Aptamer Sequence Optimization and Its Application in Food Safety Analysis
Abstract
1. Introduction
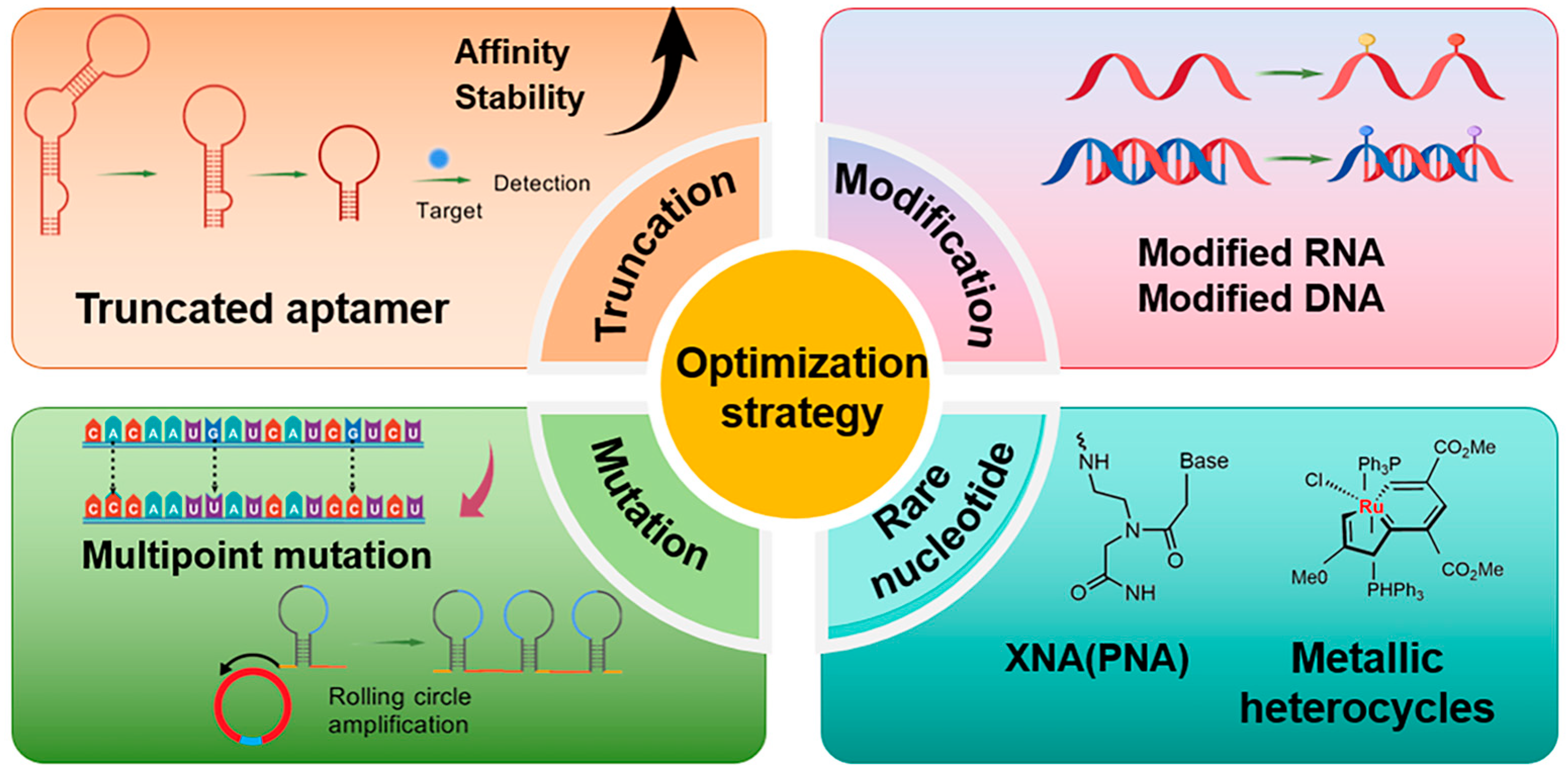
2. Aptamer Sequence Optimization Strategies
2.1. The Strategy of Truncation
2.2. The Strategy of Mutation
2.3. Non-Base Chemical Modification Strategy
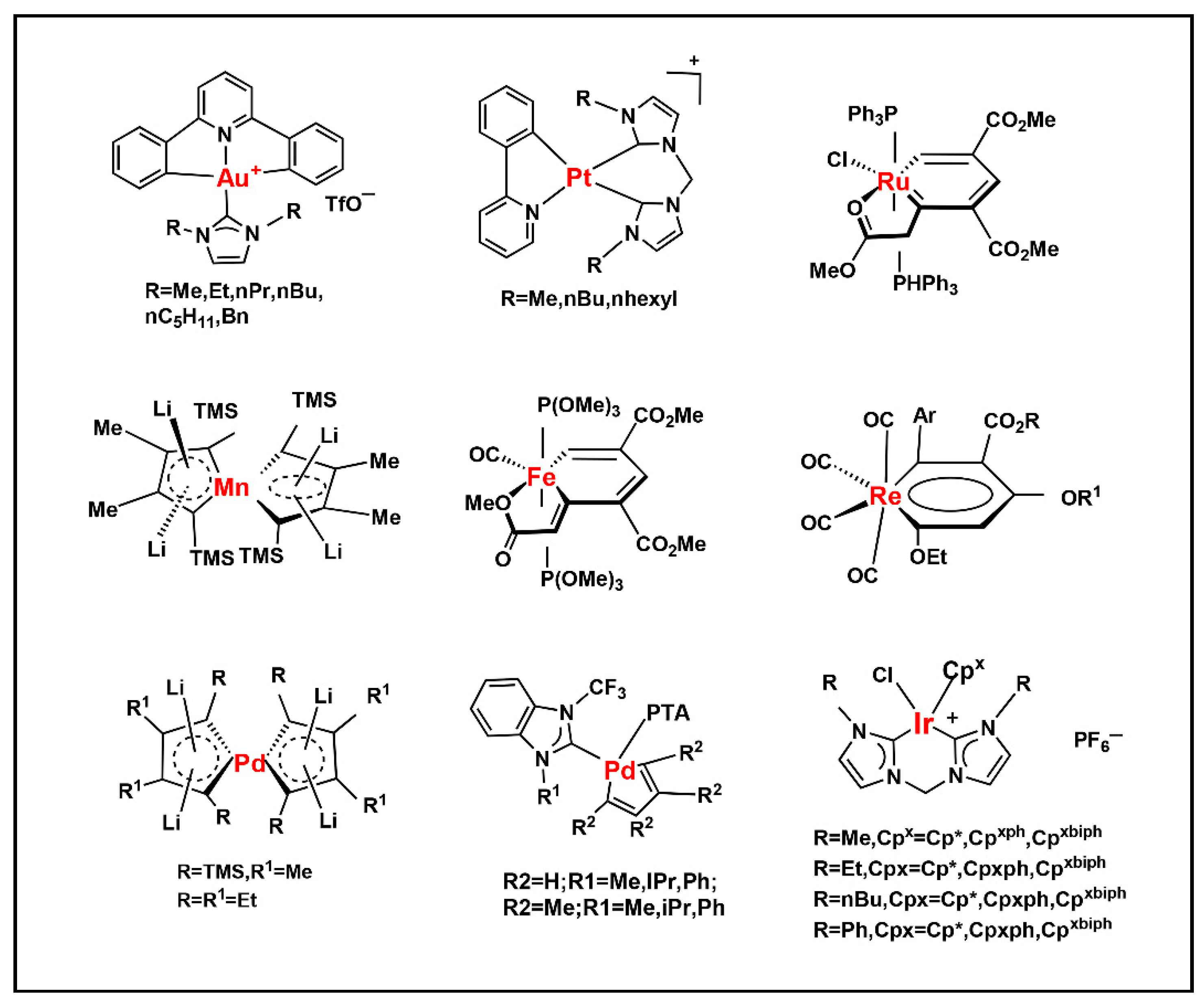
2.4. Introduction of Rare Nucleotides
2.5. Computer-Assisted Design of Aptamer
3. Evaluation Criteria for Aptamer Optimization
3.1. Experiment Methods
3.1.1. Circular Dichroism
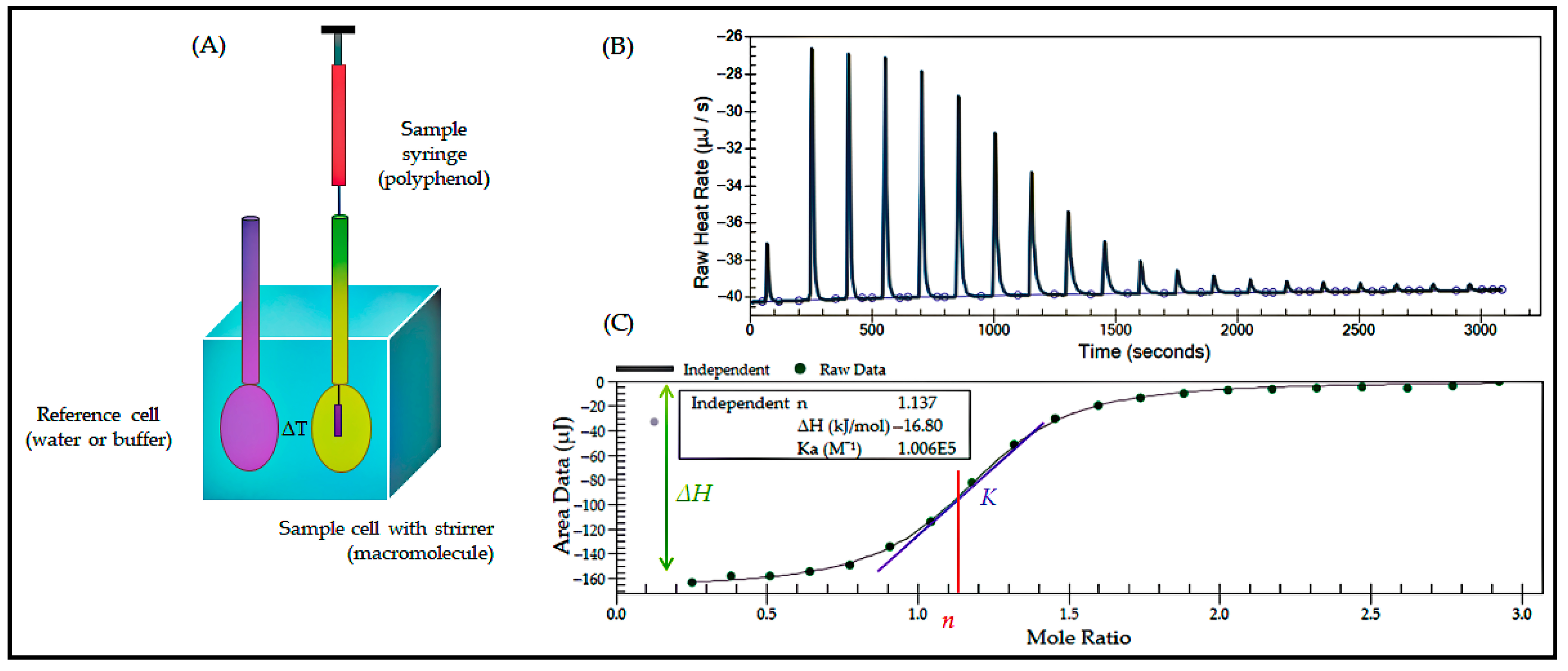
3.1.2. Isothermal Titration Calorimetry
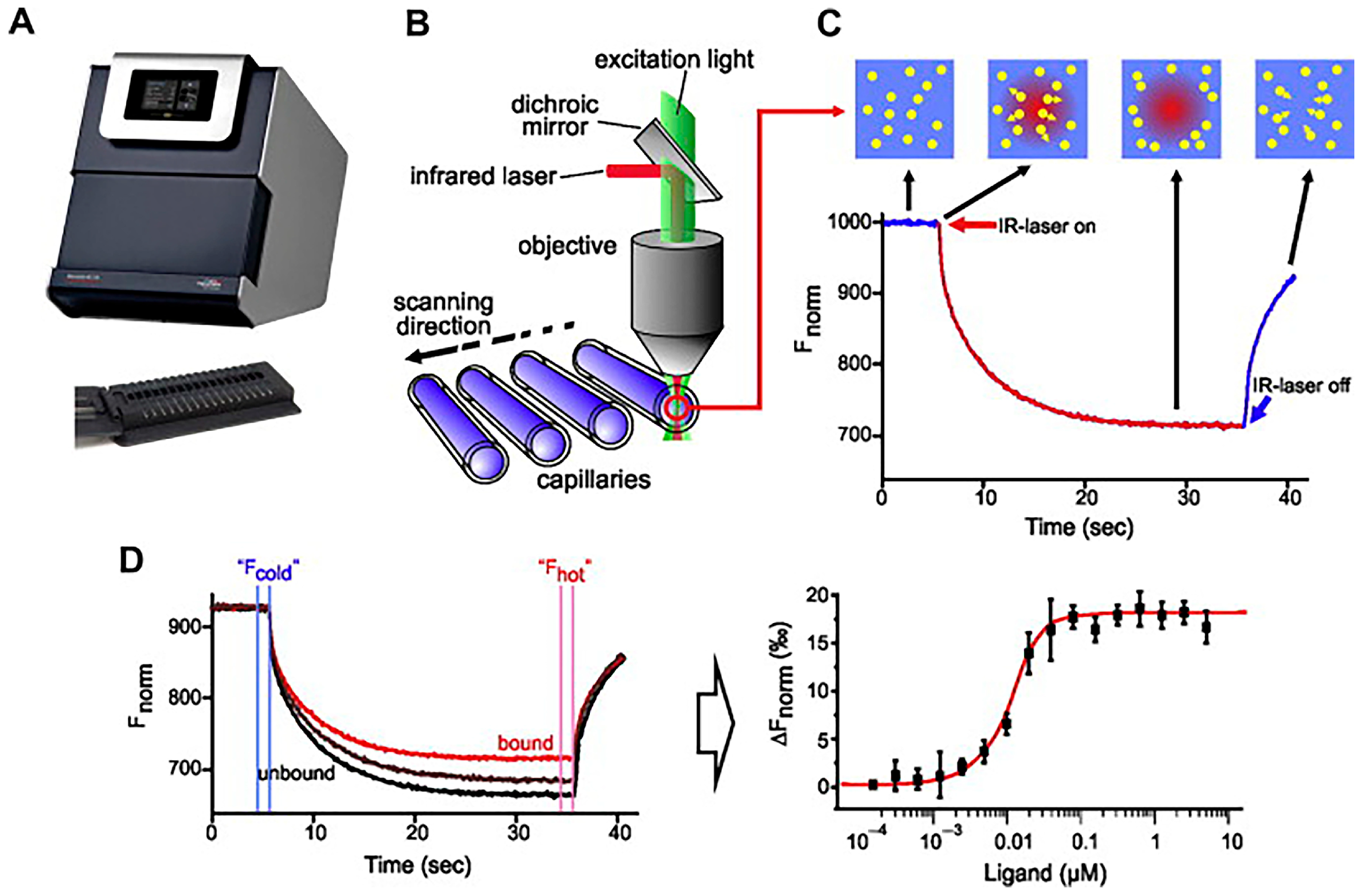
3.1.3. Microscale Thermophoresis
3.1.4. Fluorescence Analysis
3.2. Computer Simulation Methods
3.2.1. Molecular Docking
3.2.2. Molecular Dynamics
4. Examples of the Application of Aptamer Sequence Optimization in Food Analysis
4.1. Pesticide Detection
4.2. Heavy Metal Ions
4.3. Fungal Toxins
4.4. Pathogenic Bacteria
4.5. Others
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Duan, N.; Gong, W.; Wu, S.; Wang, Z. Selection and Application of ssDNA Aptamers against Clenbuterol Hydrochloride Based on ssDNA Library Immobilized SELEX. J. Agric. Food Chem. 2017, 65, 1771–1777. [Google Scholar] [CrossRef]
- Seok Kim, Y.; Ahmad Raston, N.H.; Bock Gu, M. Aptamer-Based Nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [CrossRef]
- Guo, Z.; Gao, L.; Jiang, S.; El-Seedi, H.R.; El-Garawani, I.M.; Zou, X. Sensitive Determination of Patulin by Aptamer Functionalized Magnetic Surface Enhanced Raman Spectroscopy (SERS) Sensor. J. Food Compos. Anal. 2023, 115, 104985. [Google Scholar] [CrossRef]
- Wu, S.; Liu, L.; Duan, N.; Li, Q.; Zhou, Y.; Wang, Z. Aptamer-Based Lateral Flow Test Strip for Rapid Detection of Zearalenone in Corn Samples. J. Agric. Food Chem. 2018, 66, 1949–1954. [Google Scholar] [CrossRef]
- Luo, L.; Liu, X.; Ma, S.; Li, L.; You, T. Quantification of Zearalenone in Mildewing Cereal Crops Using an Innovative Photoelectrochemical Aptamer Sensing Strategy Based on ZnO-NGQDs Composites. Food Chem. 2020, 322, 126778. [Google Scholar] [CrossRef]
- Wu, X.; Yin, L.; Gao, S.; Zhou, R.; Zhang, Y.; Xue, S.; Jayan, H.; El-Seedi, H.R.; Zou, X.; Guo, Z. Core-Satellite Nanoassembly System with Aptamer-Conjugated Au@Ag Nanoparticles for SERS Detection of Patulin in Apples. Food Control 2024, 159, 110293. [Google Scholar] [CrossRef]
- Peinetti, A.S.; Lake, R.J.; Cong, W.; Cooper, L.; Wu, Y.; Ma, Y.; Pawel, G.T.; Toimil-Molares, M.E.; Trautmann, C.; Rong, L.; et al. Direct Detection of Human Adenovirus or SARS-CoV-2 with Ability to Inform Infectivity Using DNA Aptamer-Nanopore Sensors. Sci. Adv. 2021, 7, eabh2848. [Google Scholar] [CrossRef]
- Li, H.; Ahmad, W.; Rong, Y.; Chen, Q.; Zuo, M.; Ouyang, Q.; Guo, Z. Designing an Aptamer Based Magnetic and Upconversion Nanoparticles Conjugated Fluorescence Sensor for Screening Escherichia Coli in Food. Food Control 2020, 107, 106761. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Y.; Ali, S.; Haruna, S.A.; He, P.; Li, H.; Ouyang, Q.; Chen, Q. Development of a Fluorescence Aptasensor for Rapid and Sensitive Detection of Listeria Monocytogenes in Food. Food Control 2021, 122, 107808. [Google Scholar] [CrossRef]
- Li, H.; Bei, Q.; Zhang, W.; Marimuthu, M.; Hassan, M.M.; Haruna, S.A.; Chen, Q. Ultrasensitive Fluorescence Sensor for Hg2+ in Food Based on Three-Dimensional Upconversion Nanoclusters and Aptamer-Modulated Thymine-Hg2+-Thymine Strategy. Food Chem. 2023, 422, 136202. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, P.; Yin, L.; Zuo, M.; Chen, Q.; El-Seedi, H.R.; Zou, X. Determination of Lead in Food by Surface-Enhanced Raman Spectroscopy with Aptamer Regulating Gold Nanoparticles Reduction. Food Control 2022, 132, 108498. [Google Scholar] [CrossRef]
- Song, S.-H.; Gao, Z.-F.; Guo, X.; Chen, G.-H. Aptamer-Based Detection Methodology Studies in Food Safety. Food Anal. Methods 2019, 12, 966–990. [Google Scholar] [CrossRef]
- Liu, S.; Meng, S.; Wang, M.; Li, W.; Dong, N.; Liu, D.; Li, Y.; You, T. In-Depth Interpretation of Aptamer-Based Sensing on Electrode: Dual-Mode Electrochemical-Photoelectrochemical Sensor for the Ratiometric Detection of Patulin. Food Chem. 2023, 410, 135450. [Google Scholar] [CrossRef]
- Liu, R.; Ali, S.; Haruna, S.A.; Ouyang, Q.; Li, H.; Chen, Q. Development of a Fluorescence Sensing Platform for Specific and Sensitive Detection of Pathogenic Bacteria in Food Samples. Food Control 2022, 131, 108419. [Google Scholar] [CrossRef]
- Sharma, A.S.; Ali, S.; Sabarinathan, D.; Murugavelu, M.; Li, H.; Chen, Q. Recent Progress on Graphene Quantum Dots-based Fluorescence Sensors for Food Safety and Quality Assessment Applications. Comp. Rev. Food Sci. Food Safe 2021, 20, 5765–5801. [Google Scholar] [CrossRef]
- Bi, X.; Li, L.; Luo, L.; Liu, X.; Li, J.; You, T. A Ratiometric Fluorescence Aptasensor Based on Photoinduced Electron Transfer from CdTe QDs to WS2 NTs for the Sensitive Detection of Zearalenone in Cereal Crops. Food Chem. 2022, 385, 132657. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Huang, X.; Hu, X.; Huang, X.; Yin, L.; Huang, Q.; Wen, Y.; Li, B.; Shi, J.; et al. Switchable Aptamer-Fueled Colorimetric Sensing toward Agricultural Fipronil Exposure Sensitized with Affiliative Metal-Organic Framework. Food Chem. 2023, 407, 135115. [Google Scholar] [CrossRef]
- Li, N.; Ren, C.; Hu, Q.; Wang, B.; Yang, Z.; Xiao, L.; Guan, T. Multiplex Aptamer Cluster Detection Platform and Systems Toxicology Study for 17β-Estradiol, Bisphenol A, and Diethylstilbestrol. Food Chem. 2025, 463, 141395. [Google Scholar] [CrossRef]
- Li, L.; Ma, R.; Wang, W.; Zhang, L.; Li, J.; Eltzov, E.; Wang, S.; Mao, X. Group-Targeting Aptamers and Aptasensors for Simultaneous Identification of Multiple Targets in Foods. TrAC Trends Anal. Chem. 2023, 166, 117169. [Google Scholar] [CrossRef]
- Li, W.; Pei, Y.; Wang, J. Development and Analysis of a Novel AF11–2 Aptamer Capable of Enhancing the Fluorescence of Aflatoxin B1. Chin. Chem. Lett. 2022, 33, 4096–4100. [Google Scholar] [CrossRef]
- Gao, J.; Liu, N.; Zhang, X.; Yang, E.; Song, Y.; Zhang, J.; Han, Q. Utilizing the DNA Aptamer to Determine Lethal α-Amanitin in Mushroom Samples and Urine by Magnetic Bead-ELISA (MELISA). Molecules 2022, 27, 538. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Lei, H.; Liu, J. Selection of DNA Aptamers for Detecting Metronidazole and Ibuprofen: Two Common Additives in Soft Drinks. Analyst 2024, 149, 5482–5490. [Google Scholar] [CrossRef]
- Qin, Y.; Shen, J.; Qin, Y.; Hayilati, B.; Yao, J.; Zhang, M. Research Progress on the Application of Aptamer Optimization Technology and Portable Sensing Devices in Food Safety Detection. Crit. Rev. Food Sci. Nutr. 2024, 1–33. [Google Scholar] [CrossRef]
- Li, R.; Zhu, Q.; Sun, X.; Li, Z.; Liu, X. Electrochemical Biosensor Based on the Integration of Maple Leaf-like Gold Nanocrystal and Truncated Aptamer for Detection of α-Amanitin with High Sensitivity, Selectivity and Rapidity. Food Chem. 2024, 453, 139639. [Google Scholar] [CrossRef]
- Debiais, M.; Lelievre, A.; Smietana, M.; Müller, S. Splitting Aptamers and Nucleic Acid Enzymes for the Development of Advanced Biosensors. Nucleic Acids Res. 2020, 48, 3400–3422. [Google Scholar] [CrossRef]
- Huang, T.; Chen, X.; Chen, J.; Zhang, Y.; Wang, X.; Wu, Z.; Zhou, N. In Vitro Selection and Engineering Azithromycin-Specific Aptamers and Construction of a Ratiometric Fluorescent Aptasensor for Sensitive Detection of Azithromycin. Sens. Actuators B Chem. 2024, 411, 135789. [Google Scholar] [CrossRef]
- Hu, X.; Tang, L.; Zheng, M.; Liu, J.; Zhang, Z.; Li, Z.; Yang, Q.; Xiang, S.; Fang, L.; Ren, Q.; et al. Structure-Guided Designing Pre-Organization in Bivalent Aptamers. J. Am. Chem. Soc. 2022, 144, 4507–4514. [Google Scholar] [CrossRef]
- Ji, D.; Feng, H.; Liew, S.W.; Kwok, C.K. Modified Nucleic Acid Aptamers: Development, Characterization, and Biological Applications. Trends Biotechnol. 2023, 41, 1360–1384. [Google Scholar] [CrossRef]
- Alkhamis, O.; Canoura, J.; Ly, P.T.; Xiao, Y. Using Exonucleases for Aptamer Characterization, Engineering, and Sensing. Acc. Chem. Res. 2023, 56, 1731–1743. [Google Scholar] [CrossRef]
- Thevendran, R.; Citartan, M. Assays to Estimate the Binding Affinity of Aptamers. Talanta 2022, 238, 122971. [Google Scholar] [CrossRef]
- Manceau, M.; Farre, C.; Lagarde, F.; Mathey, R.; Buhot, A.; Vidic, J.; Léguillier, V.; Hou, Y.; Chaix, C. Investigation of the Affinity of Aptamers for Bacteria by Surface Plasmon Resonance Imaging Using Nanosomes. ACS Appl. Mater. Interfaces 2024, 16, 29645–29656. [Google Scholar] [CrossRef]
- Li, M.; Qiu, Y.; Liu, G.; Xiao, Y.; Tian, Y.; Fang, S. Plasmonic Colorimetry and G-Quadruplex Fluorescence-Based Aptasensor: A Dual-Mode, Protein-Free and Label-Free Detection for OTA. Food Chem. 2024, 448, 139115. [Google Scholar] [CrossRef]
- Écija-Arenas, Á.; Kirchner, E.-M.; Hirsch, T.; Fernández-Romero, J.M. Development of an Aptamer-Based SPR-Biosensor for the Determination of Kanamycin Residues in Foods. Anal. Chim. Acta 2021, 1169, 338631. [Google Scholar] [CrossRef]
- Radi, A.-E.; Abd-Ellatief, M.R. Electrochemical Aptasensors: Current Status and Future Perspectives. Diagnostics 2021, 11, 104. [Google Scholar] [CrossRef]
- Formen, J.S.S.K.; Howard, J.R.; Anslyn, E.V.; Wolf, C. Circular Dichroism Sensing: Strategies and Applications. Angew. Chem. Int. Ed. 2024, 63, e202400767. [Google Scholar] [CrossRef]
- Bastos, M.; Abian, O.; Johnson, C.M.; Ferreira-da-Silva, F.; Vega, S.; Jimenez-Alesanco, A.; Ortega-Alarcon, D.; Velazquez-Campoy, A. Isothermal Titration Calorimetry. Nat. Rev. Methods Primers 2023, 3, 17. [Google Scholar] [CrossRef]
- Stein, J.A.C.; Ianeselli, A.; Braun, D. Kinetic Microscale Thermophoresis for Simultaneous Measurement of Binding Affinity and Kinetics. Angew. Chem. Int. Ed. 2021, 60, 13988–13995. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, T.; Wang, S.; Yan, Y. Mesoporous Silica-Based Molecularly Imprinted Fluorescence Sensor for the Ultrafast and Sensitive Recognition of Oxytetracycline. J. Food Compos. Anal. 2022, 108, 104427. [Google Scholar] [CrossRef]
- Perrier, S.; Bouilloud, P.; De Oliveira Coelho, G.; Henry, M.; Peyrin, E. Small Molecule Aptamer Assays Based on Fluorescence Anisotropy Signal-Enhancer Oligonucleotides. Biosens. Bioelectron. 2016, 82, 155–161. [Google Scholar] [CrossRef]
- Wang, S.; Ma, R.; Li, L.; Wang, L.; Li, J.; Sun, J.; Mao, X.; Tan, W. Engineering Robust Aptamers with High Affinity by Key Fragment Evolution and Terminal Fixation. Anal. Chem. 2022, 94, 16282–16289. [Google Scholar] [CrossRef]
- Chen, P.; Hu, C.; Tao, X.; Zhou, Z.; Wang, L.; Yang, X.; Che, Z.; Chen, X.; Huang, Y. Recognition Mechanism and Sequence Optimization of Organophosphorus Pesticides Aptamers for Better Monitoring Contaminations in Food. Food Sci. Hum. Wellness 2023, 12, 1708–1715. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, F.; Shi, M.; Sang, Y.; Wang, X. In Vitro Selection and Optimization of High-Affinity Aptamer for Milk Allergen α-Lactalbumin and Its Application in Dual-Mode Detection. Front. Nutr. 2022, 9, 1005230. [Google Scholar] [CrossRef]
- Han, X.; Qin, M.; Lu, X.; Wang, Q.; Zhang, Y.; Wang, Z. A Highly Sensitive SERS Aptasensor Using a Novel Truncated Aptamer for the Detection of Deoxynivalenol. Sens. Actuators B Chem. 2025, 424, 136908. [Google Scholar] [CrossRef]
- Jiang, H.; Qi, S.; Khan, I.M.; Dong, X.; Wang, Z.; Yang, J. CRISPR-Cas12a-Mediated Colorimetric Aptasensor of Netilmicin Based on Enzymes-Assisted Signal Amplification and Nanozyme Employing a Rationally Truncated Aptamer. Sens. Actuators B Chem. 2024, 414, 135969. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, K.; Du, Z.; Fang, B.; Zhan, J.; Zhu, L.; Xu, W. Magnetic Aptamer Copper Nanoclusters Fluorescent Biosensor for the Visual Detection of Zearalenone Based on Docking-Aided Rational Tailoring. Food Chem. 2024, 448, 139127. [Google Scholar] [CrossRef]
- Nguyen, M.-D.; Osborne, M.T.; Prevot, G.T.; Churcher, Z.R.; Johnson, P.E.; Simine, L.; Dauphin-Ducharme, P. Truncations and in Silico Docking to Enhance the Analytical Response of Aptamer-Based Biosensors. Biosens. Bioelectron. 2024, 265, 116680. [Google Scholar] [CrossRef]
- Chen, J.; He, J.; Bing, T.; Feng, Y.; Lyu, Y.; Lei, M.; Tan, W. Identification of the Binding Site between Aptamer Sgc8c and PTK7. Anal. Chem. 2024, 96, 10601–10611. [Google Scholar] [CrossRef]
- Gao, L.; Zhang, Y.; Chen, L.; Zhou, Q.; Zhou, N.; Xia, X. Study of Dual Binding Specificity of Aptamer to Ochratoxin A and Norfloxacin and the Development of Fluorescent Aptasensor in Milk Detection. Talanta 2024, 273, 125935. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Mao, M.; Peng, C.; Wang, Z. Accelerated CRISPR/Cas12a-Based Small Molecule Detection Using Bivalent Aptamer. Biosens. Bioelectron. 2022, 217, 114725. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Wei, D.; Zhou, H.; Zhu, J.; Yu, Q.; Luo, L.; Dai, X.; Jiang, Y.; Yu, L.; et al. Polyvalent Aptamer Nanodrug Conjugates Enable Efficient Tumor Cuproptosis Therapy Through Copper Overload and Glutathione Depletion. J. Am. Chem. Soc. 2024, 146, 30033–30045. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Erratum: Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Sun, Y.; Duan, N.; Ma, P.; Liang, Y.; Zhu, X.; Wang, Z. Colorimetric Aptasensor Based on Truncated Aptamer and Trivalent DNAzyme for Vibrio Parahemolyticus Determination. J. Agric. Food Chem. 2019, 67, 2313–2320. [Google Scholar] [CrossRef]
- Chen, H.; Li, Y.; Xiao, Z.; Li, J.; Li, T.; Wang, Z.; Liu, Y.; Tan, W. Chemical Amplification-Enabled Topological Modification of Nucleic Acid Aptamers for Enhanced Cancer-Targeted Theranostics. ACS Nano 2023, 17, 17740–17750. [Google Scholar] [CrossRef]
- Troisi, R.; Riccardi, C.; Pérez De Carvasal, K.; Smietana, M.; Morvan, F.; Del Vecchio, P.; Montesarchio, D.; Sica, F. A Terminal Functionalization Strategy Reveals Unusual Binding Abilities of Anti-Thrombin Anticoagulant Aptamers. Mol. Ther. Nucleic Acids 2022, 30, 585–594. [Google Scholar] [CrossRef]
- Das, G.; Harikrishna, S.; Gore, K.R. Influence of Sugar Modifications on the Nucleoside Conformation and Oligonucleotide Stability: A Critical Review. Chem. Rec. 2022, 22, e202200174. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. A Sensitive Aptamer Fluorescence Anisotropy Sensor for Cd2+ Using Affinity-Enhanced Aptamers with Phosphorothioate Modification. Biosensors 2022, 12, 887. [Google Scholar] [CrossRef]
- Mana, T.; Bhattacharya, B.; Lahiri, H.; Mukhopadhyay, R. XNAs: A Troubleshooter for Nucleic Acid Sensing. ACS Omega 2022, 7, 15296–15307. [Google Scholar] [CrossRef]
- Gutfreund, C.; Betz, K.; Abramov, M.; Coosemans, F.; Holliger, P.; Herdewijn, P.; Marx, A. Structural Insights into a DNA Polymerase Reading the Xeno Nucleic Acid HNA. Nucleic Acids Res. 2025, 53, gkae1156. [Google Scholar] [CrossRef]
- Alves Ferreira-Bravo, I.; DeStefano, J.J. Xeno-Nucleic Acid (XNA) 2’-Fluoro-Arabino Nucleic Acid (FANA) Aptamers to the Receptor-Binding Domain of SARS-CoV-2 S Protein Block ACE2 Binding. Viruses 2021, 13, 1983. [Google Scholar] [CrossRef]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and Challenges in Biosensor-Based Diagnosis of Infectious Diseases. Expert. Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef]
- Wang, X.; Yu, D.; Chen, L. Antimicrobial Resistance and Mechanisms of Epigenetic Regulation. Front. Cell. Infect. Microbiol. 2023, 13, 1199646. [Google Scholar] [CrossRef]
- Rangel, A.E.; Chen, Z.; Ayele, T.M.; Heemstra, J.M. In Vitro Selection of an XNA Aptamer Capable of Small-Molecule Recognition. Nucleic Acids Res. 2018, 46, 8057–8068. [Google Scholar] [CrossRef]
- Palme, D.I.; Lang, J.; Helke, D.; Kuryshko, M.; Abdelwhab, E.M. Strain-Dependent Variations in Replication of European Clade 2.3.4.4b Influenza A(H5N1) Viruses in Bovine Cells and Thermal Inactivation in Semi-Skimmed or Whole Milk. Eurosurveillance 2024, 29, 2400436. [Google Scholar] [CrossRef]
- Raymond, P.; Paul, S.; Perron, A.; Deschênes, L.; Hara, K. Extraction of Human Noroviruses from Leafy Greens and Fresh Herbs Using Magnetic Silica Beads. Food Microbiol. 2021, 99, 103827. [Google Scholar] [CrossRef]
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-Wide Variation in Potyviruses. Front. Plant Sci. 2019, 10, 1439. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, H.; Gubu, A.; Yu, S.; Zhang, H.; Dai, H.; Zhang, Y.; Zhang, B.; Ma, Y.; Lu, A.; et al. Chemically Modified Aptamers for Improving Binding Affinity to the Target Proteins via Enhanced Non-Covalent Bonding. Front. Cell Dev. Biol. 2023, 11, 1091809. [Google Scholar] [CrossRef]
- Hopfinger, M.C.; Kirkpatrick, C.C.; Znosko, B.M. Predictions and Analyses of RNA Nearest Neighbor Parameters for Modified Nucleotides. Nucleic Acids Res. 2020, 48, 8901–8913. [Google Scholar] [CrossRef]
- Shi, Y.; Lei, Y.; Chen, M.; Ma, H.; Shen, T.; Zhang, Y.; Huang, X.; Ling, W.; Liu, S.-Y.; Pan, Y.; et al. A Demethylation-Switchable Aptamer Design Enables Lag-Free Monitoring of m6 A Demethylase FTO with Energy Self-Sufficient and Structurally Integrated Features. J. Am. Chem. Soc. 2024, 146, 34638–34650. [Google Scholar] [CrossRef]
- Murray, M.T.; Wetmore, S.D. Unlocking Precision in Aptamer Engineering: A Case Study of the Thrombin Binding Aptamer Illustrates Why Modification Size, Quantity, and Position Matter. Nucleic Acids Res. 2024, 52, 10823–10835. [Google Scholar] [CrossRef]
- Saito, Y.; Hudson, R.H.E. Base-Modified Fluorescent Purine Nucleosides and Nucleotides for Use in Oligonucleotide Probes. J. Photochem. Photobiol. C Photochem. Rev. 2018, 36, 48–73. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, N.; Han, C.; Chen, Z.; Zhai, X.; Li, Z.; Zheng, K.; Zhu, J.; Wang, X.; Zou, X.; et al. Competitive Immunosensor for Sensitive and Optical Anti-Interference Detection of Imidacloprid by Surface-Enhanced Raman Scattering. Food Chem. 2021, 358, 129898. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Chen, T.; Meng, S.; Liu, D.; Dong, D.; You, T. Electric Field-Induced Specific Preconcentration to Enhance DNA-Based Electrochemical Sensing of Hg2+ via the Synergy of Enrichment and Self-Cleaning. J. Agric. Food Chem. 2022, 70, 7412–7419. [Google Scholar] [CrossRef]
- Wu, D.; Gordon, C.K.L.; Shin, J.H.; Eisenstein, M.; Soh, H.T. Directed Evolution of Aptamer Discovery Technologies. Acc. Chem. Res. 2022, 55, 685–695. [Google Scholar] [CrossRef]
- Minagawa, H.; Onodera, K.; Fujita, H.; Sakamoto, T.; Akitomi, J.; Kaneko, N.; Shiratori, I.; Kuwahara, M.; Horii, K.; Waga, I. Selection, Characterization and Application of Artificial DNA Aptamer Containing Appended Bases with Sub-Nanomolar Affinity for a Salivary Biomarker. Sci. Rep. 2017, 7, 42716. [Google Scholar] [CrossRef]
- Kong, L.-Z.; Kim, S.-M.; Wang, C.; Lee, S.Y.; Oh, S.-C.; Lee, S.; Jo, S.; Kim, T.-D. Understanding Nucleic Acid Sensing and Its Therapeutic Applications. Exp. Mol. Med. 2023, 55, 2320–2331. [Google Scholar] [CrossRef]
- Kimoto, M.; Cox, R.S.; Hirao, I. Unnatural Base Pair Systems for Sensing and Diagnostic Applications. Expert. Rev. Mol. Diagn. 2011, 11, 321–331. [Google Scholar] [CrossRef]
- Chen, Y.; Kong, D.; Qiu, L.; Wu, Y.; Dai, C.; Luo, S.; Huang, Z.; Lin, Q.; Chen, H.; Xie, S.; et al. Artificial Nucleotide Aptamer-Based Field-Effect Transistor for Ultrasensitive Detection of Hepatoma Exosomes. Anal. Chem. 2022, 95, 1446–1453. [Google Scholar] [CrossRef]
- Leconte, A.M.; Hwang, G.T.; Matsuda, S.; Capek, P.; Hari, Y.; Romesberg, F.E. Discovery, Characterization, and Optimization of an Unnatural Base Pair for Expansion of the Genetic Alphabet. J. Am. Chem. Soc. 2008, 130, 2336–2343. [Google Scholar] [CrossRef]
- Kimoto, M.; Hirao, I. Genetic Alphabet Expansion Technology by Creating Unnatural Base Pairs. Chem. Soc. Rev. 2020, 49, 7602–7626. [Google Scholar] [CrossRef]
- Matsunaga, K.; Kimoto, M.; Lim, V.W.; Tan, H.P.; Wong, Y.Q.; Sun, W.; Vasoo, S.; Leo, Y.S.; Hirao, I. High-Affinity Five/Six-Letter DNA Aptamers with Superior Specificity Enabling the Detection of Dengue NS1 Protein Variants beyond the Serotype Identification. Nucleic Acids Res. 2021, 49, 11407–11424. [Google Scholar] [CrossRef]
- Futami, K.; Kimoto, M.; Lim, Y.W.S.; Hirao, I. Genetic Alphabet Expansion Provides Versatile Specificities and Activities of Unnatural-Base DNA Aptamers Targeting Cancer Cells. Mol. Ther. Nucleic Acids 2019, 14, 158–170. [Google Scholar] [CrossRef]
- Miao, S.; Liang, Y.; Rundell, S.; Bhunia, D.; Devari, S.; Munyaradzi, O.; Bong, D. Unnatural Bases for Recognition of Noncoding Nucleic Acid Interfaces. Biopolymers 2021, 112, e23399. [Google Scholar] [CrossRef]
- Boldyrev, A.I.; Wang, L.-S. All-Metal Aromaticity and Antiaromaticity. Chem. Rev. 2005, 105, 3716–3757. [Google Scholar] [CrossRef]
- Banti, C.N.; Tsiatouras, V.; Karanicolas, K.; Panagiotou, N.; Tasiopoulos, A.J.; Kourkoumelis, N.; Hadjikakou, S.K. Antiproliferative Activity and Apoptosis Induction, of Organo-Antimony(III)–Copper(I) Conjugates, against Human Breast Cancer Cells. Mol. Divers. 2020, 24, 1095–1106. [Google Scholar] [CrossRef]
- Harlepp, S.; Chardon, E.; Bouché, M.; Dahm, G.; Maaloum, M.; Bellemin-Laponnaz, S. N-Heterocyclic Carbene-Platinum Complexes Featuring an Anthracenyl Moiety: Anti-Cancer Activity and DNA Interaction. Int. J. Mol. Sci. 2019, 20, 4198. [Google Scholar] [CrossRef]
- Zhao, Q.; Han, B.; Peng, C.; Zhang, N.; Huang, W.; He, G.; Li, J. A Promising Future of Metal-N-heterocyclic Carbene Complexes in Medicinal Chemistry: The Emerging Bioorganometallic Antitumor Agents. Med. Res. Rev. 2024, 44, 2194–2235. [Google Scholar] [CrossRef]
- Chen, D.; Hua, Y.; Xia, H. Metallaaromatic Chemistry: History and Development. Chem. Rev. 2020, 120, 12994–13086. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Zhang, W.; Li, Y.; Feng, Y.; Lv, S.; Diao, H.; Luo, Z.; Yan, P.; He, M.; et al. AptaDiff: De Novo Design and Optimization of Aptamers Based on Diffusion Models. Brief. Bioinform. 2023, 25, bbae517. [Google Scholar] [CrossRef]
- Dauparas, J.; Lee, G.R.; Pecoraro, R.; An, L.; Anishchenko, I.; Glasscock, C.; Baker, D. Atomic Context-Conditioned Protein Sequence Design Using LigandMPNN. Nat. Methods 2025, 22, 717–723. [Google Scholar] [CrossRef]
- Bashir, A.; Yang, Q.; Wang, J.; Hoyer, S.; Chou, W.; McLean, C.; Davis, G.; Gong, Q.; Armstrong, Z.; Jang, J.; et al. Machine Learning Guided Aptamer Refinement and Discovery. Nat. Commun. 2021, 12, 2366. [Google Scholar] [CrossRef]
- Mousivand, M.; Javan-Nikkhah, M.; Anfossi, L.; Di Nardo, F.; Salina, M.; Bagherzadeh, K. High Performance Aptasensing Platform Development through in Silico Aptamer Engineering for Aflatoxin B1 Monitoring. Food Control 2023, 145, 109418. [Google Scholar] [CrossRef]
- Mo, Y.; Wu, K.; Zheng, Y.; Ma, N.; Wang, Z.; Wu, S.; Duan, N. In Silico-Guided Engineering of Broad-Spectrum Aptamer for the Highly Sensitive Detection of Organophosphate Pesticides. Chem. Eng. J. 2025, 517, 164540. [Google Scholar] [CrossRef]
- Wong, F.; He, D.; Krishnan, A.; Hong, L.; Wang, A.Z.; Wang, J.; Hu, Z.; Omori, S.; Li, A.; Rao, J.; et al. Deep Generative Design of RNA Aptamers Using Structural Predictions. Nat. Comput. Sci. 2024, 4, 829–839. [Google Scholar] [CrossRef]
- Zacco, E.; Kantelberg, O.; Milanetti, E.; Armaos, A.; Panei, F.P.; Gregory, J.; Jeacock, K.; Clarke, D.J.; Chandran, S.; Ruocco, G.; et al. Probing TDP-43 Condensation Using an in Silico Designed Aptamer. Nat. Commun. 2022, 13, 3306. [Google Scholar] [CrossRef]
- Moussa, S.; Kilgour, M.; Jans, C.; Hernandez-Garcia, A.; Cuperlovic-Culf, M.; Bengio, Y.; Simine, L. Diversifying Design of Nucleic Acid Aptamers Using Unsupervised Machine Learning. J. Phys. Chem. B 2023, 127, 62–68. [Google Scholar] [CrossRef]
- Driscoll, J.; Gondaliya, P.; Ziemer, A.; Yan, I.K.; Gupta, Y.; Patel, T. In Silico Design of Novel EpCAM-Binding Aptamers for Targeted Delivery of RNA Therapeutics. Nanomaterials 2024, 14, 1727. [Google Scholar] [CrossRef]
- Yang, Y.; Qiao, L.; Jiang, Y.; Wang, Z.; Zhang, D.; Buratto, D.; Huang, L.; Zhou, R. In Silico Design and Validation of High-Affinity RNA Aptamers for SARS-CoV-2 Comparable to Neutralizing Antibodies. bioRxiv 2025. [Google Scholar] [CrossRef]
- Sun, D.; Sun, M.; Zhang, J.; Lin, X.; Zhang, Y.; Lin, F.; Zhang, P.; Yang, C.; Song, J. Computational Tools for Aptamer Identification and Optimization. TrAC Trends Anal. Chem. 2022, 157, 116767. [Google Scholar] [CrossRef]
- Hamada, M. In Silico Approaches to RNA Aptamer Design. Biochimie 2018, 145, 8–14. [Google Scholar] [CrossRef]
- Liu, Y.; Le, C.; Tyrrell, D.L.; Le, X.C.; Li, X.-F. Aptamer Binding Assay for the E Antigen of Hepatitis B Using Modified Aptamers with G-Quadruplex Structures. Anal. Chem. 2020, 92, 6495–6501. [Google Scholar] [CrossRef]
- Stangherlin, S.; Ding, Y.; Liu, J. Dissociation Constant ( Kd ) Measurement for Small-Molecule Binding Aptamers: Homogeneous Assay Methods and Critical Evaluations. Small Methods 2024, 9, 2401572. [Google Scholar] [CrossRef]
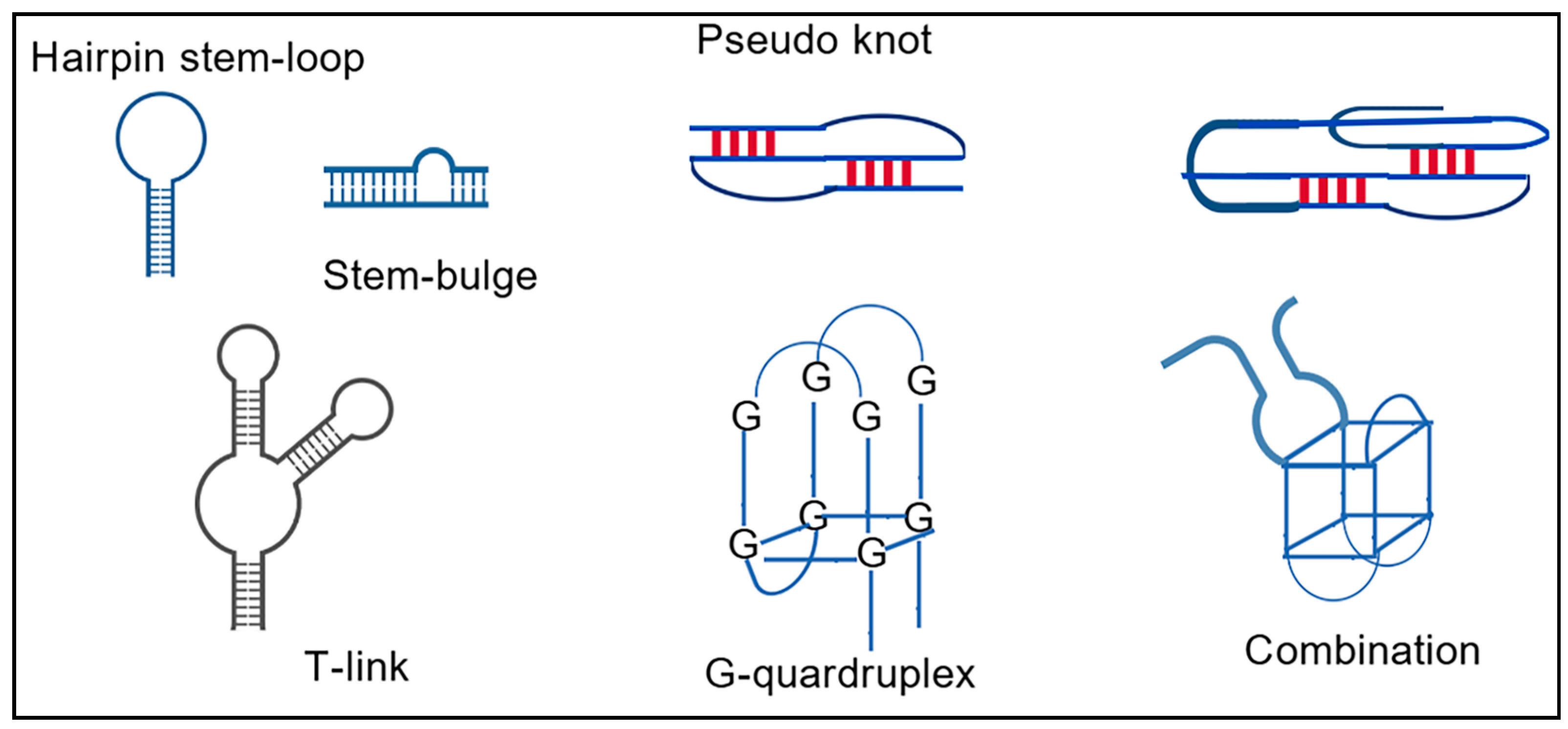
- Sullivan, R.; Adams, M.C.; Naik, R.R.; Milam, V.T. Analyzing Secondary Structure Patterns in DNA Aptamers Identified via CompELS. Molecules 2019, 24, 1572. [Google Scholar] [CrossRef]
- Gong, T.; Ju, F.; Bu, D. Accurate Prediction of RNA Secondary Structure Including Pseudoknots through Solving Minimum-Cost Flow with Learned Potentials. Commun. Biol. 2024, 7, 297. [Google Scholar] [CrossRef]
- Gao, H.; Ding, Y.; Ping, P.; Wang, D.; Ma, Y.; Li, H. Signal-on Electrogenerated Chemiluminescence Detection of Gonyautoxin 1/4 Based on Proximity Ligation-Induced an Electrode-Bound Pseudoknot DNA. Talanta 2024, 266, 124938. [Google Scholar] [CrossRef]
- de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Aptamers as Recognition Elements for Label-Free Analytical Devices. TrAC Trends Anal. Chem. 2008, 27, 437–446. [Google Scholar] [CrossRef]
- de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. TrAC Trends in Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Zhao, Q.; Yu, X.; Zhou, C.; Yagoub, A.E.A.; Ma, H. Effects of Collagen and Casein with Phenolic Compounds Interactions on Protein in Vitro Digestion and Antioxidation. LWT 2020, 124, 109192. [Google Scholar] [CrossRef]
- Zhao, D.; Dong, X.; Jiang, N.; Zhang, D.; Liu, C. Selective Recognition of Parallel and Anti-Parallel Thrombin-Binding Aptamer G-Quadruplexes by Different Fluorescent Dyes. Nucleic Acids Res. 2014, 42, 11612–11621. [Google Scholar] [CrossRef]
- Jing, M.; Bowser, M.T. Methods for Measuring Aptamer-Protein Equilibria: A Review. Anal. Chim. Acta 2011, 686, 9–18. [Google Scholar] [CrossRef]
- Troisi, R.; Napolitano, V.; Rossitto, E.; Osman, W.; Nagano, M.; Wakui, K.; Popowicz, G.M.; Yoshimoto, K.; Sica, F. Steric Hindrance and Structural Flexibility Shape the Functional Properties of a Guanine-Rich Oligonucleotide. Nucleic Acids Res. 2023, 51, 8880–8890. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, F.; Fang, L.; Chen, F.; Yang, W.; Wang, Z. Development of Folding-Based and Signal-Amplified Fluorescence Aptasensors Using G-Quadruplex Quinclorac Aptamer Variants Engineered via Exonuclease Digestion and Circular Dichroism Spectroscopy. Sens. Actuators B Chem. 2024, 415, 135996. [Google Scholar] [CrossRef]
- Wallace, B.A. Protein Characterisation by Synchrotron Radiation Circular Dichroism Spectroscopy. Quart. Rev. Biophys. 2009, 42, 317–370. [Google Scholar] [CrossRef]
- Nam, Y.; Cho, D.; Gu, B.; Rouxel, J.R.; Keefer, D.; Govind, N.; Mukamel, S. Time-Evolving Chirality Loss in Molecular Photodissociation Monitored by X-Ray Circular Dichroism Spectroscopy. J. Am. Chem. Soc. 2022, 144, 20400–20410. [Google Scholar] [CrossRef]
- Pilicer, S.L.; Dragna, J.M.; Garland, A.; Welch, C.J.; Anslyn, E.V.; Wolf, C. High-Throughput Determination of Enantiopurity by Microplate Circular Dichroism. J. Org. Chem. 2020, 85, 10858–10864. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, K.; Shi, P.; Ding, X.; Zhang, S. Rapid and Label-Free Detection of Aflatoxin B1 Using a Rationally Truncated Aptamer and via Circular Dichroism Measurement. Chem. Commun. 2022, 58, 4779–4782. [Google Scholar] [CrossRef]
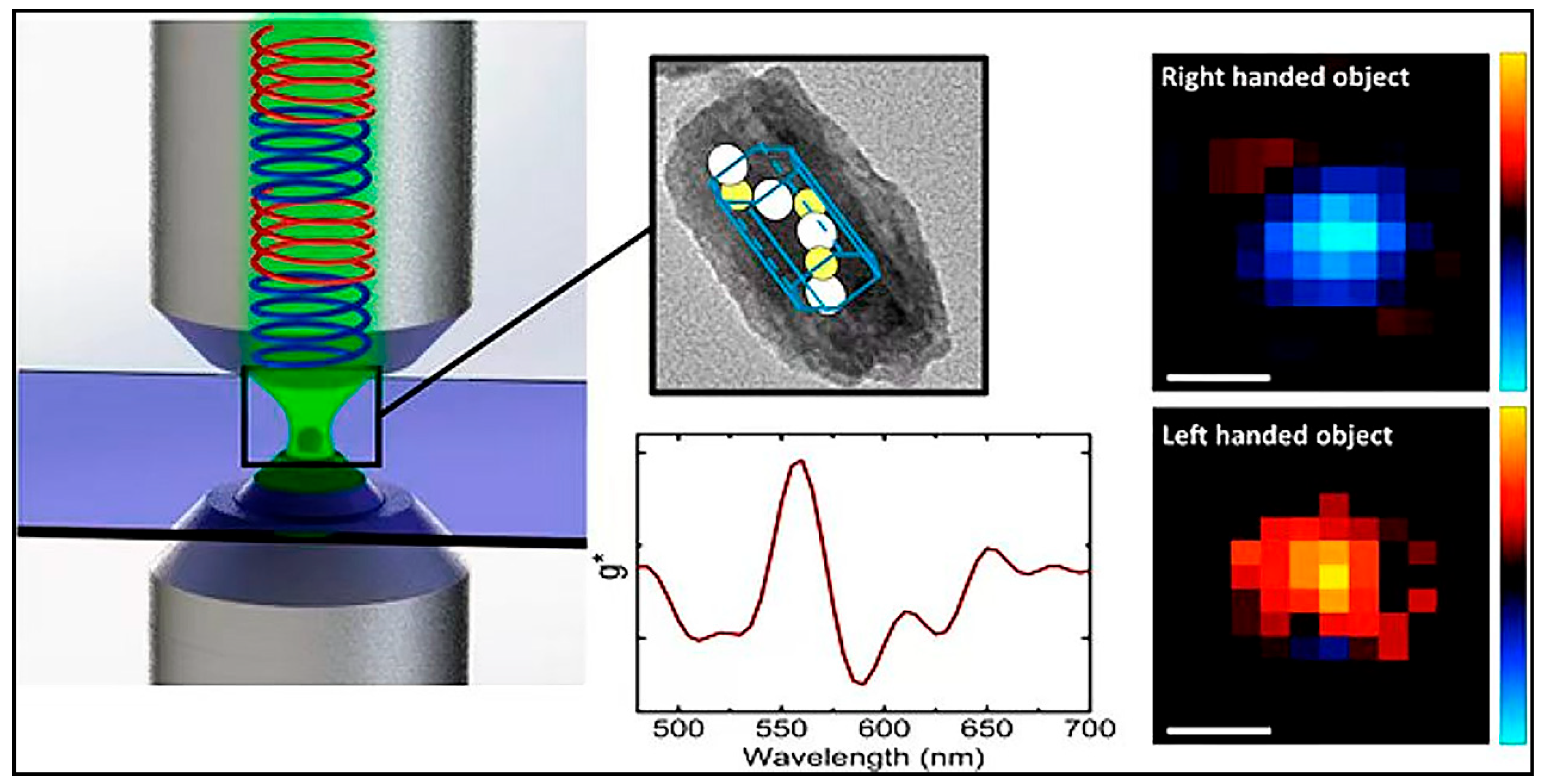
- Vinegrad, E.; Vestler, D.; Ben-Moshe, A.; Barnea, A.R.; Markovich, G.; Cheshnovsky, O. Circular Dichroism of Single Particles. ACS Photonics 2018, 5, 2151–2159. [Google Scholar] [CrossRef]
- Tai, T.; Sha, F.; Wang, X.; Wang, X.; Ma, K.; Kirlikovali, K.O.; Su, S.; Islamoglu, T.; Kato, S.; Farha, O.K. Leveraging Isothermal Titration Calorimetry to Explore Structure–Property Relationships of Protein Immobilization in Metal–Organic Frameworks. Angew. Chem. Int. Ed. 2022, 61, e202209110. [Google Scholar] [CrossRef]
- Li, W.; Sun, S.; Chen, W.; Ma, H.; Li, T.; Zhang, Z.; Wu, D.; Yan, M.; Yang, Y. Exploring the Taste Presentation and Receptor Perception Mechanism of Salty Peptides from Stropharia Rugosoannulata Based on Molecular Dynamics and Thermodynamics Simulation. Food Sci. Hum. Wellness 2024, 13, 2277–2288. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Sensitive Electrochemical Sensor for Cd2+ with Engineered Short High-Affinity Aptamer Undergoing Large Conformation Change. Talanta 2024, 271, 125642. [Google Scholar] [CrossRef]
- Zhang, S.; Ning, Z.; Zhang, Y.; Lin, X.; Duan, N.; Wang, Z.; Wu, S. Construction of a Microfluidic SELEX Platform for Efficient Screening of Advanced Glycation End Products Aptamer. Biosens. Bioelectron. 2025, 271, 117038. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Xiao, Y.; Jiang, H.; Zuo, X.; Li, X.; Fang, X. Structural Mechanisms for Binding and Activation of a Contact-Quenched Fluorophore by RhoBAST. Nat. Commun. 2024, 15, 4206. [Google Scholar] [CrossRef]
- Jia, W.; Guo, A.; Zhang, R.; Shi, L. Mechanism of Natural Antioxidants Regulating Advanced Glycosylation End Products of Maillard Reaction. Food Chem. 2023, 404, 134541. [Google Scholar] [CrossRef]
- Karonen, M. Polyphenol–Macromolecule Interactions by Isothermal Titration Calorimetry. Macromol. 2025, 5, 2. [Google Scholar] [CrossRef]
- Adam, A.A.; Michaux, F.; Dos Santos Morais, R.; Seiler, A.; Muniglia, L.; Khanji, A.N.; Jasniewski, J. Determination of the Critical Aggregation Concentration in Water of Gum Arabic Functionalized with Curcumin Oxidation Products by Micro-Scale Thermophoresis Approach. Int. J. Biol. Macromol. 2024, 271, 132510. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Z.; Shi, Q.; Wang, R.; Ji, M.; Chen, X.; Li, Y.; Liu, Y.; Yu, S. Thermal Proteome Profiling (TPP) Reveals NAMPT as the Anti-Glioma Target of Phenanthroindolizidine Alkaloid PF403. Acta Pharm. Sin. B 2025, 15, 2008–2023. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Sensitive Microscale Thermophoresis Assay for Rapid Ochratoxin A Detection with Fluorescently Labeled Engineered Aptamer. Analyst 2023, 148, 3876–3882. [Google Scholar] [CrossRef]
- Basu, S.; Gohain, N.; Kim, J.; Trinh, H.V.; Choe, M.; Joyce, M.G.; Rao, M. Determination of Binding Affinity of Antibodies to HIV-1 Recombinant Envelope Glycoproteins, Pseudoviruses, Infectious Molecular Clones, and Cell-Expressed Trimeric Gp160 Using Microscale Thermophoresis. Cells 2023, 13, 33. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction Analysis and Beyond. Journal of Molecular Structure 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Liu, J.; Ren, Z.; Sun, Y.; Xu, L.; Wei, D.; Tan, W.; Ding, D. Investigation of the Relationship between Aptamers’ Targeting Functions and Human Plasma Proteins. ACS Nano 2023, 17, 24329–24342. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Profiling Additional Effects of Aptamer Fluorophore Modification on Microscale Thermophoresis Characterization of Aptamer–Target Binding. Anal. Chem. 2023, 95, 17011–17019. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Microscale Thermophoresis Sensor for Cd2+ Using Aptamer with Tetramethylrhodamine Label at an Internal Site. Sensors and Actuators B: Chemical 2024, 402, 135102. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Sensitive Microscale Thermophoresis Assay Using Aptamer Thermal Switch. Anal. Chem. 2022, 94, 16685–16691. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. DNAzyme-Based Microscale Thermophoresis Sensor. Anal. Chem. 2023, 95, 2152–2156. [Google Scholar] [CrossRef]
- Garoli, D.; Yamazaki, H.; Maccaferri, N.; Wanunu, M. Plasmonic Nanopores for Single-Molecule Detection and Manipulation: Toward Sequencing Applications. Nano Lett. 2019, 19, 7553–7562. [Google Scholar] [CrossRef]
- Greiss, F.; Kriegel, F.; Braun, D. Probing the Cooperativity of Binding Networks with High-Throughput Thermophoresis. Anal. Chem. 2017, 89, 2592–2597. [Google Scholar] [CrossRef]
- Zhao, Q.; Tao, J.; Uppal, J.S.; Peng, H.; Wang, H.; Le, X.C. Nucleic Acid Aptamers Improving Fluorescence Anisotropy and Fluorescence Polarization Assays for Small Molecules. TrAC Trends Anal. Chem. 2019, 110, 401–409. [Google Scholar] [CrossRef]
- Zhao, Q.; Tao, J.; Feng, W.; Uppal, J.S.; Peng, H.; Le, X.C. Aptamer Binding Assays and Molecular Interaction Studies Using Fluorescence Anisotropy—A Review. Anal. Chim. Acta 2020, 1125, 267–278. [Google Scholar] [CrossRef]
- Wang, L.; Haruna, S.A.; Ahmad, W.; Wu, J.; Chen, Q.; Ouyang, Q. Tunable Multiplexed Fluorescence Biosensing Platform for Simultaneous and Selective Detection of Paraquat and Carbendazim Pesticides. Food Chem. 2022, 388, 132950. [Google Scholar] [CrossRef]
- Wang, Y.; Li, W.; Hu, X.; Zhang, X.; Huang, X.; Li, Z.; Li, M.; Zou, X.; Shi, J. Efficient Preparation of Dual-Emission Ratiometric Fluorescence Sensor System Based on Aptamer-Composite and Detection of Bis(2-Ethylhexyl) Phthalate in Pork. Food Chem. 2021, 352, 129352. [Google Scholar] [CrossRef]
- Selva Sharma, A.; Marimuthu, M.; Varghese, A.W.; Wu, J.; Xu, J.; Xiaofeng, L.; Devaraj, S.; Lan, Y.; Li, H.; Chen, Q. A Review of Biomolecules Conjugated Lanthanide Up-Conversion Nanoparticles-Based Fluorescence Probes in Food Safety and Quality Monitoring Applications. Crit. Rev. Food Sci. Nutr. 2024, 64, 6129–6159. [Google Scholar] [CrossRef]
- Megalathan, A.; Wijesinghe, K.M.; Dhakal, S. Single-Molecule FRET-Based Dynamic DNA Sensor. ACS Sens. 2021, 6, 1367–1374. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, X.; Shi, X.; Han, Y.; Guo, Z.; Liu, Y. Development of Carbon Quantum Dot–Labeled Antibody Fluorescence Immunoassays for the Detection of Morphine in Hot Pot Soup Base. Food Anal. Methods 2020, 13, 1042–1049. [Google Scholar] [CrossRef]
- Li, D.; Chen, Z.; Mei, X. Fluorescence Enhancement for Noble Metal Nanoclusters. Adv. Colloid. Interface Sci. 2017, 250, 25–39. [Google Scholar] [CrossRef]
- Wang, C.; Gu, C.; Zhao, X.; Yu, S.; Zhang, X.; Xu, F.; Ding, L.; Huang, X.; Qian, J. Self-Designed Portable Dual-Mode Fluorescence Device with Custom Python-Based Analysis Software for Rapid Detection via Dual-Color FRET Aptasensor with IoT Capabilities. Food Chem. 2024, 457, 140190. [Google Scholar] [CrossRef]
- Li, H.; Huang, X.; Mehedi Hassan, M.; Zuo, M.; Wu, X.; Chen, Y.; Chen, Q. Dual-Channel Biosensor for Hg2+ Sensing in Food Using Au@Ag/Graphene-Upconversion Nanohybrids as Metal-Enhanced Fluorescence and SERS Indicators. Microchem. J. 2020, 154, 104563. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Jäschke, A.; Sunbul, M. A Color-Shifting Near-Infrared Fluorescent Aptamer–Fluorophore Module for Live-Cell RNA Imaging. Angew. Chem. Int. Ed. 2021, 60, 21441–21448. [Google Scholar] [CrossRef]
- Gaillard, T. Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J. Chem. Inf. Model. 2018, 58, 1697–1706. [Google Scholar] [CrossRef]
- Vu, H.; Gilari, K.; Sathiyamoorthy, V.; Beckham, J. Discovery of Novel Inhibitors for Mycobacterium tuberculosis D-alanine: D-alanine Ligase through Virtual Screening. FASEB J. 2022, 36, fasebj.2022.36.S1.R4261. [Google Scholar] [CrossRef]
- Ropii, B.; Bethasari, M.; Anshori, I.; Koesoema, A.P.; Shalannanda, W.; Satriawan, A.; Setianingsih, C.; Akbar, M.R.; Aditama, R.; Fahmi, F.; et al. The Molecular Interaction of Six Single-Stranded DNA Aptamers to Cardiac Troponin I Revealed by Docking and Molecular Dynamics Simulation. PLoS ONE 2024, 19, e0302475. [Google Scholar] [CrossRef]
- Ye, H.; Wang, M.; Yu, X.; Ma, P.; Zhu, P.; Zhong, J.; He, K.; Guo, Y. Molecular Docking Insight into the Label-Free Fluorescence Aptasensor for Ochratoxin A Detection. Molecules 2023, 28, 4841. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, L.; Tu, Z.; Sun, X.; He, Q.; Lei, Z.; Xu, C.; Liu, Y.; Zhang, X.; Yang, J.; et al. Organophosphorus Pesticides Detection Using Broad-Specific Single-Stranded DNA Based Fluorescence Polarization Aptamer Assay. Biosens. Bioelectron. 2014, 55, 216–219. [Google Scholar] [CrossRef]
- Shmilovich, K.; Chen, B.; Karaletsos, T.; Sultan, M.M. DEL-Dock: Molecular Docking-Enabled Modeling of DNA-Encoded Libraries. J. Chem. Inf. Model. 2023, 63, 2719–2727. [Google Scholar] [CrossRef]
- Ivanova, A.; Mokshyna, O.; Polishchuk, P. StreaMD: The Toolkit for High-Throughput Molecular Dynamics Simulations. J. Cheminform 2024, 16, 123. [Google Scholar] [CrossRef]
- Fedulova, A.S.; Armeev, G.A.; Romanova, T.A.; Singh-Palchevskaia, L.; Kosarim, N.A.; Motorin, N.A.; Komarova, G.A.; Shaytan, A.K. Molecular Dynamics Simulations of Nucleosomes Are Coming of Age. WIREs Comput. Mol. Sci. 2024, 14, e1728. [Google Scholar] [CrossRef]
- Lappala, A. The next Revolution in Computational Simulations: Harnessing AI and Quantum Computing in Molecular Dynamics. Curr. Opin. Struct. Biol. 2024, 89, 102919. [Google Scholar] [CrossRef]
- Wang, P.; Li, H.; Hassan, M.M.; Guo, Z.; Zhang, Z.-Z.; Chen, Q. Fabricating an Acetylcholinesterase Modulated UCNPs-Cu2+ Fluorescence Biosensor for Ultrasensitive Detection of Organophosphorus Pesticides-Diazinon in Food. J. Agric. Food Chem. 2019, 67, 4071–4079. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, Y.; Wang, C.; Liu, B.; Wu, Y. Selection and Identification of a DNA Aptamer for Ultrasensitive and Selective Detection of λ-Cyhalothrin Residue in Food. Anal. Chim. Acta 2021, 1179, 338837. [Google Scholar] [CrossRef]
- Li, W.; Hu, X.; Li, Q.; Shi, Y.; Zhai, X.; Xu, Y.; Li, Z.; Huang, X.; Wang, X.; Shi, J.; et al. Copper Nanoclusters @ Nitrogen-Doped Carbon Quantum Dots-Based Ratiometric Fluorescence Probe for Lead (II) Ions Detection in Porphyra. Food Chem. 2020, 320, 126623. [Google Scholar] [CrossRef]
- Khoshbin, Z.; Housaindokht, M.R.; Izadyar, M.; Bozorgmehr, M.R.; Verdian, A. Theoretical Design and Experimental Study of New Aptamers with the Improved Target-Affinity: New Insights into the Pb2+-Specific Aptamers as a Case Study. J. Mol. Liq. 2019, 289, 111159. [Google Scholar] [CrossRef]
- Qian, S.Q.; Yuan, M.; Zuo, X.W.; Cao, H.; Yu, J.S.; Hao, L.-L.; Yang, K.L.; Xu, F. A Novel Strategy for Enhancing the Stability of Aptamer Conformations in Heavy Metal Ion Detection. Anal. Chim. Acta 2024, 1306, 342577. [Google Scholar] [CrossRef]
- Komijani, M.; Shamabadi, N.S.; Shahin, K.; Eghbalpour, F.; Tahsili, M.R.; Bahram, M. Heavy Metal Pollution Promotes Antibiotic Resistance Potential in the Aquatic Environment. Environ. Pollut. 2021, 274, 116569. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, X.; Ren, X.; Li, W.; Sun, J.; Wang, X.; Huang, Y.; Guo, Y.; Zeng, H. Novel Fluorescence Immunoassay for the Detection of Zearalenone Using HRP-Mediated Fluorescence Quenching of Gold-Silver Bimetallic Nanoclusters. Food Chem. 2021, 355, 129633. [Google Scholar] [CrossRef]
- Guan, Y.; Ma, J.; Neng, J.; Yang, B.; Wang, Y.; Xing, F. A Novel and Label-Free Chemiluminescence Detection of Zearalenone Based on a Truncated Aptamer Conjugated with a G-Quadruplex DNAzyme. Biosensors 2023, 13, 118. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, L.; Li, T.; Li, C.; Zhang, W.; Sun, X.; Yue, X.; Xu, W. In Silico Simulation-Guided Engineering of Multifunctional Bivalent Light up Aptamer for Sensitive and on-Site Detection of AFB1 Using Label-Free Ratiometric Fluorescent Aptasensor. Sens. Actuators B Chem. 2024, 420, 136425. [Google Scholar] [CrossRef]
- Gao, Y.-N.; Wu, C.-Q.; Wang, J.-Q.; Zheng, N. Metabolomic Analysis Reveals the Mechanisms of Hepatotoxicity Induced by Aflatoxin M1 and Ochratoxin A. Toxins 2022, 14, 141. [Google Scholar] [CrossRef]
- Jin, J.; Kouznetsova, V.L.; Kesari, S.; Tsigelny, I.F. Synergism in Actions of HBV with Aflatoxin in Cancer Development. Toxicology 2023, 499, 153652. [Google Scholar] [CrossRef]
- Pourakbari, R.; Shadjou, N.; Yousefi, H.; Isildak, I.; Yousefi, M.; Rashidi, M.-R.; Khalilzadeh, B. Recent Progress in Nanomaterial-Based Electrochemical Biosensors for Pathogenic Bacteria. Microchim. Acta 2019, 186, 820. [Google Scholar] [CrossRef]
- Yang, N.; Ding, N.; Qi, S.; Shang, Z.; Ma, P.; Khan, I.M.; Wang, Z.; Xia, Y.; Zhang, Y.; Zhang, L. High-Affinity Truncated Aptamers for Detection of Cronobacter Spp with Magnetic Separation-Assisted DNAzyme-Driven 3D DNA Walker. Microchim. Acta 2024, 191, 130. [Google Scholar] [CrossRef]
- Sun, M.; Ma, N.; Shi, H.; Cheong, L.-Z.; Yang, W.; Qiao, Z. A HCR Based Multivalent Aptamer Amplifier for Ultrasensitive Detection of Salmonella. Sens. Actuators B Chem. 2023, 375, 132860. [Google Scholar] [CrossRef]
- Tang, Y.; Li, Y.; Chen, P.; Zhong, S.; Yang, Y. Nucleic Acid Aptamer-Based Sensors for Bacteria Detection: A Review. BioEssays 2025, 47, e202400111. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, L.; Qin, Z.; Karges, J.; Xiao, H.; Su, X. Unraveling and Overcoming Platinum Drug-Resistant Cancer Tumors with DNA Nanostructures. Adv. Funct. Mater. 2023, 33, 2208797. [Google Scholar] [CrossRef]
- Amero, P.; Lokesh, G.L.R.; Chaudhari, R.R.; Cardenas-Zuniga, R.; Schubert, T.; Attia, Y.M.; Montalvo-Gonzalez, E.; Elsayed, A.M.; Ivan, C.; Wang, Z.; et al. Conversion of RNA Aptamer into Modified DNA Aptamers Provides for Prolonged Stability and Enhanced Antitumor Activity. J. Am. Chem. Soc. 2021, 143, 7655–7670. [Google Scholar] [CrossRef]
- Chen, X.; Duan, M.; Chang, Y.; Ye, M.; Wang, Z.; Wu, S.; Duan, N. Assembly of a Multivalent Aptamer for Efficient Inhibition of Thermostable Direct Hemolysin Toxicity Induced by Vibrio Parahaemolyticus. J. Hazard. Mater. 2024, 478, 135452. [Google Scholar] [CrossRef]
- Han, E.; Pan, Y.; Li, L.; Cai, J. Bisphenol A Detection Based on Nano Gold-Doped Molecular Imprinting Electrochemical Sensor with Enhanced Sensitivity. Food Chem. 2023, 426, 136608. [Google Scholar] [CrossRef]
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High Affinity Truncated Aptamers for Ultra-Sensitive Colorimetric Detection of Bisphenol A with Label-Free Aptasensor. Food Chem. 2020, 317, 126459. [Google Scholar] [CrossRef]
- Wu, S.; Liu, S.; Wang, Z.; Chen, Y.; Zhao, G. Comprehensive Analysis of Bisphenol Analogues in Complex Water Using a Group-Targeting Aptamer Engineered by Base Mutation. J. Hazard. Mater. 2023, 460, 132427. [Google Scholar] [CrossRef]
- Ben Aissa, S.; Mastouri, M.; Catanante, G.; Raouafi, N.; Marty, J.L. Investigation of a Truncated Aptamer for Ofloxacin Detection Using a Rapid FRET-Based Apta-Assay. Antibiotics 2020, 9, 86. [Google Scholar] [CrossRef]
- Guo, H.; Sun, Y.; Ma, P.; Khan, I.M.; Duan, N.; Wang, Z. Sensitive Detection of Patulin Based on DNase Ⅰ-Assisted Fluorescent Aptasensor by Using AuNCs-Modified Truncated Aptamer. Food Control 2022, 131, 108430. [Google Scholar] [CrossRef]
- Sun, A.; Qi, S.; Li, Y.; Wu, Y.; Zhang, Y.; Wang, Z. Ultra-Stable MAPbBr3@ZIF-8-Based Fluorescent Aptasensor for Highly Sensitive and Specific Detection of Azlocillin in Food. Food Biosci. 2025, 64, 105920. [Google Scholar] [CrossRef]
- Ma, P.; Guo, H.; Ye, H.; Zhang, Y.; Wang, Z. Aptamer-Locker Probe Coupling with Truncated Aptamer for High-Efficiency Fluorescence Polarization Detection of Zearalenone. Sens. Actuators B Chem. 2023, 380, 133356. [Google Scholar] [CrossRef]
- Ma, P.; Guo, H.; Duan, N.; Ma, X.; Yue, L.; Gu, Q.; Wang, Z. Label Free Structure-Switching Fluorescence Polarization Detection of Chloramphenicol with Truncated Aptamer. Talanta 2021, 230, 122349. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, Z.; Tian, Y.; Mou, Y.; Guo, Y.; Sun, X.; Li, F. Ratiometric Fluorescent Sensor Based on a Truncated Specific Aptamer by MGO-SELEX Screening for Streptomycin Detection. Sens. Actuators B Chem. 2024, 406, 135427. [Google Scholar] [CrossRef]
- Chen, X.; Yang, L.; Tang, J.; Wen, X.; Zheng, X.; Chen, L.; Li, J.; Xie, Y.; Le, T. An AuNPs-Based Fluorescent Sensor with Truncated Aptamer for Detection of Sulfaquinoxaline in Water. Biosensors 2022, 12, 513. [Google Scholar] [CrossRef]
- He, A.; Wan, L.; Zhang, Y.; Yan, Z.; Guo, P.; Han, D.; Tan, W. Structure-Based Investigation of a DNA Aptamer Targeting PTK7 Reveals an Intricate 3D Fold Guiding Functional Optimization. Proc. Natl. Acad. Sci. USA 2024, 121, e2404060121. [Google Scholar] [CrossRef]
- Han, J.; Ma, P.; Khan, I.M.; Zhang, Y.; Wang, Z. Study of Binding Mechanism of Aptamer to Kanamycin and the Development of Fluorescent Aptasensor in Milk Detection. Talanta 2023, 260, 124530. [Google Scholar] [CrossRef]
- Rahman, M.S.; Han, M.J.; Kim, S.W.; Kang, S.M.; Kim, B.R.; Kim, H.; Lee, C.J.; Noh, J.E.; Kim, H.; Lee, J.-O.; et al. Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection. Molecules 2023, 28, 4645. [Google Scholar] [CrossRef]
- Gawande, B.N.; Rohloff, J.C.; Carter, J.D.; Von Carlowitz, I.; Zhang, C.; Schneider, D.J.; Janjic, N. Selection of DNA Aptamers with Two Modified Bases. Proc. Natl. Acad. Sci. USA 2017, 114, 2898–2903. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Rosenthal, M.; Siegl, J.; Ewers, J.; Mayer, G. Customised Nucleic Acid Libraries for Enhanced Aptamer Selection and Performance. Curr. Opin. Biotechnol. 2017, 48, 111–118. [Google Scholar] [CrossRef]
- Kähkölä, H.; Herath, M.; Virta, P.; Lönnberg, T. Post-SELEX Modification of Quinine Aptamers through Neoacetalization. Org. Biomol. Chem. 2025, 23, 1714–1722. [Google Scholar] [CrossRef]
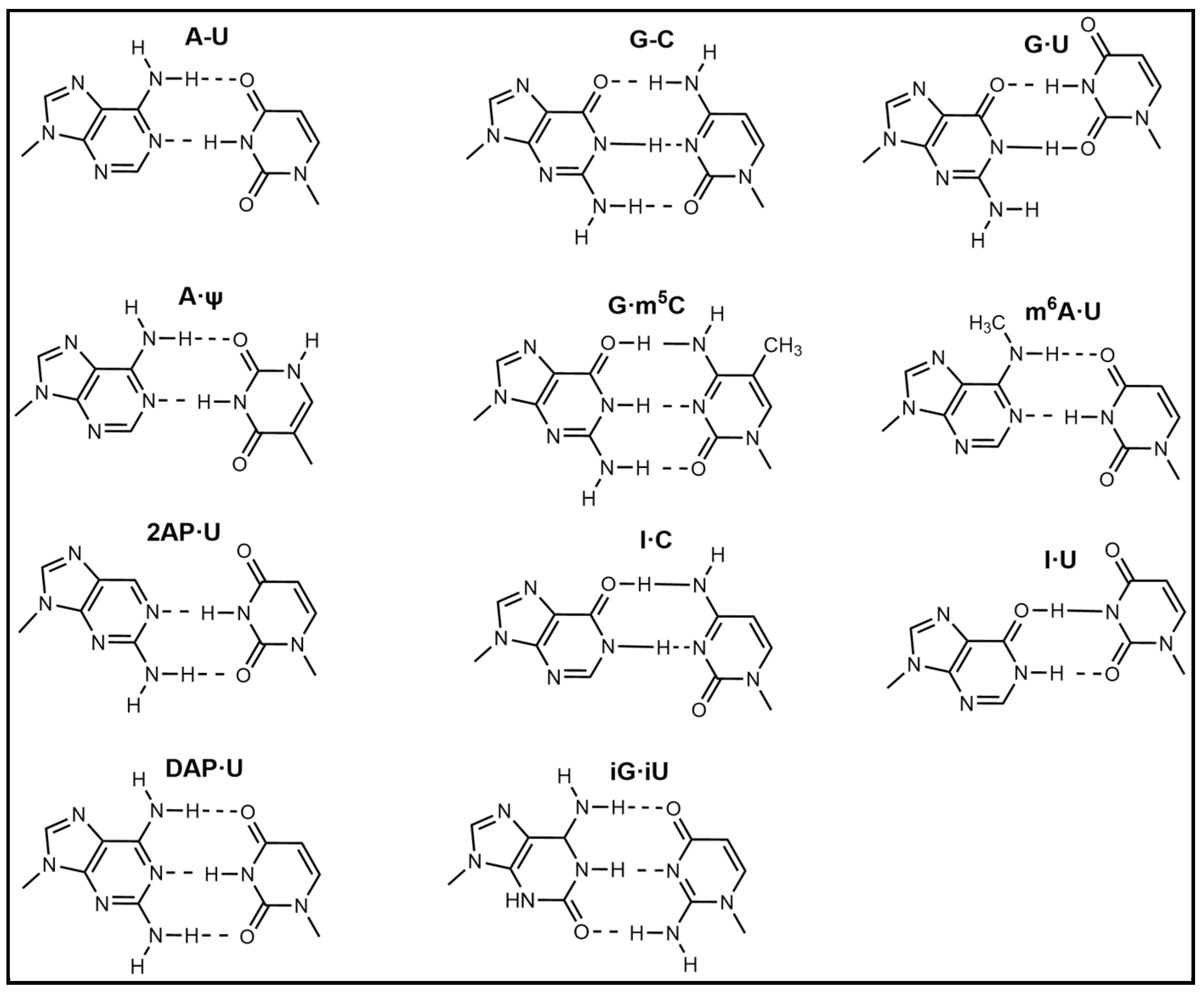
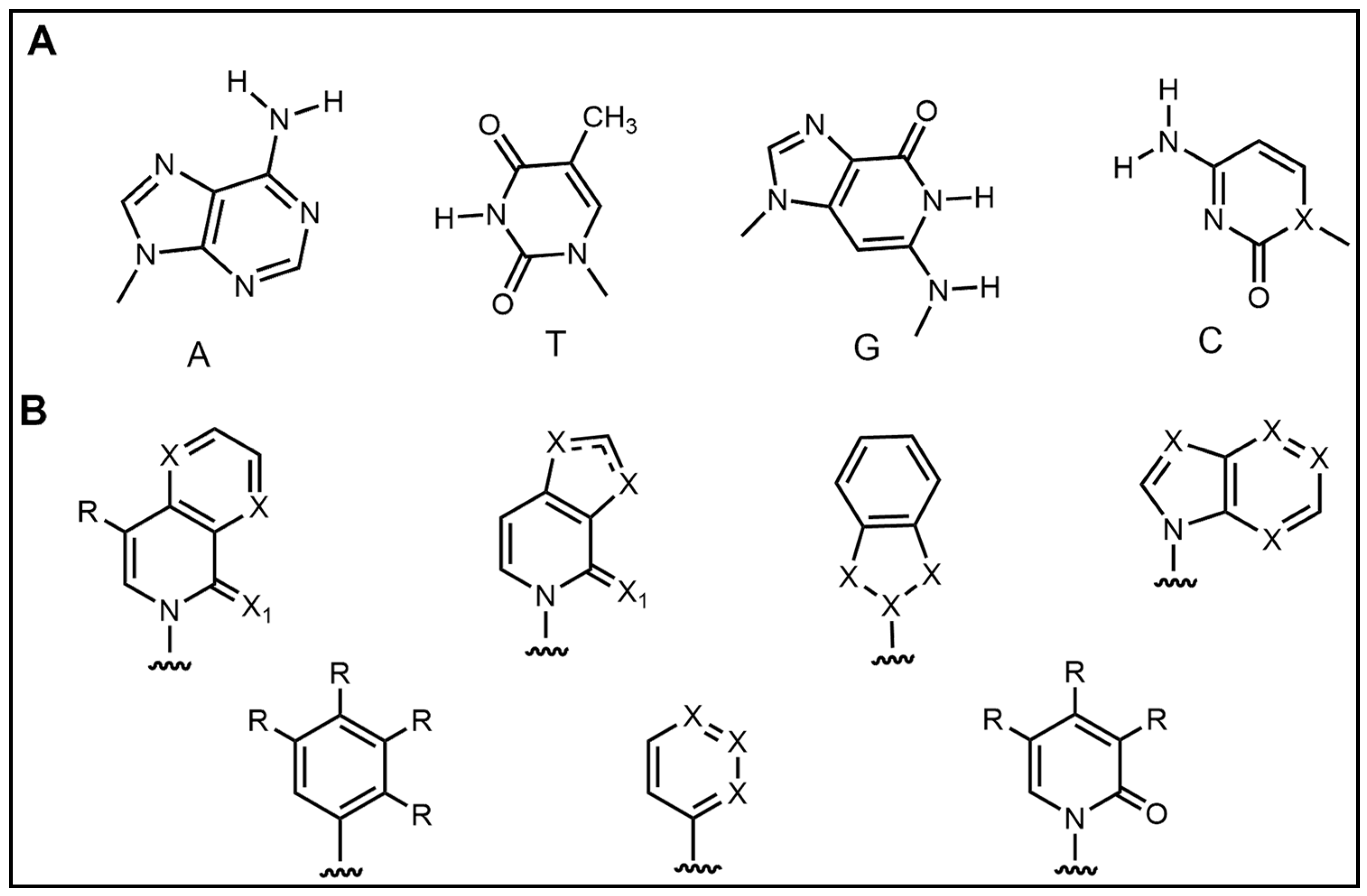
| Nucleic Acid Types | Ribose Chemical Structure | Ribose Structural Features | Nucleic Acid’s Key Features |
|---|---|---|---|
| DNA | 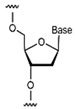 | Contains 2′-deoxyribose and lacks 2′-hydroxyl group. | Highly stable, high affinity, high specificity. |
| RNA | 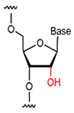 | The sugar ring structure is ribose with a hydroxyl group at the 2′ position. | Relatively unstable and prone to degradation. |
| LNA | 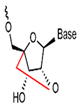 | Positions 2′ and 4′ are connected by a methyl bridge to form a rigid structure. | High affinity, high specificity, resistance to nuclease degradation, single-base mismatch discrimination. |
| PNA | 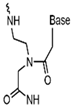 | Peptide backbone, sugar-phosphate backbone replaced by nonionic, repeating N-(2-aminoethyl)glycine units. | Resistant to nuclease and protease degradation, high thermal stability, single base mismatch identification, can be used for nucleic acid sensing. |
| TNA | 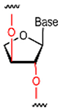 | 2′, 3′-linked sugar ring structure with acid stabilization. | High affinity, high specificity, resistant to nuclease degradation, able to stabilize in vivo. |
| HNA | 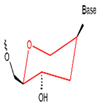 | It contains a six-carbon sugar ring for high stability and resistance to nuclease degradation. | High affinity, high specificity, able to stabilize in the physiological environment. |
| CeNA | 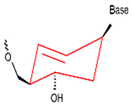 | It contains a cyclohexenose ring for high stability and resistance to nuclease degradation. | High thermal stability, RNase H activation, single base mismatch identification ability. |
| GNA | 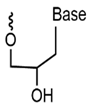 | It contains a glycol backbone for high stability and resistance to nuclease degradation. | High thermal stability, single base mismatch identification ability, can be used for nucleic acid sensing, gene detection. |
| F-ANA | 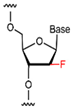 | 2′ position contains a fluorine modification for high stability and resistance to nuclease degradation. | High affinity, high specificity, able to stabilize in physiological environment. |
| ANA | 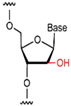 | 2′ position contains a hydroxyl group, which is resistant to nuclease degradation. | High affinity for stabilization in the physiological environment. |
| Heterocyclic Name | Heterocyclic Structure | Chemical Structure Characteristics | Application Potential |
|---|---|---|---|
| D5SICS | 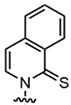 | Isoquinolone backbone, hydrophobic interactions | Expanding the Genetic Code, Biotechnology Applications |
| DMMO2 | 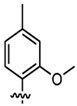 | Benzimidazole backbone, hydrophobic interactions | Expanding the Genetic Code, Biotechnology Applications |
| P | 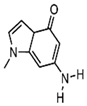 | Exhibits high fidelity in Taq DNA polymerase-mediated PCR | qPCR assay, molecular beacon technique |
| Z | 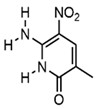 | Nitro group for improved chemical stability | RNA labeling, dynamic structure analysis |
| NaM | 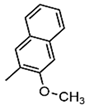 | Naphthylimidazole backbone, hydrophobic interactions | Creating Semi-Synthetic Organisms (SSOs) |
| 5SICS | 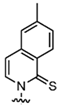 | Isoquinolone backbone, hydrophobic interactions | Creating Semi-Synthetic Organisms (SSOs) |
| Ds | 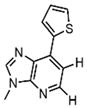 | Dibenzocycloheptane skeleton, hydrophobic interactions | RNA labeling, dynamic structure analysis |
| Px | 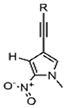 | Pyridine backbone, hydrophobic interactions | RNA labeling, dynamic structure analysis |
| Validation Methods | Principle | Core Advantages | Application Scenarios |
|---|---|---|---|
| Molecular docking | Predicting the binding mode and binding energy between two molecules. | Rapid prediction of molecular binding sites and thermodynamic analysis of docking feasibility. | Food packaging material design and development, food toxin detection and toxicology research, target and aptamer docking. |
| Molecular dynamics | Analyzing the behavior of molecules through mathematical models and computer simulation techniques. | Dynamics reveals the interaction mechanism between the aptamer and the target molecule, and it can indicate the rate of docking. The feasibility of docking is analyzed from thermodynamic and kinetic perspectives. | Process guidance and optimization in food processing, aptamer stability studies, DNA structure modeling, intermolecular interaction analysis. |
| Circular dichroism | Study of optical activity based mainly on differences in the absorption of polarized light by chiral centers in molecules. | Provides secondary structure information and unlabeled structure analysis. | Structural molecules of biomolecules, secondary structure studies of aptamers, and DNA structural stability analysis. |
| Isothermal titration | Study of intermolecular interactions by heat differences measured under isothermal conditions. | Provide thermodynamic parameters, high sensitivity, automation, and high accuracy. | Molecular interactions, aptamer-target molecule affinity determination, and food safety analysis. |
| Microscale thermophoresis | Measurement of trace thermophoretic changes to determine intermolecular affinity by changes in the hydration layer. | Broad compatibility, low sample consumption, and low running costs. | Aptamer affinity, molecular dynamics and molecular physicochemical property studies. |
| Fluorescence analysis | Based on the fluorescence of a substance when exposed to light at a specific wavelength. | High sensitivity, high selectivity, low sample volume, simple operation. | Biomedical, food safety, aptamer-target molecule interaction studies, and intermolecular interactions. |
| Target Classification | Sequence Optimization Methods | Application and Optimized Features | References |
|---|---|---|---|
| Zearalenone | Truncation | Food safety analysis. Increase sensitivity. | [180] |
| Chloromycetin | Truncation | Food safety control. High affinity and low synthesis cost. | [181] |
| Streptomycin | Truncation | Food safety control. High affinity | [182] |
| Organic Phosphorus pesticides | Truncation | Food safety analysis. Excellent recognition performance, high stability. | [41] |
| Deoxynivalenol | Truncation | Food safety analysis. High affinity and low cost. | [43] |
| Sulfaquinoxaline | Truncation | Environment and food safety. High affinity and sensitivity. | [183] |
| α-lactalbumin | Truncation | Food safety and medical diagnostics. Increased bonding affinity. | [42] |
| PTK7 | Mutation | Diagnosis of disease treatment. Improved thermal stability and binding affinity. | [184] |
| Nucleic acid group | Mutation | Treatment and diagnosis. Improved nuclease resistance and affinity. | [51] |
| Kanamycin | Mutation | Food safety analysis. Increased sensitivity. | [185] |
| Vibrio parahaemolyticus | Mutation | Food safety control. Decreased affinity. | [52] |
| SARS-CoV-2 | Mutation | Treatment and diagnosis. Enhanced viral inhibition. | [186] |
| FTO | Chemical modification | Clinical diagnosis, drug discovery and therapeutic evaluation. Reference for medical imaging analysis. | [68] |
| Thrombin | Chemical modification | Medicine and biotechnology. Enhanced stability. | [69] |
| Cytosine | Chemical modification | Research, diagnosis and treatment. Enhanced stability and rapport. | [187] |
| Uracil | Chemical modification | Treatment and diagnosis. Enhanced stability and rapport. | [188] |
| Quinine | Chemical modification | Disease treatment. Enhanced bonding affinity. | [189] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Zhao, L.; Zhang, Y.; Shi, J.; Tahir, H.E.; Xu, X.; Zheng, K.; Zou, X. Aptamer Sequence Optimization and Its Application in Food Safety Analysis. Foods 2025, 14, 2622. https://doi.org/10.3390/foods14152622
Qin X, Zhao L, Zhang Y, Shi J, Tahir HE, Xu X, Zheng K, Zou X. Aptamer Sequence Optimization and Its Application in Food Safety Analysis. Foods. 2025; 14(15):2622. https://doi.org/10.3390/foods14152622
Chicago/Turabian StyleQin, Xinna, Lina Zhao, Yang Zhang, Jiyong Shi, Haroon Elrasheid Tahir, Xuechao Xu, Kaiyi Zheng, and Xiaobo Zou. 2025. "Aptamer Sequence Optimization and Its Application in Food Safety Analysis" Foods 14, no. 15: 2622. https://doi.org/10.3390/foods14152622
APA StyleQin, X., Zhao, L., Zhang, Y., Shi, J., Tahir, H. E., Xu, X., Zheng, K., & Zou, X. (2025). Aptamer Sequence Optimization and Its Application in Food Safety Analysis. Foods, 14(15), 2622. https://doi.org/10.3390/foods14152622