection wavelength of 254 nm, an Agilent ZORBAX SB-C18 column (Agilent, Santa Clara, CA, USA) (4.6 × 250 mm 5-Micron), and a mobile phase of 25 mM KH2SO4 buffer containing 5% v/v methanol. Nucleoside concentration was calculated using standard curves (inosine: C = 2.2 × 107 A + 39,065, R2 = 0.999; guanosine: C = 2.5 × 107 A + 34,183, R2 = 0.999). Degradation ability (DA) and degradation rate (V) were calculated according to the following equations:
where C0 and C1 indicate the concentrations of inosine and guanosine (mmol/L) before and after the reaction, respectively.
DA (%) = (C0 − C1)/C0 × 100
V (mg∙L−1∙min−1) = (C0 − C1)/60 × 1000
2.4. Strain Identification
The isolated colony was dispersed in 20 µL of 20 mmol/L NaOH solution, followed by a heating process at 99 °C for 15 min to release the DNA from cells. After centrifugation, the supernatant was collected as a DNA template for gene cloning via PCR reaction. The 16S rRNA and pheS genes were amplified using the following primers: 5′-agagtttgatcctggctcag-3′ for 27F; 5′-tacgacttaaccccaatcgc-3′ for 1492R. 5′-tactatgcaaacaagggtggtaccg-3′ for pheS-F; 5′-caacggccgtccgttcaatcttttc-3′ for pheS-R. The molecular weight of the PCR product was visualized by agarose electrophoresis, and the DNA sequence was confirmed by Sangon Company (Shanghai, China). The DNA sequence was subjected to a BLAST (2.17.0) search against the NCBI nucleotide database, and species were assigned based on the criteria that both sequence similarity and sequence coverage with reference sequences exceeded 90%.
2.5. Genome Sequencing Analysis
The total genomic DNA of LAB was extracted strictly in accordance with the manufacturer’s instructions provided with the Bacterial Genomic DNA Extraction Kit (Sangon, Shanghai, China). DNA was fragmented into approximately 500 base pairs in length using an ultrasonic DNA disruptor, following the operation parameters provided by the manufacturer. A DNA library was then constructed according to the Hieff NGS® MaxUp II DNA Library Prep Kit for Illumina® (Sangon, Shanghai, China). DNA sequences of LAB were obtained using the MGI DNBSEQ-T7 platform. Raw reads were subjected to quality control using Fastp (0.11.2). Specifically, reads were discarded if they contained ≥ 40% low-quality bases or if the trimmed length of paired-end reads was <35 nt. After filtering, the average read depth of the LAB genome reached 150 bp, and the average length of the predicted genes was 860 bp. Sequence similarity searches were performed using the BLASTn tool (2.2.28) against the NCBI NT database, and results were retained if the sequence identity > 90% and the coverage > 80%. Gene function was annotated using multiple databases. In particular, the basic metabolisms of carbohydrates, amino acids, and nucleotides were analyzed based on their pathways obtained from the KEGG and COG databases.
2.6. Growth Curves
Lactobacillus was inoculated in MRS broth at a concentration of 1% v/v. Biomass was monitored every 2 h of anaerobic incubation on the first day and 24 h on the next three days. Bacterial cultures were centrifugated at 12,000 rpm for 1 min, and the supernatant was replaced with an equal volume of saline. The biomass was identified as the UV absorbance at 600 nm with saline used as a blank control.
2.7. Resistance to Bile Salts and Low pH
The growth of Lactobacillus as a function of salt concentration and pH was evaluated according to a previous study with modifications [13]. Lactobacillus (1% v/v) was inoculated in MRS broth containing different concentrations of bile salts (0, 0.03 and 0.3%, w/v.) or under different pH values (3.0 and 7.0). After 2 h of incubation, the number of viable Lactobacillus was determined using the pour plate method. The percentage survival rate was expressed as the number of viable cells incubated in harsh conditions (bile salts or low pH) against the viable counts incubated in pure MRS.
2.8. Colonization in Intestine
The colonization ability of Lactobacillus was evaluated based on chemical and cellular assessments. Auto-aggregation, coaggregation, and hydrophobicity indexes were determined following the method reported by Tuo et al. [14] to evaluate the potential colonization ability of Lactobacillus. For cellular assessment, Caco-2 cells (1 × 105 cells per well) were inoculated and incubated at 37 °C in a 5% CO2 atmosphere for 6 days. During incubation, the fresh DMEM medium in the presence of 20% v/v bovine serum and 1% v/v penicillin–streptomycin was used to replace the old one every two days. After that, the Lactobacillus pellets, obtained by centrifugation and washing twice with HBSS, were resuspended in fresh HBSS and added to the Caco-2 wells. After 2 h of incubation, free Lactobacillus was removed through four washes with HBSS. Then, the number of viable cells adhering to the Caco-2 cell line was determined using the pour plate method. Adhesion ability was calculated as follows:
where Nt and N0 represent the viable count of Lactobacillus adhering to Caco-2 cells and the total viable count of inoculated Lactobacillus, respectively.
Adhesion ability (%) = Nt/N0 × 100
2.9. Bacteriostatic Ability
The well diffusion agar method was employed to measure the antibacterial activity, as described in a previous study [15]. LB agar was melted and cooled to 48 °C. Then 0.1% v/v of the intestinal pathogen including Gram-negative (E. coli and S. enterica) and Gram-positive (S. aureus and L. monocytogenes) strains was mixed with LB agar. Wells with 8 mm diameter were made in LB–agar plates containing 0.1% v/v E. coli, S. enterica, S. aureus or L. monocytogenes, and 100 μL of Lactobacillus supernatant was added to each well. After overnight incubation at 37 °C, the diffusion diameter was recorded. The antibacterial zone was equal to the diffusion zone minus 8 mm.
2.10. Antibiotic Susceptibility
The agar disk diffusion method was used to assess the antibiotic susceptibility of the isolated Lactobacillus strains, as described previously [11]. Three groups of antibiotics were tested, including the inhibitors of nucleic acid synthesis (5 μg of ciprofloxacin and rifampicin), protein synthesis (30 μg chloramphenicol, kanamycin, tetracycline and 15 μg erythromycin), and cell wall synthesis (30 μg vancomycin, amoxicillin, and 10 μg ampicillin). Results were expressed in terms of resistant (R), intermediately sensitive (I), and sensitive (S) levels.
2.11. Hemolysis
Hemolytic activity was evaluated to assess the safety of the bacteria, as described previously [16]. In brief, Lactobacillus was inoculated onto a Columbia blood agar plate and defibrillated sheep blood. S. aureus was used as positive control. After an incubation at 37 °C for 24 h, the grow halos around the colonies were recorded and expressed in terms of γ-hemolysis (no halo), α-hemolysis (greenish halos), and β-hemolysis (bright halos). They were classified as non-hemolytic, partially hemolytic, and hemolytic.
2.12. Statistical Analysis
All experiments were performed in triplicate. Data are shown as mean ± s.d. (standard deviation) and were visualized using Prism GraphPad 8.3.0 software. The multiple-group comparisons were carried out through a one-way ANOVA analysis followed by Tukey’s honestly significant difference (HSD) post hoc test for pairwise comparisons. p < 0.05 was considered statistically significant, as indicated by * or various letters between groups.
3. Results and Discussion
3.1. LAB Diversity in Chinese Sauerkraut
As shown in Figure 1, all identified isolates belonged to four species—Lactiplantibacillus pentosus (L. pentosus), L. plantarum, Limosilactobacillus fermentum (L. fermentum), and Furfurilactobacillus rossiae (F. rossiae)—and one subspecies of L. plantarum, L. plantarum subsp. argentoratensis (L. argentoratensis). Their abundances were 64%, 31%, 3%, 0.8% and 0.8%, respectively. The results indicated that the predominant LAB species in Chinese sauerkraut are L. pentosus and L. plantarum species, which is in agreement with the literature on fermented mustard, beet, and eggplant [17]. L. pentosus was often inaccurately referred to as L. plantarum due to its high similarity in physiological (e.g., shape, growth conditions, and metabolic properties) or partial genetic traits (i.e., the conserved region of the 16S rRNA gene) [18]. Some studies have demonstrated that L. plantarum species have a higher abundance than L. pentosus or that only L. plantarum species are present in fermented vegetables [19,20,21]. This difference could be explained by the fact that the microbial composition and population of traditional fermented foods differ by geographic position, materials, and fermentation stages [22]. However, Lactobacillus sp. is the dominant LAB genus that plays essential roles in the fermentation of carbohydrates [23] and contributes to food’s organolepticity, quality, and safety [24].
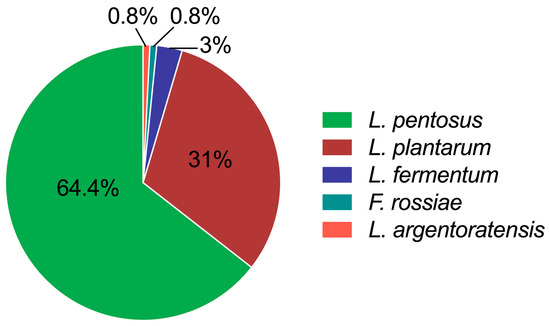
Figure 1.
Diversity of LAB in 50-day-fermented sauerkraut collected from Guangzhou markets. Data were calculated as the isolation frequencies of the species or subspecies relative to the total number of identified isolates.
L. pentosus, L. plantarum, and L. fermentum belonging to Lactobacillus sp. have been frequently found in various native fermented vegetables. In particular, L. plantarum and L. fermentum species have been approved as common edible probiotics by China’s National Health Commission. Their wide range of applications have been explored in the food industry as natural preservatives and texture and flavor enhancers [25]. Furthermore, they can be found as active ingredients in probiotic supplements or functional foods due to their promising health benefits, e.g., enhancing gut health by maintaining a balanced intestinal microbiota, improving digestion, and modulating immunity [26]. Although L. pentosus has not yet been officially approved as a probiotic in China, its potential beneficial effects on host health—such as the regulation of gut microbiota [27], protection against viral infection, and attenuation of alcoholic liver injury—have attracted considerable attention [28]. F. rossiae species, as an obligately hetero-fermentative LAB like L. fermentum but unlike L. plantarum, were scarcely found in traditional fermented vegetables [29]. However, F. rossiae has been reported as a starter culture of cocoa fermentation to produce D-L lactic acid by utilizing glucose and fructose, leading to the resistance of cocoa-related stress conditions (e.g., low pH, high temperature, and high osmotic pressure) [30]. L. argentoratensis is a subspecies of the L. plantarum species; however, its functionality and applications in the food industry have rarely been studied.
3.2. Nucleoside-Degrading Activity of LAB
Lactobacillus species exhibit species-specific differences in their ability to degrade nucleosides [31]. As a result, the top fifty strains isolated from Chinese sauerkraut exhibited various abilities and rates of degradation for inosine and guanosine (Table S1). Notably, nine strains of LAB were capable of complete degradation of inosine and guanosine (Table 1), and they are as follows: L. plantarum F42, L. pentosus P38, L. plantarum 44, L. argentoratensis 15, L. fermentum S8641, L. pentosus b21, L. pentosus S856s, L. fermentum SI823, and L. plantarum b8643. In contrast, no nucleoside-degrading ability was found in L. casei and L. paracasei, which were both isolated from commercial dairy products. Previous studies have reported that the nucleoside-degrading reaction is probably catalyzed by the intracellular nucleoside N-ribohydrolase for RNA and DNA synthesis and energy supply for cellular processes, whereby the enzyme is widely present in LAB, including L. plantarum [9], L. brevis [32], L. gasseri [33] and L. fermentum [34]. Here, four different strains of LAB (L. plantarum F42, L. pentosus P38, L. argentoratensis 15, and L. fermentum S8641) with the highest degradation ability for nucleosides were selected for further evaluation of their probiotic properties based on genotypic and phenotypic traits.

Table 1.
The nucleoside-degrading ability of nine LAB strains.
3.3. Genomic Characterization of LAB
3.3.1. General Genomic Features
The GC content of the genome of all Lactobacillus species is in the range of 43–51% (Table 2), which is in accordance with the Lactobacillus genus [29,32]. The number of annotated genes in Lactobacillus was consistently over 3000, except for the L. fermentum S8641, which contained only 1859 genes. This could imply that genomic research on L. fermentum is relatively less extensive or that its annotation has not reached a depth comparable to that of other Lactobacillus species [35]. The percentage of annotated genes in Lactobacillus differed between databases (Figure S1). Due to the high homology between L. plantarum and L. argentoratensis, their percentage of annotated genes in each database was nearly identical. In comparison, L. pentosus exhibited the highest percentage of genes annotated by CDD, PFAM, GO and KEGG database, suggesting that a considerable level of depth has been achieved in genomic research on this strain. Overall, a substantial number of annotated genes in all Lactobacillus were found to be associated with carbohydrate and amino acid metabolism pathways (Figures S2 and S3).

Table 2.
General genome features of Lactobacillus.
3.3.2. Carbohydrate Metabolism
The genome sequence analysis revealed a substantial number of genes involved in PEP-PTS systems, ABC transporters, and permeases for carbohydrate utilization in various Lactobacillus strains (Table S2). Specifically, all Lactobacillus strains displayed conserved carbohydrate specificity predictions, with the exception of L. fermentum S8641, which notably lacked sorbitol recognition and maltose transport capabilities (Figure 2). This gene deficiency resulted in poor growth and proliferation when sorbitol or maltose served as the sole carbon source (Figure 3). Despite the absence of the pfkA gene encoding 6-phosphofructokinase, L. fermentum S8641 demonstrated robust growth when sucrose, glucose, or fructose was supplied as the sole carbon source. This phenotype could be attributed to its phosphoketolase (PK) pathway, in which hexoses are cleaved, ultimately generating CO2, ethanol, acetate, and lactate as fermentation end-products [36]. Conversely, L. plantarum F42, L. pentosus 38, and L. argentoratensis 15 were capable of utilizing maltose and sorbitol. Moreover, their metabolism of glucose, sucrose, and fructose was conducted via the glycolysis pathway. This highlights the metabolic versatility within the genus, where different strains employ distinct pathways to adapt to varying carbohydrate substrates. In addition, all Lactobacillus could utilize cellobiose, galactooligosaccharide, and fructooligosaccharide for proliferation due to the presence of genes encoding a series of enzymes (e.g., α-amylase, α-galactosidase, β-galactosidase genes cluster, Endo-beta-N-acetylglucosaminidase D, glycosyl hydrolases) which are responsible for the degradation of di-, tri-, and even polysaccharides [37].
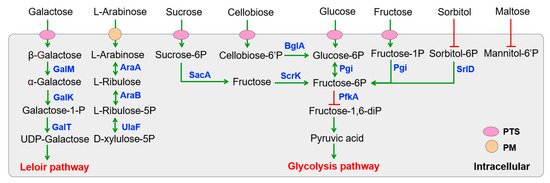
Figure 2.
KEGG pathway-based sugar metabolism in Lactobacillus isolated from Chinese sauerkraut. PTS (phosphotransferase systems) and PM (plasma membrane transporter) are involved in sugar transport. Genes encoded enzymes are shown in blue. Red lines with bars indicate a potential missing reaction in L. fermentum S8641.
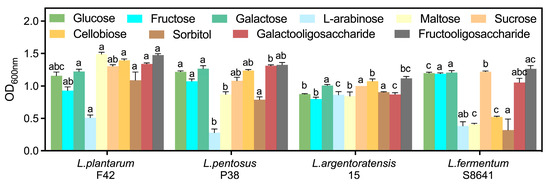
Figure 3.
Growth activity (OD600nm) of Lactobacillus in MRS broth with glucose replaced by different carbohydrates. Different letters indicate significant difference (p < 0.05) between Lactobacillus in the same medium.
3.3.3. Amino Acid Metabolism
All Lactobacillus strains putatively dedicated the glnA enzyme to the conversion of glutamate into glutamine (Figure 4 and Table S3), which is associated with the beginning of arginine biosynthesis and glutamate degradation. Furthermore, previous studies have evidenced that the non-essential amino acid had positive effects on the intestinal mucosal system in the host [37]. Additionally, a series of arg and car enzymes potentially converted glutamate into aspartate, which ultimately led to the formation of arginine. Glycerate could be converted into serine through a series of metabolic reactions. Serine then serves as a precursor for the synthesis of glycine and cysteine. However, the pathway for cysteine synthesis from serine may be absent in L. fermentum S8641 due to the lack of the enzyme serine O-acetyltransferase (cysE). Additionally, conversion of threonine into glycine was not universal among Lactobacillus species. Nevertheless, they possessed an almost complete pathway for generating histidine from PRPP, which involves the pentose phosphate pathway. Conversely, only L. fermentum S8641 was capable of synthesizing tyrosine as it possessed the enzyme EC 5.4.99.5. This metabolic diversity highlights the species-specific metabolic capabilities and limitations within the genus, underscoring the genetic and biochemical adaptations that enable these bacteria to synthesize essential amino acids.
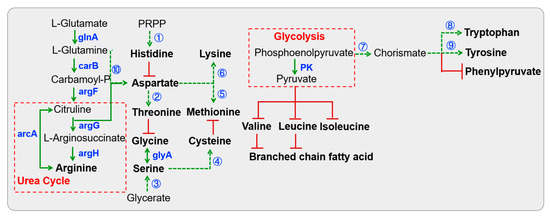
Figure 4.
KEGG pathway-based amino acid biosynthesis in Lactobacillus isolated from Chinese sauerkraut. Red lines with bars indicate a potential missing reaction. Dotted arrows indicate a multi-step reaction. The first multi-step reaction is catalyzed by a series of his enzymes including hisA, hisB, hisC, hisD, hisE, hisF, hisG, hisI and hisZ. The second multi-step reaction is catalyzed by lysC, asd, thrB1 and thrC. The third multi-step reaction is catalyzed by garK, gpmA, serA and serC. Enzymes cysE and cysK participate in the fourth reaction. The fifth reaction is catalyzed by lysC, asd, metA, metB, metC and mmuM. Numerous enzymes (lysA, lysC, asd, dapA, dapB, dapE, dapF, dapG, dapH and mtnE) are involved in the sixth reaction. Enzymes aroA, aroB, aroC, aroD, aroE, aroK and EC:2.5.1.19 participate in the seventh reaction. Enzymes trpA, trpB, trpC, trpD, trpE, trpF and trpG participate in the eighth reaction. Enzymes EC:5.4.99.5, tyrA2 and hisC participate in the ninth reaction, and the tenth reaction is catalyzed by the three enzymes (carA, caiB and pyrB).
3.3.4. Nucleotide Metabolism
The first step in purine metabolism involves de novo purine biosynthesis, which initiates with a phosphorylation reaction of ribose-5-phosphate and culminates in the formation of IMP (inosine monophosphate) (Figure 5). All Lactobacillus putatively possessed the metabolic capability to engage in the de novo purine biosynthesis (Table S4). However, they were unable to autonomously complete a series of reactions in de novo purine biosynthesis due to the absence of two essential enzymes (phosphoribosylformylglycinamidine synthase, EC:6.3.5.3; 5-(carboxyamino) imidazole ribonucleotide mutase, EC:5.499.18).
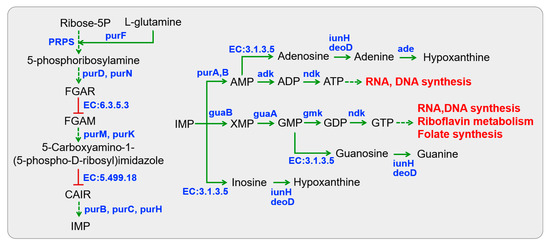
Figure 5.
KEGG pathway-based nucleotide metabolism in Lactobacillus isolated from Chinese sauerkraut. Red lines with bars indicate a potential missing reaction in all Lactobacillus species.
IMP acts as a substrate for the subsequent synthesis of AMP (adenosine monophosphate) and GMP (guanosine monophosphate). All Lactobacillus were predicted to be capable of participating in ATP and GTP metabolism through reversible conversions from AMP and GMP, respectively, represented by enzymes adenylate kinase (adk, EC:2.7.4.3), nucleoside-diphosphate kinase (ndk, EC:2.7.4.6), and guanylate kinase (gmk, EC:2.7.4.8). This process predominantly provides energy for cellular activities, and the products can be utilized in RNA and DNA synthesis, riboflavin metabolism, and folate biosynthesis. Only L. pentosus P38 was capable of synthesizing nucleosides (e.g., inosine, guanosine, and adenosine) from IMP, GMP, and AMP through dephosphorylation (5′-nucleotidase, EC:3.1.3.5). For nucleoside degradation, two specific enzyme-encoding genes (purine nucleosidase, iunH, EC:3.2.2.1; adenine deaminase, ade, EC:3.5.4.2) were present in all Lactobacillus, which convert nucleosides into purine bases such as inosine to hypoxanthine, guanosine to guanine, and adenosine to adenine and hypoxanthine. By comparison, the presence of an isoenzyme urine-nucleoside phosphorylase (deoD, EC:2.4.2.1) was a peculiarity of L. fermentum S8641, leading to nucleoside degradation. This observation was in accordance with the abovementioned results, which indicated a strong ability to degrade inosine and guanosine in all tested Lactobacillus strains (Table 1). Finally, these nucleosides and their degraded products could be metabolized as the substrates of DNA synthesis or participate in the metabolism of clycine, serine, and threonine.
Overall, the metabolic pathways of four Lactobacillus strains were characterized by the identification of gene presence based on genotypic analysis. However, these genotype-based results are susceptible to potential artifacts from gene annotation and the genome assembly processes. Therefore, phenotypic validation, such as targeted metabolic profiling, would be critical in future work to confirm the metabolic pathways in these strains.
3.4. Phenotypic Characterization of LAB
3.4.1. Growth Curves
As shown in Figure 6, L. plantarum F42 exhibited a comparable pattern of growth kinetics to L. pentosus P38, with equivalent periods allocated for the lag phase (from 0 to 2 h), logarithmic (log) growth phase (at 4 h until 16 h), and stationary phase (after 24 h). This strongly suggests that both bacterial species had similar temporal dynamics during their respective growth cycles under identical or closely matched environmental conditions. In addition, the lag phase duration of L. argentoratensis 15 was found to be longer than that of L. fermentum S8641. Specifically, the lag phase for L. argentoratensis 15 extended from 0 to 12 h, whereas L. fermentum S8641 exhibited a lag phase lasting from 0 to 8 h. This indicates a difference in their initial adaptation periods before entering exponential growth. Finally, both species started the stationary phase between 24 and 48 h of cultivation after undergoing a different duration of log phase. Overall, the precise timing for entering each growth phase was probably variable in different Lactobacillus strains due to their specific growth requirements and metabolic capabilities.
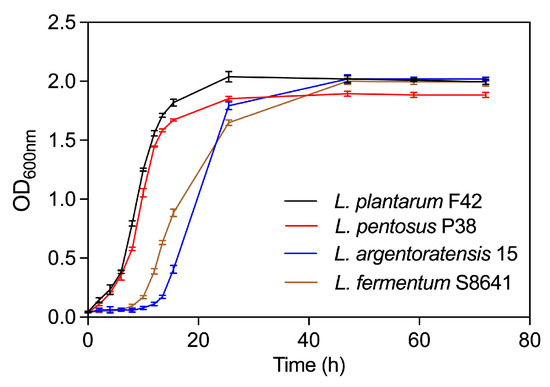
Figure 6.
Growth curves of four Lactobacillus species derived from Chinese sauerkraut.
3.4.2. Resistance to Bile Salts and Low pH
The resistance of LAB to bile salts and low pH during the simulated digestion is a pivotal criterion for assessing their potential as probiotic candidates. As shown in Table 3, the initial viable count of all tested Lactobacillus was in the range of 6.79–7.19 log CFU mL−1, with no significant differences between strains (p > 0.05). After 2 h of incubation in a simulated environment with a low pH level of 3.0, which represented postprandial human stomach acidity, a significant decrease in the survival rate of all tested Lactobacillus strains to 61.79–77.51% was observed. Specifically, L. plantarum F42 (77.51 ± 1.29%) and L. argentoratensis 15 (72.47 ± 1.72%) exhibited a significantly higher survival rate than L. pentosus P38 (66.78 ± 3.26%, p < 0.05) and L. fermentum S8641 (61.79 ± 1.25%, p < 0.05).

Table 3.
The viability (log CFU mL−1) and survival rate of four Lactobacillus species at low pH (3.0) and in the presence of 0.03% porcine bile salts (BS) at pH 7.0.
In addition, the viability of all Lactobacillus significantly decreased in the presence of 0.03% bile salts, which mimicked the small intestine at its lowest concentration of bile salts. Finally, the survival rate was in the following order: L. plantarum F42 > L. argentoratensis 15 > L. pentosus P38 > L. fermentum S8641. The decrease in cell viability underscores the fact that low pH and bile salts threaten Lactobacillus. Furthermore, numerous studies have consistently demonstrated a reduction in the viability of different Lactobacillus strain under harsh gastrointestinal conditions, but with strain-dependent resistance or sensitivity [38,39]. By comparison, L. plantarum SF4722 exhibited a higher survival rate than the other three strains under such conditions (p < 0.05), indicating its potential to remain alive until reaching the intestine, where it can exert beneficial effects on the host’s health. Rzepkowska et al. reported that the differences in the resistance of LAB to bile salts could be attributed to their innate bile salt hydrolase, which deconjugates bile salts [40]. According to the genomic features, a gene encoding the specific enzyme bile salt hydrolase (EC:3.5.1.24), which is involved in bile acid biosynthesis, was found in all tested Lactobacillus, and the presence of this gene was in the following order, from highest to lowest: L. plantarum F42 (8) = L. argentoratensis 15 (8) > L. pentosus P38 (6) > L. fermentum S8641 (4). However, the hypothesis that the correlation between the quantity of bile salt hydrolase/genes in Lactobacillus and its resistance to bile salts requires further investigation.
3.4.3. Potential Colonization in the Intestine
All Lactobacillus strains presented an auto-aggregation capacity ranging from 8.7 to 26.8% after 4 h incubation (Figure 7A), where the L. fermentum S8641 strain exhibited the lowest value, but L. plantarum F42 displayed the highest value. The auto-aggregation capacity of L. fermentum S8641 at 8.7% is consistent with the literature showing the auto-aggregation percentage of eleven strains of L. fermentum was in the range of 3.5 to 14.2% under similar incubation conditions [13]. In addition, the auto-aggregation of all tested Lactobacillus gradually increased by incubation time. Notably, no significant differences were detected among L. plantarum F42, L. pentosus P38, and L. argentoratensis 15 at each incubation time; however, their auto-aggregation values were significantly higher than that of L. fermentum S8641. Previous studies have illustrated that bacterial aggregation ability, including auto- and co-aggregation, is potentially linked to their adhesion to intestinal epithelial cells, which in turn affects their survival and persistence within the gastrointestinal tract [14]. All tested strains had a greater coaggregation with E. coli compared to the other three pathogenic species (Figure 7B), which is in accordance with previous publications [39,41].
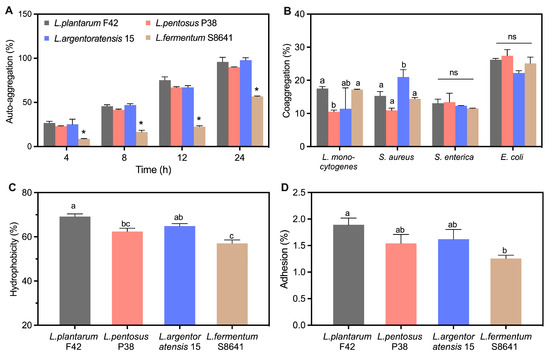
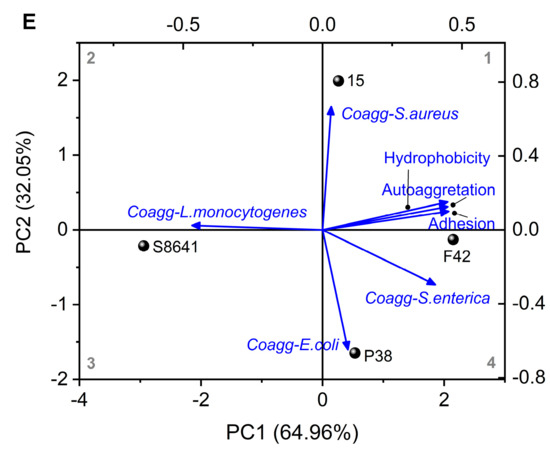
Figure 7.
Auto-aggregation (A), coaggregation with four pathogens (L. monocytogenes, S. aureus, S. enterica and E. coli) (B), hydrophobicity (C), adhesion ability to Caco-2 cells (D), and the principal component analysis (PCA) (E) integrating the results of auto-aggregation, coaggregation, hydrophobicity and adhesion ability of Lactobacillus derived from Chinese sauerkraut. Vectors represent trait loadings; longer vectors indicate stronger contributions to principal components (PC1: 64.96% variance, PC2: 32.05% variance). Different letters indicate significant difference (p < 0.05) between groups. *: p < 0.05.
The hydrophobicity of the bacterial surface has been recognized as an important factor in promoting their ability to adhere to host tissues [14]. This attachment mechanism is often mediated by the affinity between the hydrophobic regions of bacterial cell surfaces and the similar regions on host tissue cells or mucus layers lining the gastrointestinal tract. The hydrophobicity was also strain-specific (Figure 7C). The mean value of hydrophobicity for all tested strains followed the order L. plantarum F42 > L. argentoratensis 15 > L. pentosus P38 > L. fermentum S8641, with the highest value for L. plantarum F42 (69.2%) and the lowest value for L. fermentum S8641 (57.0%). However, a similar trend to that observed in hydrophobicity was found in the adhesion properties of these tested strains (Figure 7D). L. plantarum F42 showed the highest adhesion percentage compared to other three strains, which is consistent with previous studies showing that the adhesion of L. plantarum species to Caco-2 cell line was higher than L. fermentum species [41]. The components on the surface of bacterial cell walls play an important role in the adhesive properties they exhibited. Tuo et al. summarized that the surface-bound protein of bacteria contributes to its adhesive properties, as indicated by the fact that the adhesion of Lactobacillus to Caco-2 cells was significantly decreased after treating with protein denaturant [14]. Lebeer et al. illustrated that the surface proteins and mucin-binding proteins on probiotics play a critical role in their interaction with the intestinal mucus layer [42]. Specifically, the Leu-Pro-any-Thr-Gly sequence at the C-terminus of mucin-binding proteins can link to peptidoglycan in the cell wall, functioning as a bridge between the probiotic and gut mucus layer, ultimately facilitating bacterial adhesion and colonization [43]. Furthermore, bacterial adhesion characteristics could also be attributed to the glycoproteins and teichoic and lipoteichoic acids present on the surface of the bacterial cell wall [44].
The results obtained from the assays (regarding the hydrophobicity, auto-aggregation, and coaggregation ability of Lactobacillus with various pathogens) were subjected to PCA analysis to identify the potential correlations between these bacterial characteristics and their adhesive properties. As shown in Figure 7E, the first two principal components (PC1: 64.96%; PC2: 32.05%), which accounted for 97.01% of the overall variance, were sufficient for further discussion (p < 0.05). The circle points representing the four tested Lactobacillus strains were distributed separately in different quadrants, indicating a diversity of bacterial properties among them. The vectors displayed bacterial characteristics. Notably, the hydrophobicity and auto-aggregation abilities were close to the adhesion properties. As shown in Table 4, their positive loading values on PC1 (> 0.44) were larger than coaggregation with different pathogens (−0.469–0.404). These results indicate a strong positive correlation between both traits and adhesion ability. In addition, the point L. plantarum F42 was closer to the adhesion properties than other points/strains, implying its superior potential to adhere to the intestine.

Table 4.
Trait loadings on PC1 and PC2.
3.4.4. Bacteriostatic Ability
As shown in Table 5, different strains of Lactobacillus displayed distinct inhibitory capacities against various pathogens. L. plantarum F42, L. argentoratensis 15, and L. fermentum S8641 exhibited antibacterial activity against the four tested pathogens, with a low inhibition against E.coli (+) and a moderate inhibition against S. aureus (++). This result is consistent with previous publications showing that Lactobacillus strains were more active against Gram-positive bacteria [40]. Antibacterial activity against S. aureus was not found in L. pentosus P38, although inhibition of the other three pathogens occurred. Notably, L. plantarum F42 had stronger anti-listeria (L. monocytogenes) activity than other Lactobacillus strains. Previous studies have demonstrated that Lactobacillus exhibits antibacterial activity against a variety of pathogenic microorganisms (e.g., L. monocytogenes, S. aureus, S. enterica, and E.coli), which can be attributed to their competitive adhesion to the epithelium cells or to the secretion of antibacterial substances (e.g., organic acids, bacteriocin, and hydrogen peroxide) [11,45]. It has been evidenced that bacteriocin production is a general feature of Lactobacillus. Bacteriocins, especially the class IIa type, have strong inhibitory activity against L. monocytogenes strains [40]. L. plantarum strains are known to produce a diversity of bacteriocins, which could induce membrane leakage and cell death by specifically binding the mannose phosphotransferase system (man-PTS) on their target pathogenic cells [46]. According to the genetic features, genes encoding two specific enzymes (LciA and LagD) which involve the formation and transport of bacteriocin were found in L. plantarum F42 and L. argentoratensis 15 strains. In particular, the presence of genes encoding a variety of plantaricin systems (plnE, plnF, plnJ and plnK) putatively contributed to the production of at least two types of bacteriocins. However, those genes were not detected in the L. fermentum S8641 strain, implying that the low-pH environment resulting from its organic acid is a crucial factor in inhibiting pathogen growth.

Table 5.
Antimicrobial activity of four Lactobacillus species against four pathogens.
3.4.5. Antibiotic Susceptibility
All tested strains were resistant to the ciprofloxacin, an inhibitor of nucleic acid synthesis (Table 6), which is in accordance with previous publications showing the resistance of L. fermentum, L. plantarum, and its subsp. strains to ciprofloxacin [9,11]. The tested strains exhibited various degrees of sensitivity to rifampicin, with L. plantarum F42 and L. argentoratensis 15 showing resistance, while L. pentosus P38 and L. fermentum S8641 demonstrated intermediate sensitivity. All Lactobacillus were susceptible to chloramphenicol and tetracycline, the inhibitors of protein synthesis, while none of the strains were susceptible to kanamycin. All tested strains were intermediately susceptible to erythromycin except for L. fermentum S8641, which was resistant to erythromycin. Similar observations have been demonstrated by previous studies [16,47].

Table 6.
Antibiotic susceptibility of four Lactobacillus species.
In addition, resistance to the inhibitors of cell wall synthesis, vancomycin and amoxicillin, was found in all tested strains; however, the strains were consistently susceptible to ampicillin. Inherent non-acquired resistance to vancomycin in a variety of Lactobacillus species has been consistently reported [45]. Overall, numerous studies have shown that most probiotics are naturally resistant to ciprofloxacin, kanamycin, and vancomycin; however, these resistances are suspected to encode within their chromosomes and are neither inducible nor horizontally transferable among bacteria [48,49]. Although no probiotic health claims related to antibiotic resistance have been positively assessed by the European Food and Safety Agency (EFSA), Lactobacillus species still can be recommended for inclusion in the qualified presumption of safety (QPS) list [50] as they have a long history of safe use and have never been implicated in human or animal diseases.
3.4.6. Safety Evaluation
According to WHO and EFSA guidelines, the absence of hemolytic activity is a fundamental criterion for selecting probiotic strains to guarantee their non-virulent nature. Therefore, hemolytic activity was employed to evaluate the safety of isolated Lactobacillus strains for applications as bio-preservatives or probiotics in food industry. As a result, the positive control S. aureus exhibited β-hemolytic activity (Table 7). In contrast, all tested Lactobacillus strains were classified as γ-hemolytic, demonstrating that they are non-hemolytic and therefore safe for use.

Table 7.
Hemolytic activity of four Lactobacillus species.
4. Conclusions
In this study, Lactobacillus species including L. pentosus, L. plantarum, L. fermentum and L. argentoratensis were successfully isolated from Chinese sauerkraut. These strains exhibited different nucleoside-degrading activities, which are potentially associated with urate-lowering effects in vivo. Four different strains (L. plantarum F42, L. pentosus P38, L. argentoratensis 15, and L. fermentum S8641) demonstrated complete degradation of inosine and guanosine (100%) and were selected for further evaluation of probiotic potential. Genomic analysis demonstrated their similar metabolic pathways related to carbohydrates, amino acids and nucleotides, except for L. fermentum S8641. Among them, L. plantarum F42 showed higher tolerance to low pH (survival rate: 77.51 ± 1.29%) and bile salts (survival rate: 19.46 ± 4.33%). PCA analysis illustrated that the adhesion ability (PC1 loading = 0.452) of Lactobacillus is positively correlated with its surface hydrophobicity (0.449) and auto-aggregation abilities (0.449), supporting the superior gut colonization potential of L. plantarum F42 (1.89 ± 0.12%). Furthermore, L. plantarum F42 displayed broad antimicrobial activity against both Gram-positive (L. monocytogenes: 6.9 ± 0.3 mm; S. aureus: 4.4 ± 0.5 mm) and Gram-negative bacteria (S. enterica: 4.2 ± 0.3 mm, E. coli: 1.9 ± 0.1 mm). Its safety for use in the food industry was further supported by the lack of hemolytic activity on blood agar plates. Considering these advantageous characteristics, L. plantarum F42 was identified as a promising probiotic candidate for anti-hyperuricemia. Moreover, the strain could be incorporated into fermented foods (e.g., yogurt, probiotic drinks) or as a therapeutic candidate for individuals at risk of hyperuricemia. This study might open up opportunities to develop targeted probiotic isolation strategies aimed at managing hyperuricemia, although their in vivo anti-hyperuricemia effects should be confirmed using animal and clinical tests in the future in order to advance their application in the fields of functional foods and therapeutics.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods14152690/s1, Table S1: The top fifty strains of LAB ranked by the ability to degrade nucleosides; Table S2: The number of genes and their predicted specificity for carbohydrate transport and metabolism in various Lactobacillus; Table S3: Putative classification of amino acid metabolism; Table S4: Putative classification of nucleotide metabolism; Figure S1: Percentage of annotated genes in different databases; Figure S2: KEGG pathway annotated gene categories for different Lactobacillus species; Figure S3: COG classification of Lactobacillus.
Author Contributions
Conceptualization, W.-Y.L. and M.-F.L.; methodology, M.-F.L. and M.-Y.H.; validation, M.-F.L.; formal analysis, M.-Y.H.; investigation, M.-F.L.; resources, W.-Y.L. and M.-F.L.; data curation, M.-Y.H.; writing—original draft preparation, M.-Y.H.; writing—review and editing, M.-F.L. and W.-Y.L.; visualization, M.-Y.H.; supervision, W.-Y.L.; project administration, W.-Y.L.; funding acquisition, W.-Y.L. and M.-F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2022YFD2101402), the National Natural Science Foundation of China (22378146, 22278161, 22078111), the Guangdong Basic and Applied Basic Research Foundation (2022B1515020013), Guangzhou Science and Technology Projects (2023B03J1317), and the Research Group of Functional Food Development for Specific Populations (2024ZZ12).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Xia, Y.; Wu, Q.; Wang, H.; Zhang, S.; Jiang, Y.; Gong, T.; Xu, X.; Chang, Q.; Niu, K.; Zhao, Y. Global, regional and national burden of gout, 1990–2017: A systematic analysis of the Global Burden of Disease Study. Rheumatology 2020, 59, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Helget, L.N.; England, R.B.; Roul, P.; Sayles, H.; Petro, A.D.; Michaud, K.; Mikuls, T.R. Incidence, prevalence, and burden of gout in the Veterans health administration. Arthritis Care Res. 2021, 73, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Yang, Y.-S.; Chang, D.-J.; Chuang, Y.W.; Kim, H.; Ko, S.J.; Yoo, S.; Oh, J.S.; Kang, D.Y.; Yang, H.-J.; et al. Association between the use of allopurinol and risk of increased thyroid-stimulating hormone level. Sci. Rep. 2021, 11, 20305. [Google Scholar] [CrossRef]
- Stamp, L.K.; Day, R.O.; Yun, J. Allopurinol hypersensitivity: Investigating the cause and minimizing the risk. Nat. Rev. Rheumatol. 2016, 12, 235–242. [Google Scholar] [CrossRef]
- Guan, X.; Zhang, S.; Liu, J.; Wu, F.; Zhou, L.; Liu, Y.; Su, N. Cardiovascular safety of febuxostat and allopurinol in patients with gout: A meta-analysis. Front. Pharmacol. 2022, 13, 998441. [Google Scholar] [CrossRef]
- Yamada, N.; Lwamoto, C.; Kano, H.; Yamaoka, N.; Fukuuchi, T.; Kaneko, K.; Asami, Y. Evaluation of purine utilization by Lactobacillus gasseri strains with potential to decrease the absorption of food-derived purines in the human intestine. Nucleosides Nucleotides Nucleic Acids 2016, 35, 670–676. [Google Scholar] [CrossRef]
- Yamada, N.; Saito, C.; Murayama-Chiba, Y.; Kano, H.; Asami, Y.; Ito, H. Lactobacillus gasseri PA-3 utilizes the purines GMP and guanosine and decreases their absorption in rats. Nucleosides Nucleotides Nucleic Acids 2018, 37, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, X.; Meng, F.; Zhou, L.; Pang, X.; Lu, Z.; Lu, Y. Ameliorative effect of Lacticaseibacillus rhamnosus Fmb14 from Chinese yogurt on hyperuricemia. Food Sci. Hum. Wellness 2023, 12, 1379–1390. [Google Scholar] [CrossRef]
- Lin, J.; Xiong, T.; Peng, Z.; Xie, M.; Peng, F. Novel lactic acid bacteria with anti-hyperuricemia ability: Screening and in vitro probiotic characteristics. Food Biosci. 2022, 49, 101840. [Google Scholar] [CrossRef]
- Lu, L.; Liu, T.; Liu, X.; Wang, C. Screening and identification of purine degrading Lactobacillus fermentum 9-4 from Chinese fermented rice-flour noodles. Food Sci. Hum. Wellness 2022, 11, 1402–1408. [Google Scholar] [CrossRef]
- Joice, F.M.S.; Bruna, V.A.; Paula, A.A.A.; Isabela, V.B.; Otávio, A.B.; Letícia, R.B.; Laura, M.B.; Liliane, D.M.M.; Márcio, R.S.; da Geraldo, M.C.; et al. Searching for antibiotic-susceptible bioprotective lactic acid bacteria to control dangerous biological agents in artisanal cheese. Food Microbiol. 2025, 130, 104762. [Google Scholar] [CrossRef]
- Kuo, Y.; Hsieh, S.; Chen, J.; Liu, C.; Chen, C.; Huang, Y.; Ho, H. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ 2021, 9, e11209. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef]
- Roy, S.; Mandal, S. Lactiplantibacillus plantarum isolates from natural honey (Malda, India): Probiotic potentiality and antibacterial activity analysis by in vitro methods. Food Humanit. 2024, 3, 100348. [Google Scholar] [CrossRef]
- Margalho, L.P.; Feliciano, M.D.; Silva, C.E.; Abreu, J.S.; Piran, M.; Sant’Ana, A.S. Brazilian artisanal cheeses are rich and diverse sources of nonstarter lactic acid bacteria regarding technological, biopreservative, and safety properties—Insights through multivariate analysis. J. Dairy Sci. 2020, 103, 7908–7926. [Google Scholar] [CrossRef]
- Nguyen, D.T.L.; Van Hoorde, K.; Cnockaert, M.; De Brandt, E.; Aerts, M.; Binh Thanh, L.; Vandamme, P. A description of the lactic acid bacteria microbiota associated with the production of traditional fermented vegetables in Vietnam. Int. J. Food Microbiol. 2013, 163, 19–27. [Google Scholar] [CrossRef]
- Khemariya, P.; Singh, S.; Jaiswal, N.; Chaurasia, S.N.S. Isolation and identification of Lactobacillus plantarum from vegetable samples. Food Biotechnol. 2016, 30, 49–62. [Google Scholar] [CrossRef]
- Oyedeji, O.; Ogunbanwo, S.T.; Onilude, A.A. Predominant lactic acid bacteria involved in the traditional fermentation of Fufu and Ogi, two Nigerian fermented food products. Food Nutr. Sci. 2013, 4, 40–46. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, Y.; Liu, Y.; Tao, D.; Yue, X.; Cao, X.; Wu, R.; Wu, J. Diversity and screening of lactic acid bacteria in la-baicai made by Korean-Chinese in northeastern China. Food Biotechnol. 2017, 3, 193–209. [Google Scholar] [CrossRef]
- Peng, Q.; Jiang, S.; Chen, J.; Ma, C.; Huo, D.; Shao, Y.; Zhang, J. Unique microbial diversity and metabolic pathway features of fermented vegetables from Hainan, China. Front. Microbiol. 2018, 9, 399. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Peng, Z.; Huang, T.; Xiao, Y.; Li, J.; Xie, M.; Xiong, T. Comparison of bacterial diversity in traditionally homemade Paocai and Chinese spicy cabbage. Food Microbiol. 2019, 83, 141–149. [Google Scholar] [CrossRef]
- Jhamb, V.; Swaminathan, P. Role and importance of lactic acid bacteria in different Indian fermented foods. Biologia 2023, 78, 3609–3623. [Google Scholar] [CrossRef]
- Bautista-Gallego, J.; Medina, E.; Sánchez, B.; Benítez-Cabello, A.; Arroyo-López, F.N. Role of lactic acid bacteria in fermented vegetables. Grasas Aceites 2020, 71, e358. [Google Scholar] [CrossRef]
- Patrick, O.A.; Great, I.E.; Ali, B.M.; Ali, E.Y.; Khalid, Z.; Joseph, O.O.; Endurance, F.I.; Ufuoma, A.I.; Arthur, E.A.E.; Raghda, S.M.; et al. Lactic acid bacteria: Nature, characterization, mode of action, products and applications. Process Biochem. 2025, 152, 1–28. [Google Scholar] [CrossRef]
- Lim, S.-M.; Lee, N.-K.; Kim, K.-T.; Paik, H.-D. Probiotic lactobacillus fermentum KU200060 isolated from watery kimchi and its application in probiotic yogurt for oral health. Microb. Pathog. 2020, 147, 104430. [Google Scholar] [CrossRef]
- López-García, E.; Benítez-Cabello, A.; Arenas-de Larriva, A.P.; Gutierrez-Mariscal, F.M.; Pérez-Martínez, P.; Yubero-Serrano, E.M.; Garrido-Fernández, A.; Arroyo-López, F.N. Oral intake of Lactiplantibacillus pentosus LPG1 produces a beneficial regulation of gut microbiota in healthy persons: A randomised, Placebo-controlled, single-blind trial. Nutrients 2023, 15, 1931. [Google Scholar] [CrossRef]
- Gan, Y.; Peng, J.; Zhang, Y.; Feng, X.; Qian, Y.; Long, X.; Zhao, X.; Li, Q. Protective effect of Lactiplantibacillus pentosus CQZC01 in Kunming mice of subacute alcoholic liver injury. J. Food Sci. 2023, 88, 2642–2654. [Google Scholar] [CrossRef]
- De Angelis, M.; Bottacini, F.; Fosso, B.; Kelleher, P.; Calasso, M.; Di Cagno, R.; Ventura, M.; Picardi, E.; Van Sinderen, D.; Gobbetti, M. Lactobacillus rossiae, a vitamin B12 producer, represents a metabolically versatile species within the genus Lactobacillus. PLoS ONE 2014, 9, e107232. [Google Scholar] [CrossRef] [PubMed]
- Dea, K.; Giovanni, R.; Alberto, F.; Davide, E.; Alberto, B.; Maria Grazia, F. Exploration of Lactiplantibacillus fabifermentans and Furfurilactobacillus rossiae as potential cocoa fermentation starters. J. Appl. Microbiol. 2022, 133, 1769–1780. [Google Scholar] [CrossRef]
- Kano, H.; Saito, C.; Yamada, N.; Fukuuchi, T.; Yamaoka, N.; Kaneko, K.; Asami, Y. Species-dependent patterns of incorporation of purine mononucleotides and nucleosides by lactic acid bacteria. Nucleosides Nucleotides Nucleic Acids 2020, 39, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mei, L.; Deng, Y.; Liu, Y.; Wei, X.; Liu, M.; Zhou, J.; Ma, H.; Zheng, P.; Yuan, J. Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutrition 2019, 62, 63–73. [Google Scholar] [CrossRef]
- Yamada, N.; Saito-Iwamoto, C.; Nakamura, M.; Soeda, M. Lactobacillus gasseri PA-3 uses the purines IMP, inosine and hypoxanthine and reduces their absorption in rats. Microorganisms 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhao, C.; Li, X.; Wang, H.; Huang, Q.; Sun, Y.; Zhou, Y. Discovery of lactic acid bacteria with high nucleoside degradation and low purine production in tomato sour soup. Int. J. Food Microbiol. 2025, 434, 111139. [Google Scholar] [CrossRef]
- Brandt, K.; Nethery, M.A.; O’Flaherty, S.; Barrangou, R. Genomic characterization of Lactobacillus fermentum DSM 20052. BMC Genom. 2020, 21, 328. [Google Scholar] [CrossRef]
- Okoye, C.O.; Dong, K.; Wang, Y.; Gao, L.; Li, X.; Wu, Y.; Jiang, J. Comparative genomics reveals the organic acid biosynthesis metabolic pathways among five lactic acid bacterial species isolated from fermented vegetables. New Biotechnol. 2022, 25, 73–83. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Altermann, E.; Goh, Y.J.; Tallon, R.; Sanozky-Dawes, R.B.; Pfeiler, E.A.; O’ Flaherty, S.; Buck, B.L.; Dobson, A.; Duong, T.; et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 2008, 74, 4610–4625. [Google Scholar] [CrossRef] [PubMed]
- Fuochi, V.; Petronio, G.P.; Lissandrello, E.; Furneri, P.M. Evaluation of resistance to low pH and bile salts of human Lactobacillus spp. isolates. Int. J. Immunopathol. Pharmacol. 2015, 28, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.L.; Li, M.F.; Liu, S.Y.; Yu, M.; Lou, W.Y. Regulation of hepatocellular cholesterol metabolism by Lactobacillus paracasei by2 and its embedding delivery. Probiotics Antimicrob. Proteins 2022, 16, 181–195. [Google Scholar] [CrossRef]
- Rzepkowska, A.; Zielińska, D.; Ołdak, A.; Kołożyn-Krajewska, D. Safety assessment and antimicrobial properties of the lactic acid bacteria strains isolated from polish raw fermented meat products. Int. J. Food Prop. 2017, 20, 2736–2747. [Google Scholar] [CrossRef]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-specific probiotics properties of Lact. fermentum. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C. Host interactions of probiotic bacterial surface molecules: Comparison with commensals and pathogens. Nat. Rev. Microbiol. 2010, 8, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef] [PubMed]
- Darilmaz, D.O.; Beyatli, Y. Investigating hydrophobicity and the effect of exopolysaccharide on aggregation properties of dairy propionibacteria isolated from Turkish homemade cheeses. J. Food Prot. 2012, 75, 359–365. [Google Scholar] [CrossRef]
- Tavakoli, M.; Esfahani, Z.H.; Hejazi, M.A.; Azizi, M.H.; Abbasi, S. Characterization of probiotic abilities of Lactobacilli isolated from Iranian Koozeh traditional cheese. Pol. J. Food Nutr. Sci. 2017, 67, 41–48. [Google Scholar] [CrossRef]
- Cho, G.-S.; Huch, M.; Hanak, A.; Holzapfel, W.H.; Franz, C.M.A.P. Genetic analysis of the plantaricin EFI locus of Lactobacillus plantarum PCS20 reveals an unusual plantaricin E gene sequence as a result of mutation. Int. J. Food Microbiol. 2010, 141 (Suppl. 1), S117–S124. [Google Scholar] [CrossRef]
- Abushelaibi, A.; Ai-Mahadin, S.; Ei-Tarabily, K.; Shah, N.P.; Ayyash, M. Characterization of potential probiotic lactic acid bacteria isolated from camel milk. LWT—Food Sci. Technol. 2017, 79, 316–325. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Zavišić, G.; Popović, M.; Stojkov, S.; Medić, D.; Gusman, V.; Lješković, N.J.; Galović, A.J. Antibiotic resistance and probiotics: Knowledge gaps, market overview and preliminary screening. Antibiotics 2023, 12, 1281. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ). Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 4: Suitability of taxonomic units notified to EFSA until March 2016. EFSA J. 2016, 14, e04522. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).