The Natural Fermentation of Greek Tsounati Olives: Microbiome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Olives Fermentation
2.2. Physicochemical Analysis of Olives and Brines
2.2.1. pH and Salt Content
2.2.2. HPLC Analysis of Sugars, Organic Acids, Alcohols, and Triterpenic Acids
2.2.3. Total Phenolic Content
2.3. Microbiological Analysis of Olives and Brines
2.4. DNA Extraction and Rep-PCR Fingerprinting of Bacterial and Fungal Isolates
2.5. Identification of Bacterial and Fungal Isolates by 16S and ITS rDNA Sequencing
2.6. Total DNA Extraction and Metataxonomic Analysis
2.7. Bioinformatic and Statistical Analysis
2.8. Sensory Evaluation of Olives
2.9. Statistical Analysis of Experimental Values
3. Results and Discussion
3.1. Physicochemical Analysis of Olives and Brines
3.1.1. pH and Salt Content of Brines
3.1.2. HPLC Analysis of Sugars, Organic Acids, Alcohols, and Triterpenic Acids
3.1.3. Total Phenol Content of Fermented Olive and Brine Samples
3.2. Microbiological Analysis of Olives and Brines
3.3. Rep-PCR Fingerprinting and Identification of Bacterial Isolates
3.4. Rep-PCR Fingerprinting and Identification of Fungal Isolates
3.5. Metataxonomic Analysis
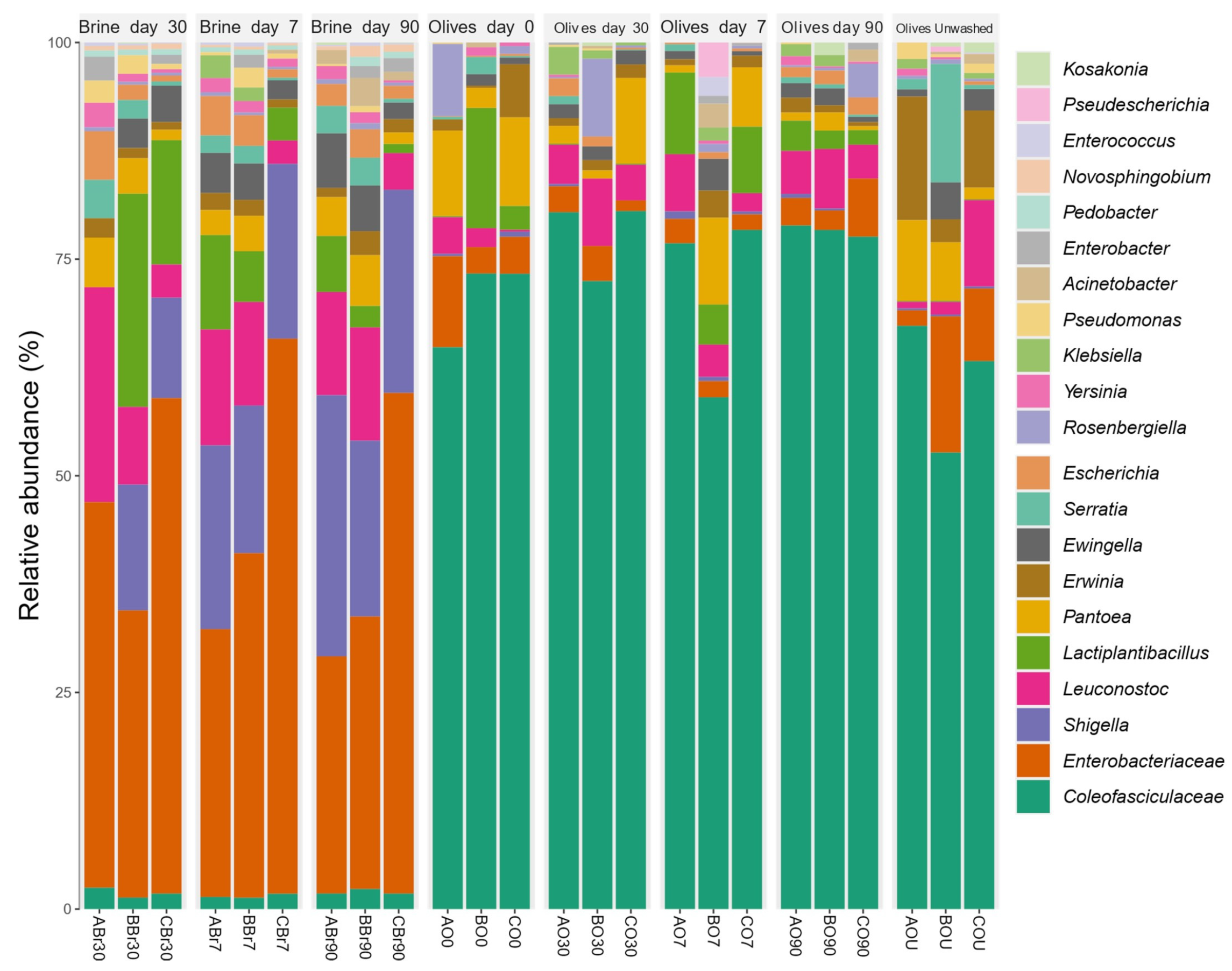
3.5.1. Bacterial Microbiota Composition
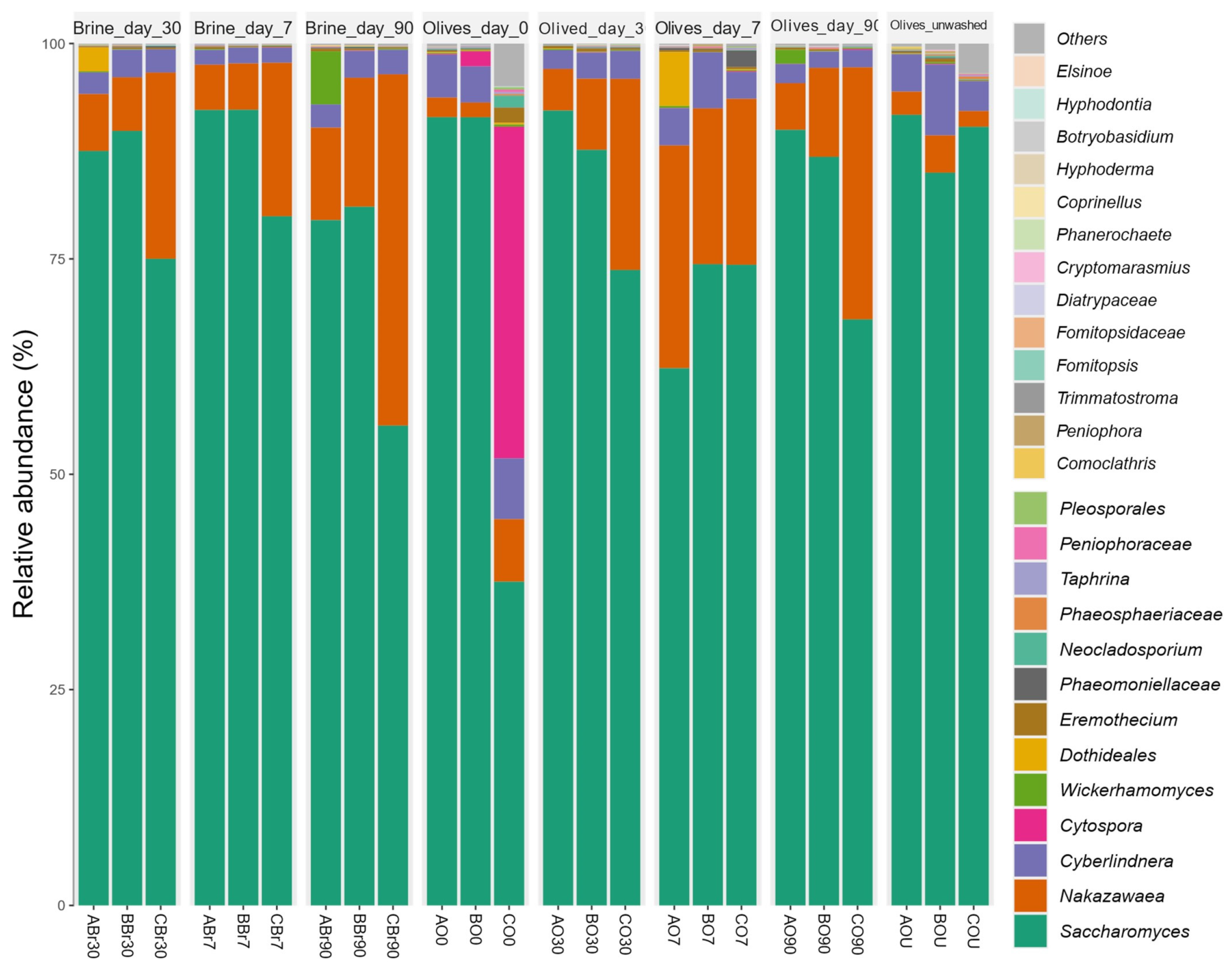
3.5.2. Mycobiota Composition
3.6. Sensory Evaluation of Fermented Tsounati Table Olives
3.7. Overall Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NGS | Next-Generation Sequencing |
| IOC | International Olive Council |
| NOGB | National Olive Germplasm Bank of Greece |
| WOGBC | World Olive Germplasm Bank of IFAPA, Córdoba |
| PDO | Protected Designation of Origin |
| PGI | Protected Geographical Indication |
| LAB | Lactic Acid Bacteria |
| TPC | Total Polyphenol Content |
| HPLC | High-Performance Liquid Chromatography |
| GAE | Gallic Acid Equivalents |
| PCA | Plate Count agar |
| MRS | De Man–Rogosa–Sharpe |
| VRBG | Violet Red Bile Glucose agar |
| YGC | Yeast Glucose Chloramphenicol agar |
| CFU | Colony Forming Units |
| SD | Standard Deviation |
| TE | Tris-EDTA |
| PCR | Polymerase Chain Reaction |
| Rep | Repetitive Extragenic Palindromic Elements |
| NCBI | National Center for Biotechnology Information |
| NADH | Nicotinamide Adenine Dinucleotide Hydrogen |
| WHO | World Health Organization |
| ASV | Amplicon Sequence Variant |
| LDA | Linear Discriminant Analysis |
| PCoA | Principal Component Analysis |
References
- Green, P.S. Oleaceae. In The Families and Genera of Vascular Plants; Kubitzki, K., Kadereit, J.W., Eds.; Springer: Berlin,/Heidelberg, Germany, 2004; Volume 7, pp. 296–306. ISBN 978-3-540-40593-1. [Google Scholar]
- Huxley, A.; Griffiths, M.; Levy, M. The New Royal Horticultural Society Dictionary of Gardening; The Macmillan Press, Ltd.: London, UK, 1992; ISBN 978-0-333-47494-5. [Google Scholar]
- Green, P.S. A revision of Olea L. (Oleaceae). Kew Bull. 2002, 57, 91–140. [Google Scholar] [CrossRef]
- Cimato, A.; Attilio, C. Worldwide diffusion and relevance of olive culture. In Olive Diseases and Disorders; Schena, L., Agosteo, G.E., Cacciola, S.O., Eds.; Transworld Research Network: Trivandrum, India, 2011; Chapter 1; pp. 1–21. [Google Scholar]
- Barazani, O.; Dag, A.; Dunseth, Z. The history of olive cultivation in the southern Levant. Front. Plant Sci. 2023, 14, 1131557. [Google Scholar] [CrossRef]
- Foley, B.P.; Hansson, M.C.; Kourkoumelis, D.P.; Theodoulou, T.A. Aspects of ancient Greek trade re-evaluated with amphora DNA evidence. J. Archaeol. Sci. 2012, 39, 389–398. [Google Scholar] [CrossRef]
- Mascagni, F.; Barghini, E.; Ceccarelli, M.; Baldoni, L.; Trapero, C.; Díez, C.M.; Natali, L.; Cavallini, A.; Giordani, T. The singular evolution of Olea genome structure. Front. Plant Sci. 2022, 13, 869048. [Google Scholar] [CrossRef] [PubMed]
- Barranco, D.; Cimato, A.; Fiorino, P.; Rallo, L.; Touzani, A.; Castañeda, C.; Serafín, F.; Trujillo, I. World Catalogue of Olive Varieties. International Olive Oil Council, Madrid. 2000. Available online: https://www.internationaloliveoil.org/product/world-catalogue-of-olive-varieties/ (accessed on 22 March 2025).
- International Olive Council (IOC). The IOC Network of Germplasm Banks and the True Healthy Olive Cultivars Project. In Book of the IOC Network of Germplasm Banks, International Seminar, Córdoba, Argentina, 21–24 October 2019; IOC: Córdoba, Argentina, 2019; Available online: https://www.internationaloliveoil.org/the-iocs-book-of-germplasm-banks/ (accessed on 5 February 2025).
- Conte, P.; Fadda, C.; Del Caro, A.; Urgeghe, P.P.; Piga, A. Table olives: An overview on effects of processing on nutritional and sensory quality. Foods 2020, 9, 514. [Google Scholar] [CrossRef]
- Gómez-Gálvez, F.J.; Ninot, A.; Rodríguez, J.C.; Compañ, S.P.; Andreva, J.U.; Rubio, J.A.G.; Aragón, I.P.; Viñuales-Andreu, J.; Casanova-Gascón, J.; Ŝatović, Z.; et al. New insights in the Spanish gene pool of olive (Olea europaea L.) preserved ex situ and in situ based on high-throughput molecular markers. Front. Plant Sci. 2024, 14, 1267601. [Google Scholar] [CrossRef]
- FEK 4460. 2016. Available online: http://www.minagric.gr/images/stories/docs/agrotis/Pollaplasiastiko_Yliko/nomothesia_pollaplasiastiko_yliko/fek4460_301216.pdf (accessed on 25 September 2024).
- Hagidimitriou, M.; Katsiotis, A.; Menexes, G.; Pontikis, C.; Loukas, M. Genetic diversity of major Greek olive cultivars using molecular (AFLPs and RAPDs) markers and morphological traits. J. Am. Soc. Hortic. Sci. 2005, 130, 211–217. [Google Scholar] [CrossRef]
- Agriopoulou, S.; Tarapoulouzi, M.; Bedine Boat, M.A.; Rébufa, C.; Dupuy, N.; Theocharis, C.R.; Varzakas, T.; Roussos, S.; Artaud, J. Authentication and chemometric discrimination of six Greek PDO table olive varieties through morphological characteristics of their stones. Foods 2021, 10, 1829. [Google Scholar] [CrossRef]
- Stefanoudaki, E.; Kotsifaki, F.; Koutsaftakis, A. Classification of virgin olive oils of the two major cretan cultivars based on their fatty acid composition. J. Am. Oil Chem. Soc. 1999, 76, 623–626. [Google Scholar] [CrossRef]
- Andreou, V.; Psarianos, M.; Dimopoulos, G.; Tsimogiannis, D.; Taoukis, P. Effect of pulsed electric fields and high pressure on improved recovery of high-added-value compounds from olive pomace. J. Food Sci. 2020, 85, 1500–1512. [Google Scholar] [CrossRef]
- Skiada, V.; Tsarouhas, P.; Varzakas, T. Comparison and discrimination of two major monocultivar extra virgin olive oils in the Southern region of Peloponnese, according to specific compositional/traceability markers. Foods 2020, 9, 155. [Google Scholar] [CrossRef]
- Mousa, Y.M.; Gerasopoulos, D.; Metzidakis, I.; Kiritsakis, A. Effect of altitude on fruit and oil quality characteristics of “Mastoides” olives. J. Sci. Food Agric. 1996, 71, 345–350. [Google Scholar] [CrossRef]
- International Olive Council (IOC). (2024/2025 Crop Year). Export Figures of Table Olives in the European Union (EU-27). Available online: https://www.internationaloliveoil.org/wp-content/uploads/2025/03/IOC-Export-figures-TO-EU.html (accessed on 20 May 2025).
- Alexandraki, V.; Georgalaki, M.; Papadimitriou, K.; Anastasiou, R.; Zoumpopoulou, G.; Chatzipavlidis, I.; Papadelli, M.; Vallis, N.; Moschochoritis, K.; Tsakalidou, E. Determination of triterpenic acids in natural and alkaline-treated Greek table olives throughout the fermentation process. Food Sci. Technol. 2014, 58, 609–613. [Google Scholar] [CrossRef]
- Rocha, J.; Borges, N.; Pinho, O. Table olives and health: A review. J. Nutr. Sci. 2020, 2, e57. [Google Scholar] [CrossRef]
- Mioc, M.; Prodea, A.; Racoviceanu, R.; Mioc, A.; Ghiulai, R.; Milan, A.; Voicu, M.; Mardale, G.; Șoica, C. Recent advances regarding the molecular mechanisms of triterpenic acids: A review (Part II). Int. J. Mol. Sci. 2022, 23, 8896. [Google Scholar] [CrossRef] [PubMed]
- Rufino-Palomares, E.E.; Pérez-Jiménez, A.; García-Salguero, L.; Mokhtari, K.; Reyes-Zurita, F.J.; Peragón-Sánchez, J.; Lupiáñez, J.A. Nutraceutical role of polyphenols and triterpenes Present in the extracts of fruits and leaves of Olea europaea as antioxidants, anti-infectives and anticancer agents on healthy growth. Molecules 2022, 5, 2341. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Alessandria, V.; Botta, C.; Gorra, R.; Filippis, F.; Ercolini, D.; Rantsiou, K. NaOH-debittering induces changes in bacterial ecology during table olives fermentation. PLoS ONE 2013, 31, e69074. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Todaro, A.; Pino, A.; Corona, O.; Caggia, C. Microbiota and metabalome during controlled and spontaneous fermentation of Nocellara Etnea table olives. Food Microbiol. 2017, 65, 136–148. [Google Scholar] [CrossRef]
- De Angelis, M.; Campanella, D.; Cosmai, L.; Summo, C.; Giuseppe Rizzello, C.; Caponio, F. Microbiota and metabolome of un-started and started Greek-type fermentation of Bella di Cerignola table olives. Food Microbiol. 2015, 52, 18–30. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Medina, E.; Ruiz-Bellido, M.A.; Romero-Gil, V.; Montes-Borrego, M.; Landa, B.B. Enhancement of the knowledge on fungal communities in directly brines Alorena de Malaga green olive fermentations by metabarcoding analysis. PLoS ONE 2016, 11, e0163135. [Google Scholar] [CrossRef]
- Medina, E.; Ruiz-Bellido, M.A.; Romero-Gil, V.; Rodriguez-Gomez, F.; Montes-Borrego, M.; Landa, B.B.; Arroyo-Lopez, F.N. Assessment of bacterial community in directly brined Alorena de Malaga table olive fermentations by metagenetic analysis. Int. J. Food Microbiol. 2016, 236, 47–55. [Google Scholar] [CrossRef]
- Argyri, K.; Doulgeraki, A.I.; Manthou, E.; Grounta, A.; Argyri, A.A.; Nychas, G.-J.E.; Tassou, C. Microbial diversity of fermented Greek table olives of Halkidiki and Konservolia varieties from different regions as revealed by metagenomic analysis. Microorganisms 2020, 8, 1241. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Tsaltas, D. Fermented table olives from Cyprus: Microbiota profile of three varieties from different regions through metabarcoding sequencing. Front. Microbiol. 2023, 13, 1101515. [Google Scholar] [CrossRef]
- Ruiz-Barba, J.L.; Sánchez, A.H.; López-López, A.; Cortés-Delgado, A.; Montaño, A. Microbial and chemical characterization of natural-style green table olives from the Gordal, Hojiblanca and Manzanilla cultivars. Foods 2023, 12, 2386. [Google Scholar] [CrossRef] [PubMed]
- Penland, M.; Deutsch, S.-M.; Falentin, H.; Pawtowski, A.; Poirier, E.; Visenti, G.; Le Meur, C.; Maillard, M.-B.; Thierry, A.; Mounier, J.; et al. Deciphering microbial community dynamics and biochemical changes during Nyons black olive natural fermentations. Front. Microbiol. 2020, 11, 586614. [Google Scholar] [CrossRef] [PubMed]
- Kazou, M.; Tzamourani, A.; Panagou, E.Z.; Tsakalidou, E. Unraveling the microbiota of natural black cv. Kalamata fermented olives through 16S and ITS metagenomic analysis. Microorganisms 2020, 8, 672. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Cabello, A.; Romero-Gil, V.; Medina-Pradas, E.; Garrido-Fernández, A.; Arroyo-López, F.N. Exploring bacteria diversity in commercialized table olive biofilms by metataxonomic and compositional data analysis. Sci. Rep. 2020, 10, 11381. [Google Scholar] [CrossRef]
- RES-2/91-IV/04; Trade Standard Applying to Table Olives. Document COI/OT/NC No. 1. International Olive Oil Council: Madrid, Spain, 2004.
- Angelopoulou, A.; Alexandraki, V.; Georgalaki, M.; Anastasiou, R.; Manolopoulou, E.; Tsakalidou, E.; Papadimitriou, K. Production of probiotic Feta cheese using Propionibacterium freudenreichii subsp. shermanii as adjunct. Int. Dairy J. 2017, 66, 135–139. [Google Scholar] [CrossRef]
- Romero, C.; García, A.; Medina, E.; Ruíz-Méndez, M.V.; de Castro, A.; Brenes, M. Triterpenic acids in table olives. Food Chem. 2010, 118, 670–674. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Marsilio, V.; Seghetti, L.; Iannucci, E.; Russi, F.; Lanza, B.; Felicioni, M. Use of a lactic acid bacteria starter culture during green olive (Olea europaea L cv Ascolana tenera) processing. J. Sci. Food Agric. 2005, 85, 1084–1090. [Google Scholar] [CrossRef]
- Georgalaki, M.; Zoumpopoulou, G.; Mavrogonatou, E.; Van Driessche, G.; Alexandraki, V.; Anastasiou, R.; Papadelli, M.; Kazou, M.; Manolopoulou, E.; Kletsas, D. Evaluation of the antihypertensive angiotensin-converting enzyme inhibitory (ACE-I) activity and other probiotic properties of lactic acid bacteria isolated from traditional Greek dairy products. Int. Dairy J. 2017, 75, 10–21. [Google Scholar] [CrossRef]
- Kopsahelis, N.; Nisiotou, A.; Kourkoutas, Y.; Panas, P.; Nychas, G.-J.E.; Kanellaki, M. Molecular characterization and molasses fermentation performance of a wild yeast strain operating in an extremely wide temperature range. Bioresour. Technol. 2009, 100, 4854–4862. [Google Scholar] [CrossRef] [PubMed]
- da Silva-Filho, E.A.; dos Santos, S.K.B.; do Monte Resende, A.; de Morais, J.O.F.; de Morais, M.A.; Simões, D.A. Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR-fingerprinting. Antonie Van Leeuwenhoek 2005, 88, 13–23. [Google Scholar] [CrossRef]
- Ntougias, S.; Zervakis, G.I.; Ehaliotis, C.; Kavroulakis, N.; Papadopoulou, K.K. Ecophysiology and molecular phylogeny of bacteria isolated from alkaline two-phase olive mill wastes. Res. Microbiol. 2006, 157, 376–385. [Google Scholar] [CrossRef]
- Korabecna, M. The Variability in the fungal ribosomal DNA (ITS1, ITS2, and 5.8 S rRNA Gene): Its biological meaning and application in medical mycology. In Communicating Current Research and Educational Topics and Trends in Applied Microbiology; Mendez-Vilas, A., Ed.; FORMATEX: Badajoz, Spain, 2007; pp. 783–787. [Google Scholar]
- Papademas, P.; Aspri, M.; Mariou, M.; Dowd, S.E.; Kazou, M.; Tsakalidou, E. Conventional and omics approaches shed light on Halitzia cheese, a long-forgotten white-brined cheese from Cyprus. Int. Dairy J. 2019, 98, 72–83. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High resolution sample inference from amplicon data. BioRxiv 2015, 7, 024034. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2024, 42, 715–718. [Google Scholar] [CrossRef]
- International Olive Council (IOC). Sensory Analysis of Table Olives; Document COI/OT/MO No 1/Rev. 2; International Olive Council: Madrid, Spain, 2011; Available online: https://www.internationaloliveoil.org/wp-content/uploads/2022/10/COI-OT-MO.-1-Rev.2-2011-Eng.pdf (accessed on 17 October 2024).
- Portilha-Cunha, M.F.; Macedo, A.C.; Malcata, F.X. A Review on adventitious lactic acid bacteria from table olives. Foods 2020, 9, 948. [Google Scholar] [CrossRef]
- Garrido-Fernández, A.; Fernández Díaz, M.J.; Adams, R.M. Table Olives: Production and Processing; Chapman & Hall: London, UK, 1997; pp. 289–367. [Google Scholar]
- Arroyo-López, F.N.; Benítez-Cabello, A.; Romero-Gil, V.; Rodríguez-Gomez, F.; Garrido-Fernandez, A. Delving into the bacterial diversity of spoiled green Manzanilla Spanish-style table olive fermentations. Int. J. Food Microbiol. 2021, 359, 109415. [Google Scholar] [CrossRef]
- Campus, M.; Corrias, F.; Angioni, A.; Arru, N.; Sedda, P.; Addis, M.; Fiori, M.; Paba, A.; Chessa, L.; Comunian, R. Efficacy of a native microbial starter in promoting table olive fermentation: An industrial-scale trial at controlled and ambient temperature. Foods 2025, 14, 2159. [Google Scholar] [CrossRef] [PubMed]
- López-García, E.; Benítez-Cabello, A.; Rodríguez-Gómez, F.; Romero-Gil, V.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. Bacterial metataxonomic analysis of industrial Spanish-style green table olive fermentations. Food Control 2022, 137, 108969. [Google Scholar] [CrossRef]
- EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA); Turck, D.; Castenmiller, J.; de Henauw, S.; de Henauw, S.; Hirsch-Ernst, K.I.; Kearney, J.; Knutsen, H.K.; Maciuk, A.; Mangelsdorf, I. Dietary reference values for sodium. EFSA J. 2019, 17, 5778. [Google Scholar] [CrossRef] [PubMed]
- Papadelli, M.; Zoumpopoulou, G.; Georgalaki, M.; Anastasiou, R.; Manolopoulou, E.; Lytra, I.; Papadimitriou, K.; Tsakalidou, E. Evaluation of two lactic acid bacteria starter cultures for the fermentation of natural black table olives (Olea europaea L cv Kalamon). Pol. J. Microbiol. 2015, 64, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Anagnostopoulos, D.A.; Goulas, V.; Xenofontos, E.; Vouras, C.; Nikoloudakis, N.; Tsaltas, D. Benefits of the use of lactic acid bacteria starter in green cracked Cypriot table olives fermentation. Foods 2020, 9, 17. [Google Scholar] [CrossRef]
- Lee, J. Exploring sucrose fermentation: Microorganisms, biochemical pathways, and applications. Ferment. Technol. 2023, 12, 166. [Google Scholar] [CrossRef]
- Anagnostopoulos, D.; Bozoudi, D.; Tsaltas, D. Yeast Ecology of Fermented Table Olives: A Tool for Biotechnological Applications. In Yeast—Industrial Applications; InTech Open: London, UK, 2017. [Google Scholar] [CrossRef]
- Ge, Y.; Wu, Y.; Aihaiti, A.; Wang, L.; Wang, Y.; Xing, J.; Zhu, M.; Hong, J. The metabolic pathways of yeast and acetic acid bacteria during fruit vinegar fermentation and their influence on favor development. Microorganisms 2025, 13, 477. [Google Scholar] [CrossRef]
- Oude Elferink, S.J.; Krooneman, J.; Gottschal, J.C.; Spoelstra, S.F.; Faber, F.; Driehuis, F. Anaerobic conversion of lactic acid to acetic acid and 1, 2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 2001, 67, 125–132. [Google Scholar] [CrossRef]
- Silva, T.; Reto, M.; Sol, M.; Peito, A.; Peres, C.M.; Peres, C.; Malcata, F.X. Characterization of yeasts from Portuguese brined olives, with a focus on their potentially probiotic behavior. Food Sci. Technol. 2011, 44, 1349–1354. [Google Scholar] [CrossRef]
- Eram, M.; Ma, K. Decarboxylation of pyruvate to acetaldehyde for ethanol production by hyperthermophiles. Biomolecules 2013, 3, 578–596. [Google Scholar] [CrossRef]
- Vustin, M.M. The biological role of glycerol in yeast cells. Yeast as glycerol producers. Appl. Biochem. Microbiol. 2021, 57, 907–916. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhan, M.; Yang, Z.; Zumstein, K.; Chen, H.; Huang, Q. The major qualitative characteristics of olive (Olea europaea L.) cultivated in Southwest China. Front. Plant Sci. 2017, 8, 559. [Google Scholar] [CrossRef]
- Sidari, R.; Martorana, A.; De Bruno, A. Effect of brine composition on yeast biota associated with naturally fermented Nocellara messinese table olives. Food Sci. Technol. 2019, 109, 163–170. [Google Scholar] [CrossRef]
- Sorrentino, G.; Muzzalupo, I.; Muccilli, S.; Timpanaro, N.; Russo, M.P.; Guardo, M.; Rapisarda, P.; Romeo, F.V. New accessions of Italian table olives (Olea europaea): Characterization of genotypes and quality of brined products. Sci. Hortic. 2016, 213, 34–41. [Google Scholar] [CrossRef]
- Sánchez, A.H.; De Castro, A.; Rejano, L.; Montaño, A. Comparative study on chemical changes in olive juice and brine during green olive fermentation. J. Agric. Food Chem. 2000, 48, 5975–5980. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Tufariello, M.; Tommasi, L.; Lenucci, M.S.; Bleve, G.; Mita, G. Evaluation of bioactive compounds in black table olives fermented with selected microbial starters. J. Sci. Food Agric. 2017, 98, 96–103. [Google Scholar] [CrossRef]
- Moreno-González, R.; Juan, M.E.; Planas, J.M. Profiling of pentacyclic triterpenes and polyphenols by LC-MS in Arbequina and Empeltre table olives. LWT Food Sci. Technol. 2020, 126, 109310. [Google Scholar] [CrossRef]
- Boskou, D. Table Olives: A vehicle for the delivery of bioactive compounds. J. Exp. Food Chem. 2017, 3, 123. [Google Scholar] [CrossRef]
- Medina, E.; Morales-Sillero, A.; Ramírez, E.M.; Rallo, P.; Brenes, M.; Romero, C. New genotypes of table olives: Profile of bioactive compounds. Int. J. Food Sci. Technol. 2012, 47, 2334–2341. [Google Scholar] [CrossRef]
- Han, N.; Bakovic, M. Biologically active triterpenoids and their cardioprotective and anti-inflammatory effects. J. Bioanal. Biomed. S 2015, 12, 1948–1959. [Google Scholar] [CrossRef]
- Lin, C.; Wen, X.; Sun, H. Oleanolic acid derivatives for pharmaceutical use: A patent review. Expert Opin. Ther. Pat. 2016, 26, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G. Lipids and phenols in table olives. Eur. J. Lipid Sci. Technol. 2003, 105, 229–242. [Google Scholar] [CrossRef]
- Kiai, H.; Hafidi, A. Chemical composition changes in four green olive cultivars during spontaneous fermentation. Food Sci. Technol. 2014, 57, 663–670. [Google Scholar] [CrossRef]
- Malheiro, R.; Mendes, P.; Fernandes, F.; Rodrigues, N.; Bento, A.; Pereira, J.A. Bioactivity and phenolic composition from natural fermented table olives. Food Funct. 2014, 5, 3132–3142. [Google Scholar] [CrossRef]
- Faghim, J.; Mohamed, M.B.; Bagues, M.; Guasmi, F.; Triki, T.; Nagaz, K. Irrigation effects on phenolic profile and extra virgin olive oil quality of “Chemlali’’ variety grown in South Tunisia. S. Afr. Bot. 2021, 141, 322–329. [Google Scholar] [CrossRef]
- García, P.; Romero, C.; Brenes, M. Influence of olive tree irrigation and the preservation system on the fruit characteristics of Hojiblanca black ripe olives. Food Sci. Technol. 2014, 55, 403–407. [Google Scholar] [CrossRef]
- Tataridou, M.; Kotzekidou, P. Fermentation of table olives by oleuropeinolytic starter culture in reduced salt brines and inactivation of Escherichia coli O157:H7 and Listeria monocytogenes. Int. J. Food Microbiol. 2015, 208, 122–130. [Google Scholar] [CrossRef]
- Georgiou, S.M.; Kosma, I.S.; Badeka, A.V.; Kontominas, M.G. Characterization and geographical differentiation of Kalamata table olives using physical, chemical, mechanical and sensory properties: A chemometric approach. Microchem. J. 2024, 199, 110085. [Google Scholar] [CrossRef]
- Rodríguez, H.; Landete, J.M.; de las Rivas, B.; Muñoz, R. Metabolism of food phenolic acids by Lactobacillus plantarum CECT 748T. Food Chem. 2008, 107, 1393–1398. [Google Scholar] [CrossRef]
- Bleve, G.; Tufariello, M.; Durante, M.; Perbellini, E.; Ramires, F.A.; Grieco, F.; Cappello, M.S.; De Domenico, S.; Mita, G.; Tasioula-Margari, M.; et al. Physico-chemical and microbiological characterization of spontaneous fermentation of Cellina di Nardò and Leccino table olives. Front. Microbiol. 2014, 5, 570. [Google Scholar] [CrossRef]
- Ramírez, E.; Brenes, M.; García, P.; Medina, E.; Romero, C. Oleuropein hydrolysis in natural green olives: Importance of the endogenous enzymes. Food Chem. 2016, 206, 204–209. [Google Scholar] [CrossRef]
- Piscopo, A.; De Bruno, A.; Zappia, A.; Poiana, M. Antioxidant activity of dried green olives (Carolea cv.). Food Sci. Technol. 2014, 58, 49–54. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Romero, C.; Durán Quintana, M.; López López, A.; García García, P.; Garrido Fernández, A. Kinetic study of the physicochemical and microbiological changes in seasoned olives during the shelf period. J. Agric. Food Chem. 2005, 53, 5285–5292. [Google Scholar] [CrossRef]
- Maicas, S. The role of yeasts in fermentation processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef]
- Giannattasio, S.; Guaragnella, N.; Ždralević, M.; Marra, E. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front. Microbiol. 2013, 4, 33. [Google Scholar] [CrossRef]
- Adeboye, P.T.; Bettiga, M.; Olsson, L. The chemical nature of phenolic compounds determines their toxicity and induces distinct physiological responses in Saccharomyces cerevisiae in lignocellulose hydrolysates. AMB Express 2014, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Panagou, E.Z.; Tassou, C.C.; Katsaboxakis, C.Z. Induced lactic acid fermentation of untreated green olives of the Conservolea cultivar by Lactobacillus pentosus. J. Sci. Food Agric. 2003, 83, 667–674. [Google Scholar] [CrossRef]
- Abriouel, H.; Benomar, N.; Lucas, R.; Gálvez, A. Culture-independent study of the diversity of microbial populations in brines during fermentation of naturally-fermented Aloreña green table olives. Int. J. Food Microbiol. 2011, 144, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Bonatsou, S.; Panagou, E.Z. Fermentation of cv. Kalamata natural black olives with potential multifunctional yeast starters. Foods 2022, 11, 3106. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Naser, S.; Thompson, F.L.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Thompson, C.C.; Vancanneyt, M.; Swings, J. Phylogeny and identification of enterococci by atpA gene sequence analysis. J. Clin. Microbiol. 2005, 43, 2224–2230. [Google Scholar] [CrossRef]
- Olsen, K.N.; Brockmann, E.; Molin, S. Quantification of Leuconostoc populations in mixed dairy starter cultures using fluorescence in situ hybridization. J. Appl. Microbiol. 2007, 103, 855–863. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Querol, A.; Bautista-Gallego, J.; Garrido Fernández, A. Role of yeasts in table olive production. Int. J. Food Microbiol. 2008, 128, 189–196. [Google Scholar] [CrossRef]
- Bautista-Gallego, J.; Arroyo-López, F.N.; Rantsiou, K.; Jiménez-Díaz, R.; Garrido-Fernández, A.; Cocolin, L. Screening of lactic acid bacteria isolated from fermented table olives with probiotic potential. Food Res. Int. 2013, 50, 135–142. [Google Scholar] [CrossRef]
- Bonatsou, S.; Tassou, C.; Panagou, E.; Nychas, G.-J.E. Table olive fermentation using starter cultures with multifunctional potential. Microorganisms 2017, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Perpetuini, G.; Prete, R.; Garcia-Gonzalez, N.; Khairul Alam, M.; Corsetti, A. Table olives more than a fermented food. Foods 2020, 9, 178. [Google Scholar] [CrossRef]
- Hurtado, A.; Reguant, C.; Bordons, A.; Rozès, N. Lactic acid bacteria from fermented table olives. Food Microbiol. 2012, 31, 1–8. [Google Scholar] [CrossRef]
- Bleve, G.; Tufariello, M.; Durante, M.; Grieco, F.; Ramires, F.A.; Mita, G.; Tasioula-Margari, M.; Logrieco, A.F. Physico-chemical characterization of natural fermentation process of Conservolea and Kalamàta table olives and developement of a protocol for the pre-selection of fermentation starters. Food Microbiol. 2015, 46, 368–382. [Google Scholar] [CrossRef]
- Heperkan, D. Microbiota of table olive fermentations and criteria of selection for their use as starters. Front. Microbiol. 2013, 4, 143. [Google Scholar] [CrossRef]
- Abriouel, H.; Benomar, N.; Cobo, A.; Caballero, N.; Fernández Fuentes, M.Á.; Pérez-Pulido, R.; Gálvez, A. Characterization of lactic acid bacteria from naturally-fermented Manzanilla Aloreña green table olives. Food Microbiol. 2012, 32, 308–316. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Ribbera, A.; Pitino, I.; Romeo, F.V.; Caggia, C. Diversity of bacterial population of table olives assessed by PCR- DGGE analysis. Food Microbiol. 2012, 32, 87–96. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Rajendram, R.; Caggia, C. Lactic acid bacteria in table olive fermentation. In Olives and Olive Oil in Health and Disease Prevention; Pretty, V.R., Watson, R.S., Eds.; Elsevier: London, UK, 2010; pp. 371–379. [Google Scholar]
- De Castro, A.; Montaño, A.; Casado, F.-J.; Sánchez, A.-H.; Rejano, L. Utilization of Enterococcus casseliflavus and Lactobacillus pentosus as starter cultures for Spanish-style green olive fermentation. Food Microbiol. 2002, 19, 637–644. [Google Scholar] [CrossRef]
- Alves, M.; Gonçalves, T.; Quintas, C. Microbial quality and yeast population dynamics in cracked green table olives’ fermentations. Food Control 2012, 23, 363–368. [Google Scholar] [CrossRef]
- Rameshkumar, N.; Lang, E.; Nair, S. Mangrovibacter plantisponsor gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from a mangrove-associated wild rice (Porteresia coarctata Tateoka). Int. J. Syst. Evol. Microbiol. 2010, 60, 179–186. [Google Scholar] [CrossRef]
- Chin, H.S.; Ravi Varadharajulu, N.; Lin, Z.-H.; Chen, W.-Y.; Zhang, Z.-H.; Arumugam, S.; Lai, C.-Y.; Yu, S.S.-F. Isolation, molecular identification, and genomic analysis of Mangrovibacter phragmitis strain ASIOC01 from activated sludge harboring the bioremediation prowess of glycerol and organic pollutants in high-salinity. Front. Microbiol. 2024, 15, 1415723. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.; Palombo, E.A.; Jaimes Castillo, A.; Zaferanloo, B. Endophytes in agriculture: Potential to improve yields and tolerances of agricultural crops. Microorganisms 2023, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- López-García, E.; Benítez-Cabello, A.; Rodríguez-Gómez, F.; Martín-Arranz, V.; Garrido-Fernández, A.; Arroyo-López, F.N. Influence of 1-methylcyclopropene (1-MCP) on the processing and microbial communities of Spanish-style and directly brined green table olive fermentations. Fermentation 2022, 8, 441. [Google Scholar] [CrossRef]
- Bouhia, Y.; Hafidi, M.; Ouhdouch, Y.; Soulaimani, A.; Zeroual, Y.; Lyamlouli, K. Microbial intervention improves pollutant removal and semi-liquid organo-mineral fertilizer production from olive mill wastewater sludge and rock phosphate. J. Environ. Manag. 2024, 354, 120317. [Google Scholar] [CrossRef]
- Mennane, Z.; Abrini, J.; Elmtili, N. Hygienic quality, study of enterobacteria isolated from table olives and antimicrobial activity of Olea europaea L from Tetouan, Morocco. E3S Web Conf. 2021, 319, 01088. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Cannarsi, M.; Gallo, M.; Sinigaglia, M.; Corbo, M.R. Characterization and implications of Enterobacter cloacae strains, isolated from Italian table olives “Bella di Cerignola”. J. Food Sci. 2010, 75, 53–60. [Google Scholar] [CrossRef]
- van der Aa Kühle, A.; Jespersen, L. The taxonomic position of Saccharomyces boulardii as evaluated by sequence analysis of the D1/D2 domain of 26S rDNA, the ITS1-5.8S rDNA-ITS2 region and the mitochondrial cytochrome-c oxidase II gene. Syst. Appl. Microbiol. 2003, 26, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Dashko, S.; Liu, P.; Volk, H.; Butinar, L.; Piškur, J.; Fay, J.C. Changes in the relative abundance of two Saccharomyces species from oak forests to wine fermentations. Front. Microbiol. 2016, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Pontes, A.; Čadež, N.; Gonçalves, P.; Sampaio, J.P. A quasi-domesticate relic hybrid population of Saccharomyces cerevisiae × S. paradoxus adapted to olive brine. Front. Genet. 2019, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Pontes, A.; Hutzler, M.; Brito, P.H.; Sampaio, J.P. Revisiting the taxonomic synonyms and populations of Saccharomyces cerevisiae—Phylogeny, phenotypes, ecology and domestication. Microorganisms 2020, 8, 903. [Google Scholar] [CrossRef]
- Çelik, Z.D.; Erten, H.; Darici, M.; Cabaroğlu, T. Molecular characterization and technological properties of wine yeasts isolated during spontaneous fermentation of Vitis vinifera L.cv. Narince grape must grown in ancient wine making area Tokat, Anatolia. BIO Web Conf. 2017, 9, 02017. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Chorianopoulos, N.; Nychas, G.-J.E.; Panagou, E.Z. Yeast heterogeneity during spontaneous fermentation of black Conservolea olives in different brine solutions. J. Appl. Microbiol. 2010, 108, 396–405. [Google Scholar] [CrossRef]
- Bonatsou, S.; Paramithiotis, S.; Panagou, E.Z. Evolution of yeast consortia during the fermentation of Kalamata natural black olives upon two initial acidification treatments. Front. Microbiol. 2018, 8, 2673. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Durán-Quintana, M.C.; Ruiz-Barba, J.L.; Querol, A.; Garrido-Fernández, A. Use of molecular methods for the identification of yeast associated with table olives. Food Microbiol. 2006, 23, 791–796. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Romero-Gil, V.; Bautista-Gallego, J.; Rodríguez-Gómez, F.; Jiménez-Díaz, R.; García-García, P.; Querol, A.; Garrido-Fernández, A. Yeasts in table olive processing: Desirable or spoilage microorganisms? Int. J. Food Microbiol. 2012, 160, 42–49. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Selection of yeasts as starter cultures for table olives: A step-by-step procedure. Front. Microbiol. 2012, 31, 194. [Google Scholar] [CrossRef]
- Ruiz-Barba, J.L.; Cortés-Delgado, A.; Sánchez, A.H.; López-López, A.; Montaño, A. Impact of selected wild yeasts starters on the volatilome and phenolic contents of Gordal, Manzanilla and Hojiblanca naturally fermented green olives. Food Sci. Technol. 2024, 195, 115811. [Google Scholar] [CrossRef]
- Traina, C.; Ferrocino, I.; Bonciolini, A.; Cardenia, V.; Lin, X.; Rantsiou, K.; Cocolin, L. Monitoring the yeasts ecology and volatiles profile throughout the spontaneous fermentation of Taggiasca cv. table olives through culture-dependent and independent methods. Int. J. Food Microbiol. 2024, 417, 110688. [Google Scholar] [CrossRef] [PubMed]
- Ciafardini, G.; Zullo, B.A. Use of selected yeast starter cultures in industrial-scale processing of brined Taggiasca black table olives. Food Microbiol. 2019, 84, 103250. [Google Scholar] [CrossRef] [PubMed]
- Camiolo, S.; Porru, C.; Benítez-Cabello, A.; Rodríguez-Gómez, F.; Calero-Delgado, B.; Porceddu, A.; Budroni, M.; Mannazzu, I.; Jiménez-Díaz, R.; Arroyo-López, F.N. Genome overview of eight Candida boidinii strains isolated from human activities and wild environments. Stand. Genom. Sci. 2017, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef]
- Ciafardini, G.; Zullo, B.A. Microbiological activity in stored olive oil. Int. J. Food Microbiol. 2002, 75, 111–118. [Google Scholar] [CrossRef]
- Kong, C.; Zhang, Q.; Wang, Y.; Huang, J.; Li, A.; Tao, Y. Decoding polysaccharides from two Pichia yeasts and their molecular interaction with wine fruity esters. J. Agric. Food Chem. 2024, 72, 12707–12718. [Google Scholar] [CrossRef]
- Colautti, A.; Orecchia, E.; Coppola, F.; Iacumin, L.; Comi, G. Cyberlindnera fabianii, an uncommon yeast responsible for gluten bread spoilage. Foods 2024, 13, 2381. [Google Scholar] [CrossRef]
- Viljoen, B.C. Yeast ecological interactions. Yeast-yeast, yeast-bacteria, yeast-fungi interactions and yeasts as biocontrol agents. In Yeasts in Food and Beverages; Querol, A., Fleet, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 83–110. [Google Scholar]
- Hurtado, A.; Ben Othman, N.; Chammem, N.; Hamdi, M.; Ferrer, S. Characterization of Lactobacillus isolates from fermented olives and their bacteriocin gene profiles. Food Microbiol. 2011, 28, 1514–1518. [Google Scholar] [CrossRef]
- Domínguez-Manzano, J.; Olmo-Ruiz, C.; Bautista-Gallego, J.; Arroyo-López, F.N.; Garrido-Fernández, A.; Jiménez-Díaz, R. Biofilm formation on abiotic and biotic surfaces during Spanish style green table olive fermentation. Int. J. Food Microbiol. 2012, 157, 230–238. [Google Scholar] [CrossRef]
- Benincasa, C.; Muccilli, S.; Amenta, M.; Perri, E.; Romeo, F.V. Phenolic trend and hygienic quality of green table olives fermented with Lactobacillus plantarum starter culture. Food Chem. 2015, 186, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.; Martín, A.; Aranda, E.; Pérez-Nevado, F.; Córdoba, M.G. Identification and characterization of yeast isolated from the elaboration of seasoned green table olives. Food Microbiol. 2007, 24, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Sariguzel, F.M.; Unuvar, G.K.; Kucukoglu, O.; Parkan, O.M.; Koc, A.N. Identification, molecular characterization, and antifungal susceptibility of Cyberlindnera fabianii strains isolated from urinary tract. J. Med. Mycol. 2023, 33, 101429. [Google Scholar] [CrossRef] [PubMed]
- Ciafardini, G.; Zullo, B.A.; Iride, A. Lipase production by yeasts from extra virgin olive oil. Food Microbiol. 2006, 23, 60–67. [Google Scholar] [CrossRef]
- Koidis, A.; Triantafillou, E.; Boskou, D. Endogenous microflora in turbid virgin olive oils and the physicochemical characteristics of these oils. Eur. J. Lipid Sci. Technol. 2008, 110, 164–171. [Google Scholar] [CrossRef]
- Ciafardini, G.; Zullo, B.A. Effect of lipolytic activity of Candida adriatica, Candida diddensiae and Yamadazyma terventina on the acidity of extra-virgin olive oil with a different polyphenol and water content. Food Microbiol. 2015, 47, 12–20. [Google Scholar] [CrossRef]
- Vichi, S.; Romero, A.; Tous, J.; Caixach, J. The activity of healthy olive microbiota during virgin olive oil extraction influences oil chemical composition. J. Agric. Food Chem. 2011, 59, 4705–4714. [Google Scholar] [CrossRef]
- Taylor, J.W. One Fungus = One Name: DNA and Fungal Nomenclature Twenty Years After PCR. IMA Fungus 2011, 2, 113–120. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Shang, Y.J.; Wei, X.Y.; Groenewald, M.; Robert, V.; Zhang, R.P.; Li, A.H.; Han, P.J.; Ji, F.; Li, J.N.; et al. Taxonomic revision of Geotrichum and Magnusiomyces, with the descriptions of five new Geotrichum species from China. Mycology 2024, 15, 400–423. [Google Scholar] [CrossRef]
- Kamilari, E.; Stanton, C.; Reen, F.J.; Ross, R.P. Uncovering the biotechnological importance of Geotrichum candidum. Foods 2023, 12, 1124. [Google Scholar] [CrossRef]
- Giannoutsou, E.P.; Meintanis, C.; Karagouni, A.D. Identification of yeast strains isolated from a two-phase decanter system olive oil waste and investigation of their ability for its fermentation. Bioresour. Technol. 2004, 93, 301–306. [Google Scholar] [CrossRef]
- Asses, N.; Ayed, L.; Bouallagui, H.; Benrejeb, I.; Gargouri, M.; Hamdi, M. Use of Geotrichum candidum for olive mill wastewater treatment in submerged and static culture. Bioresour. Technol. 2009, 100, 2182–2188. [Google Scholar] [CrossRef] [PubMed]
- Bouhia, Y.; Hafidi, M.; Ouhdouch, Y.; El Boukhari, M.E.M.; El Fels, L.; Zeroual, Y.; Lyamlouli, K. Microbial community succession and organic pollutants removal during olive mill waste sludge and green waste co-composting. Front. Microbiol. 2022, 12, 814553. [Google Scholar] [CrossRef] [PubMed]
- Zalar, P.; Gostinčar, C.; de Hoog, G.S.; Uršič, V.; Sudhadham, M.; Gunde-Cimerman, N. Redefinition of Aureobasidium pullulans and its varieties. Stud. Mycol. 2008, 61, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Lei, Y.; Wang, C.; Wei, Y.; Wang, C.; Sun, Y. Patterns of yeast diversity distribution and its drivers in rhizosphere soil of Hami melon orchards in different regions of Xinjiang. BMC Microbiol. 2021, 6, 170. [Google Scholar] [CrossRef]
- Preto, G.; Martins, F.; Pereira, J.A.; Baptista, P. Fungal community in olive fruits of cultivars with different susceptibilities to anthracnose and selection of isolates to be used as biocontrol agents. Biol. Control 2017, 110, 1–9. [Google Scholar] [CrossRef]
- López-García, E.; Benítez-Cabello, A.; Ramiro-García, J.; Romero-Gil, V.; Rodríguez-Gómez, F.; Arroyo-López, F.N. New insights into microbial diversity of the traditional packed table olives Aloreña de Málaga through metataxonomic analysis. Microorganisms 2021, 9, 561. [Google Scholar] [CrossRef]
- Perez, B.A.; Farinon, O.M.; Berretta, M.F. First report of Fusarium solani causing root rot of olive in Southeastern Argentina. Plant Dis. 2011, 95, 1476. [Google Scholar] [CrossRef]
- Chliyeh, M.; Rhimini, Y.; Selmaoui, K.; Touhami, A.O.; Filali-Maltouf, A.; Modafar, C.E.; Moukhli, A.; Oukabli, A.; Benkirane, R.; Douira, A. Survey of the fungal species associated to oive-tree (Olea europaea L.) in Morocco. Int. J. Recent Biotechnol. 2014, 2, 15–32. [Google Scholar]
- Markakis, E.A.; Roditakis, E.N.; Kalantzakis, G.S.; Chatzaki, A.; Soultatos, S.K.; Stavrakaki, M.; Tavlaki, G.I.; Koubouris, G.C.; Bagkis, N.; Goumas, D.E. Characterization of fungi associated with olive fruit rot and olive oil degradation in Crete, Southern Greece. Plant Dis. 2021, 105, 3623–3635. [Google Scholar] [CrossRef]
- Qiao, G.; Zhao, J.; Liu, J.; Tan, X.; Qin, W. Two novel Lasiodiplodia species from blighted stems of Acer truncatum and Cotinus coggygria in China. Biology 2022, 11, 1459. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.; Fernandes, V.; Giraldo-Silva, A.; Roush, D.; Garcia-Pichel, F. Coleofasciculaceae, a monophyletic home for the Microcoleus steenstrupii complex and other desiccation-tolerant filamentous cyanobacteria. J. Phycol. 2021, 57, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, D.O.; Andreote, A.P.D.; Branco, L.H.Z.; Fiore, M.F. Kryptousia macronema gen. nov., sp. nov. and Kryptousia microlepis sp. nov., nostocalean cyanobacteria isolated from phyllospheres. Int. J. Syst. Evol. Microbiol. 2017, 67, 3301–3309. [Google Scholar] [CrossRef] [PubMed]
- Kaštovský, J. Welcome to the jungle!: An overview of modern taxonomy of cyanobacteria. Hydrobiologia 2024, 851, 1063–1077. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.; Hao, H.; et al. Advances in metagenomics and its application in environmental microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef]
- Liu, S.; Moon, C.D.; Zheng, N.; Huws, S.; Zhao, S.; Wang, J. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 2022, 12, 76. [Google Scholar] [CrossRef]
- Petrović, E.; Vrandečić, K.; Ivić, D.; Ćosić, J.; Godena, S. First report of olive branch dieback in Croatia caused by Cytospora pruinosa Défago. Microorganisms 2023, 11, 1679. [Google Scholar] [CrossRef]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A guide from sampling to data analysis. Microb. Inform. Exp. 2012, 9, 3. [Google Scholar] [CrossRef]
- Srinivas, M.; O’Sullivan, O.; Cotter, P.D.; Sinderen, D.v.; Kenny, J.G. The application of metagenomics to study microbial communities and develop desirable traits in fermented foods. Foods 2022, 11, 3297. [Google Scholar] [CrossRef]
- Martins, F.; Rodrigues, N.; Ramalhosa, E. A review of the microbial dynamics of natural and traditional fermentations of table olive. Appl. Microbiol. 2025, 5, 52. [Google Scholar] [CrossRef]
- Cardoni, M.; Mercado-Blanco, J. Confronting stresses affecting olive cultivation from the holobiont perspective. Front. Plant Sci. 2023, 14, 1261754. [Google Scholar] [CrossRef]
- Melloni, R.; Cardoso, E.J.B.N. Microbiome associated with olive cultivation: A review. Plants 2023, 12, 897. [Google Scholar] [CrossRef]
- Ferrocino, I.; Buzzanca, D.; Pagiati, L.; Kazou, M.; Georgalaki, M.; Hatzopoulos, I.; Tsakalidou, E. The microbial terroir of the Greek olive varieties. Int. J. Food Microbiol. 2025, 441, 111332. [Google Scholar] [CrossRef]


| Identification | OU | O0 | O4 | O7 | O15 | O30 | O60 | O90 | Br4 | Br7 | Br15 | Br30 | Br60 | Br90 | Nr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactic Acid Bacteria | 85 | ||||||||||||||
| Lactiplantibacillus spp. | 0 | 0 | 5 | 8 | 9 | 3 | 6 | 28 | 0 | 1 | 2 | 0 | 0 | 2 | 64 |
| Leuconostoc spp. | 3 | 1 | 6 | 5 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 |
| Lactococcus lactis | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Enterococcus spp. | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Enterobacteriaceae | 63 | ||||||||||||||
| Mangrovibacter spp. | 2 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 14 | 11 | 8 | 0 | 0 | 0 | 39 |
| Enterobacter/Raoultella/Klebsiella spp. | 7 | 14 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 |
| Kluyvera intermedia | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Pseudocitrobacter spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Number of isolates per sample/ Total number | 13 | 15 | 13 | 15 | 13 | 6 | 6 | 28 | 15 | 12 | 10 | 0 | 0 | 2 | 148 |
| Identification | OU | O0 | O4 | O7 | O15 | O30 | O60 | O90 | Br4 | Br7 | Br15 | Br30 | Br60 | Br90 | Nr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yeasts | 136 | ||||||||||||||
| Candida spp. | 2 | 4 | 2 | 5 | 6 | 4 | 8 | 4 | 2 | 3 | 3 | 2 | 0 | 3 | 48 |
| Nakazawaea molendinolei | 1 | 0 | 2 | 4 | 4 | 4 | 9 | 6 | 3 | 3 | 2 | 2 | 0 | 3 | 43 |
| Saccharomyces cerevisiae/boulardii/paradoxus | 0 | 0 | 0 | 1 | 3 | 3 | 5 | 3 | 1 | 3 | 0 | 2 | 0 | 0 | 21 |
| Pichia kluyveri/fermentans | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 16 |
| Barnettozymacalifornica | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 6 |
| Rhodotorula glutinis/graminis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Cyberlindnerafabianii | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Yeast-like or non-yeast Fungi | 42 | ||||||||||||||
| Geotrichum candidum/australiense/galactomycetum | 2 | 1 | 3 | 3 | 3 | 3 | 6 | 2 | 0 | 3 | 2 | 3 | 0 | 0 | 31 |
| Aureobasidium pullulans | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| Filobasidium magnum | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Cladosporium halotolerans | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Cladosporium tenellum/herbarum/ramnotenellum/iridis | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Aspergillus amstelodami/cristatus/chevalieri/montevidensis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Fusarium solani/hoffmannii/ambrosium/ensiforme | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Lasiodiplodia macrospora | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Number of isolates per sample/Total number | 9 | 11 | 13 | 15 | 20 | 15 | 29 | 15 | 11 | 14 | 11 | 9 | 0 | 6 | 178 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgalaki, M.; Ferrocino, I.; Buzzanca, D.; Anastasiou, R.; Zoumpopoulou, G.; Giabasakou, D.; Ziova, D.; Kokkali, A.; Paraskevakos, G.; Tsakalidou, E. The Natural Fermentation of Greek Tsounati Olives: Microbiome Analysis. Foods 2025, 14, 2568. https://doi.org/10.3390/foods14152568
Georgalaki M, Ferrocino I, Buzzanca D, Anastasiou R, Zoumpopoulou G, Giabasakou D, Ziova D, Kokkali A, Paraskevakos G, Tsakalidou E. The Natural Fermentation of Greek Tsounati Olives: Microbiome Analysis. Foods. 2025; 14(15):2568. https://doi.org/10.3390/foods14152568
Chicago/Turabian StyleGeorgalaki, Marina, Ilario Ferrocino, Davide Buzzanca, Rania Anastasiou, Georgia Zoumpopoulou, Despoina Giabasakou, Danai Ziova, Alexandra Kokkali, George Paraskevakos, and Effie Tsakalidou. 2025. "The Natural Fermentation of Greek Tsounati Olives: Microbiome Analysis" Foods 14, no. 15: 2568. https://doi.org/10.3390/foods14152568
APA StyleGeorgalaki, M., Ferrocino, I., Buzzanca, D., Anastasiou, R., Zoumpopoulou, G., Giabasakou, D., Ziova, D., Kokkali, A., Paraskevakos, G., & Tsakalidou, E. (2025). The Natural Fermentation of Greek Tsounati Olives: Microbiome Analysis. Foods, 14(15), 2568. https://doi.org/10.3390/foods14152568






