Food Allergenicity Evaluation Methods: Classification, Principle, and Applications
Abstract
1. Introduction
2. The Mechanisms Underlying Food Allergy Development
2.1. Allergenic Epitope and Food Allergy
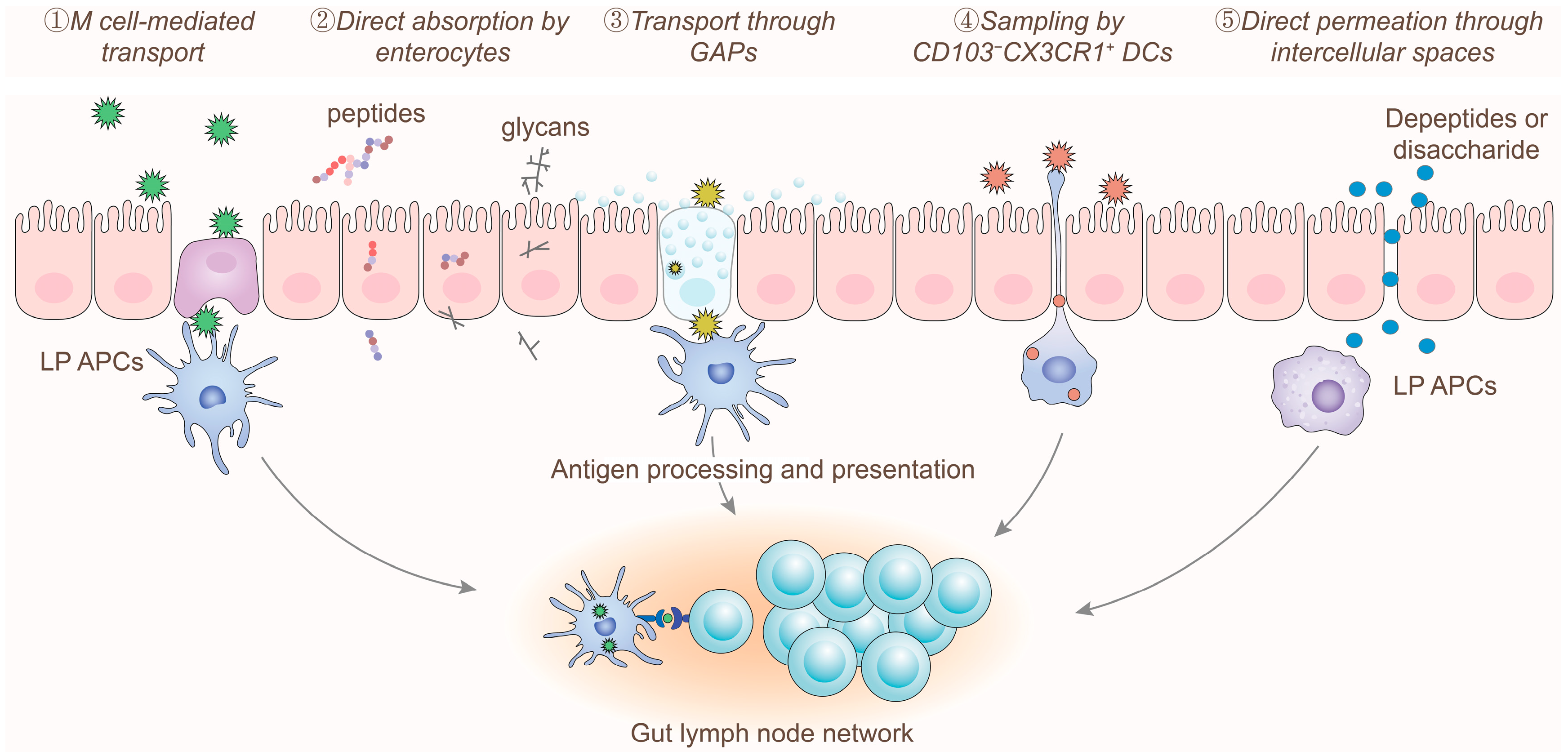
2.2. Digestion and Absorption of Allergens
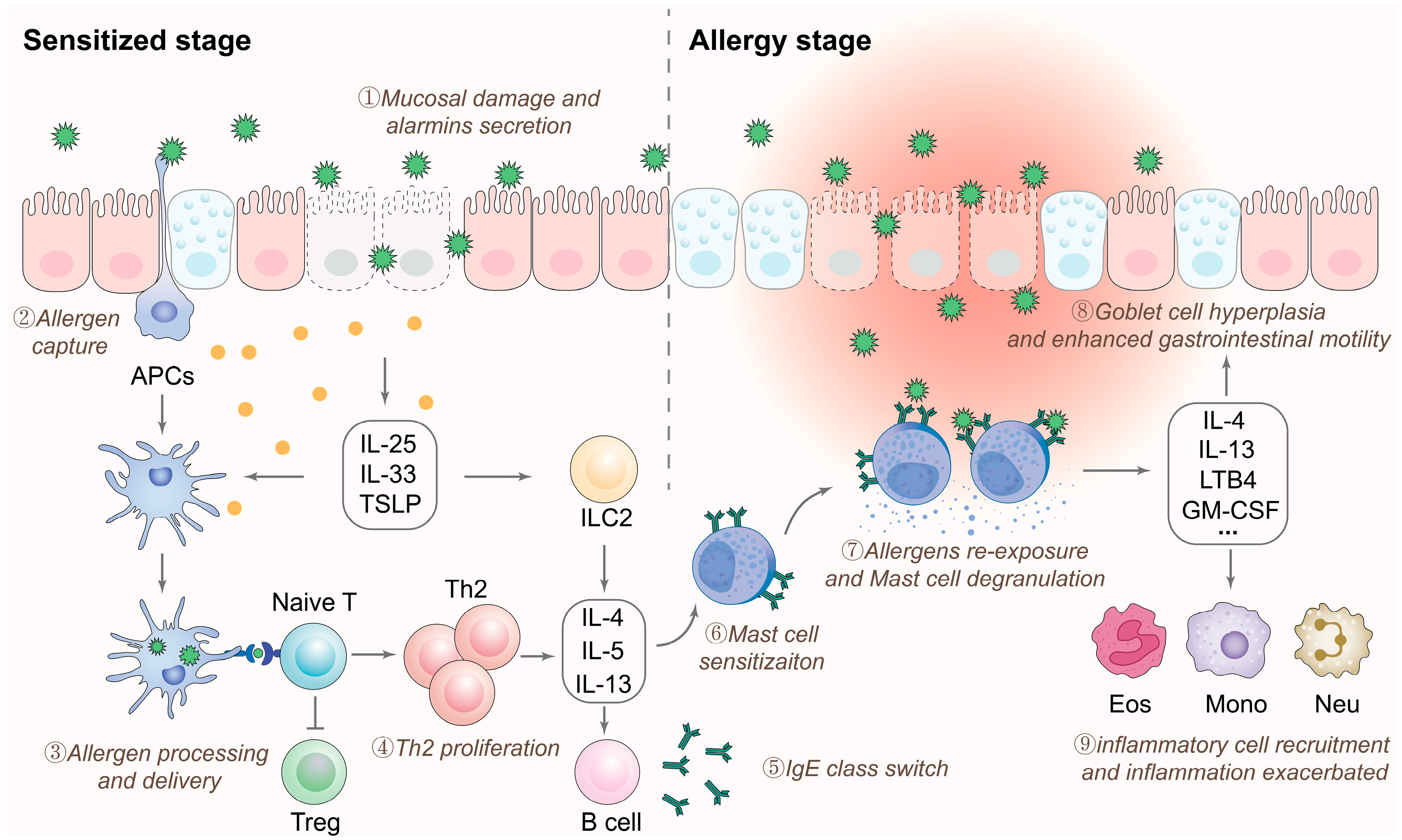
2.3. Immune Responses to Allergens
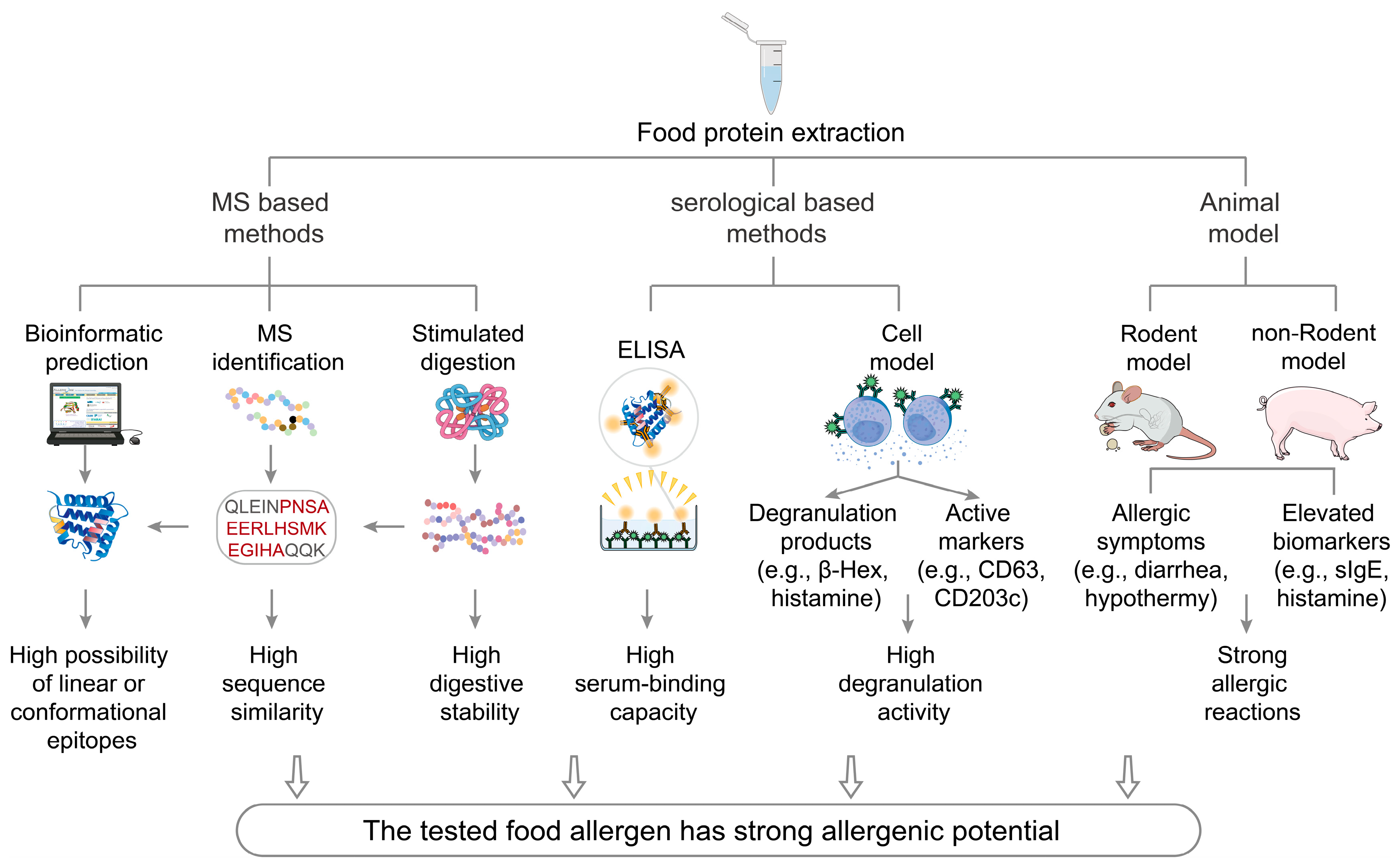
3. Assessment Methods for the Allergenicity of Food Components
3.1. Mass Spectrometry-Based Methods
| Methods | Detection Principle | Advantages | Shortcomings | Applicable Range | Predictive Accuracy |
|---|---|---|---|---|---|
| Mass spectrometry [41,42,43] | Comparing the peptide segments with known epitopes that can be recognized by T/B cells | The procedure is relatively simple and allows for quick results | Large-scale instruments and a reliable database are required, and only linear epitope information can be obtained | Suitable for almost all allergenic foods | Relatively low |
| Bioinformatic prediction [47,48,49,50] | Comparing peptide segments with allergic epitopes and making predictions of potential linear and conformational epitopes | Computational simulation can identify a wide range of potential epitopes | This process demands proficient operational skills and a reliable database; otherwise, inaccurate results may occur | Suitable for almost all allergenic foods | Relatively low |
| Simulated digestion [15,21,22] | Detecting the structure, molecular weight, and allergic epitopes in residual digestion products | It can reflect the effects of different digestion conditions and interactions between food components and allergic epitopes after digestion | The operation is relatively complex, and the experimental results may not reflect the actual situation accurately | Suitable for almost all allergenic foods | Relatively low |
| ELISA examination [51,52,53] | Detecting the allergic epitopes based on the antigen–antibody interaction | Can quickly determine the components responsible for potential allergic reactions in patients | Obtaining suitable serum can be challenging | Suitable for allergenic foods tested with human or animal serum | Medium |
| Cell model [54,55] | The interaction between allergens and allergen-specific IgE can induce mast cell or basophil degranulation | Can accurately determine the presence of substances that trigger mast cell or basophil activation | Obtaining suitable serum can be challenging | Suitable for allergenic foods tested with human or animal serum | Relatively high |
| Animal model [56,57,58] | Sensitizing animals with allergens and observing changes in allergic symptoms and biomarkers after allergen challenge | The experimental results best capture the actual allergenic potential of the tested food components through multiple aspects | The experiment is costly, time-consuming, and may involve ethical issues | Suitable for allergenic foods tested with protein reference material | Relatively high |
3.2. Bioinformatic Prediction
3.3. Simulated Digestion
| Methods | Food Matrix | Experimental Design | Epitope Features | Reference | |
|---|---|---|---|---|---|
| Mass spectrometry-based methods | LC-MS/MS | Cookies | Establishing a fast sample-handling procedure and using the signature peptide sequences to evaluate the allergenic potential. | Linear epitope | [61] |
| LC-MS/MS | Insects | Proteins extracted from cricket and black soldier fly were sequenced and their cross-reactivity to allergens from shrimps was examined. | Linear epitope | [76] | |
| RPLC-ESI-HRMS | Chocolate | Utilizing the signature peptide sequences to search for and align the target allergens. | Linear epitope | [77] | |
| LC-QQQ-MS | Meat-based foodstuffs like cooked meat and sausages | Utilizing the signature peptide and its isotope-labeled peptide as standards and internal standards, respectively. | Linear epitope | [78] | |
| LC-MS/MS | Shrimps | The signature peptides were confirmed and synthesized as the quantitative peptides, and the relative isotope-labeled internal standards were used in the quantitative analysis. | Linear epitope | [79] | |
| Bioinformatic prediction | Machine learning | Microalgae | DIA-NN was used to create a peptide spectrum library from PBQC DIA-MS data, with the “Deep learning-based spectra” option enabled to predict potential allergenicity. | Linear epitope | [80] |
| Database-based searching and prediction | Milk | Utilizing the UniProt protein database to predict key homology sequences and use DNAMAN software to identify allergic sequence alignments. | Linear epitope | [65] | |
| Database-based searching and prediction | Egg products | The linear epitope of potential allergens was predicted based on computational approaches including Immunomedicine Group, ABCpred, BCEpred, BepiPred-3.0 Server, and DNAStar. | Linear epitope | [81] | |
| Machine learning | Sesame | Utilizing AlphaFold2 to predict the 3D structure of potential allergens based on known sequences and evaluate the accuracy of the predicted results using Psi/Phi Ramachandran plots and the WHATCHECK Complete suite. | Conformational epitope | [82] | |
| Machine learning | Fruits and nuts | Peptides similar to known IgE epitopes can be identified with the property distance tool, and conformational epitopes can be identified by the cross–react method, both based on the SDAP webserver. | Conformational epitope | [83] | |
| Simulated digestion | Static simulated digestion | Peanuts | Verifying the correlation between allergenicity and resistance to proteolysis by pepsin. | Linear epitope | [75] |
| Static simulated digestion | Milk | Verifying the contribution of protein phosphorylation to food allergenicity before and after simulated digestion. | Linear epitope | [68] | |
| Dynamic simulated digestion | Shrimps | Examining the effects of digestion stability on food allergenicity during dynamic digestion. | Conformational epitope | [84] | |
| Dynamic simulated digestion | Milk | Evaluating the effects of thermal processing and lactosylation on digestibility and allergenicity. | Conformational epitope | [85] | |
| ELISA examination | Indirect ELISA | Insects | Evaluating the risk of food allergy from eating Gryllus bimaculatus in patients allergic to shrimps, using ELISA and IgE crosslinking luciferase assays. | Conformational epitope | [86] |
| Sandwich ELISA | Crustacean products | Designing a system using polyclonal and monoclonal antibodies to capture and recognize potential allergens. | Linear epitope and Conformational epitope | [87] | |
| Sandwich ELISA | Fish products | Polyclonal antibodies were raised against parvalbumins from 13 fish species of seven fish orders, selected for molecular diversity and immunoreactivity. | Conformational epitope | [88] | |
| Sandwich ELISA | Almonds | Combining sandwich ELISA with lateral flow immunoassay to identify potential allergens. | Conformational epitope | [89] | |
| Cell model | Human peripheral blood basophils | Peanuts | Assessing diagnostic application of natural and recombinant allergens by BAT and identifying IgE-binding epitopes using oligopeptide microarrays. | Linear epitope | [90] |
| KU812 cell model | Soymilk | Utilizing a sensitized cell model to evaluate the changes in spatial structure and sequences after bacterial fermentation. | Linear epitope | [91] | |
| LAD2 cell model | Shrimps | The LAD2 cell degranulation assay was used to assess the binding affinity and antigenicity of the allergenic epitopes. | Linear epitope | [92] | |
| RBL-2H3 cell model | Mealworm | The RBL-2H3 cell model was used to evaluate the allergenicity of mealworm tropomyosin in comparison with shrimp and chicken tropomyosin. | Conformational epitope | [93] | |
| Animal model | BALB/c mice model | Soybean meal | Utilizing the BALB/c mice model to determine whether solid-state fermentation could degrade major allergens and reduce the potential allergenicity of soybean meal. | Linear and conformational epitope | [94] |
| C3H/HeNCrl mice model | Milk | Using the C3H/HeNCrl mice model to examine the susceptibility of potential allergens. | Conformational epitope | [95] | |
| BN rat model | Egg and potato components | Utilizing the BN rat model to determine the potential allergenicity of novel proteins in genetically modified food. | Conformational epitope | [96] | |
| Pig model | Soybean products | Using the pig model to evaluate the allergenicity of soybean protein after deglycosylation and pepsin digestion. | Linear and conformational epitope | [97] | |
| Monkey model | Transgenic rice | Evaluating the allergenicity of transgenic crops by examining the hematological, biochemical, pathological, and histopathological changes in sensitized monkeys. | Linear epitope | [98] | |
3.4. ELISA
3.5. Cell Model
3.6. Animal Model
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| APCs | Antigen-presenting cells | LAD2 | Laboratory of Allergic Diseases 2 |
| BAT | Basophil activation test | LP | Lamina propria |
| β-Hex | β-hexosaminidase | LTB4 | Leukotriene B4 |
| DCs | Dendritic cells | Mono | Monocytes |
| ELISA | Enzyme-linked immunosorbent assays | MS | Mass spectrometry |
| Eos | Eosinophils | Neu | Neutrophils |
| FcεRI | high-affinity IgE receptor | RBL-2H3 | Rat Basophilic Leukemia 2H3 |
| GAPs | Goblet cell-associated antigen passages | Th2 cells | T helper 2 cells |
| GM-CSF | Stimulating factor | Tregs | Regulatory T cells |
| HMC-1 | Human Mast Cell Line 1 | TSLP | Thymic stromal lymphopoietin |
| ILC2s | Lymphoid cells |
References
- Sampath, V.; Abrams, E.M.; Adlou, B.; Akdis, C.; Akdis, M.; Brough, H.A.; Chan, S.; Chatchatee, P.; Chinthrajah, R.S.; Cocco, R.R. Food allergy across the globe. J. Allergy Clin. Immunol. 2021, 148, 1347–1364. [Google Scholar] [CrossRef] [PubMed]
- Sindher, S.B.; Long, A.; Chin, A.R.; Hy, A.; Sampath, V.; Nadeau, K.C.; Chinthrajah, R.S. Food allergy, mechanisms, diagnosis and treatment: Innovation through a multi-targeted approach. Allergy 2022, 77, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Warren, C.M.; Smith, B.M.; Blumenstock, J.A.; Jiang, J.; Davis, M.M.; Nadeau, K.C. The public health impact of parent-reported childhood food allergies in the United States. Pediatrics 2018, 142, e20181235. [Google Scholar] [CrossRef] [PubMed]
- Branum, A.M.; Lukacs, S.L. Food allergy among children in the United States. Pediatrics 2009, 124, 1549–1555. [Google Scholar] [CrossRef]
- Conrado, A.B.; Ierodiakonou, D.; Gowland, M.H.; Boyle, R.J.; Turner, P.J. Food anaphylaxis in the United Kingdom: Analysis of national data, 1998–2018. BMJ 2021, 372, n251. [Google Scholar] [CrossRef]
- Ma, Z.; Chen, L.; Xian, R.; Fang, H.; Wang, J.; Hu, Y. Time trends of childhood food allergy in China: Three cross-sectional surveys in 1999, 2009, and 2019. Pediatr. Allergy Immunol. 2021, 32, 1073–1079. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef]
- Renz, H.; Allen, K.J.; Sicherer, S.H.; Sampson, H.A.; Lack, G.; Beyer, K.; Oettgen, H.C. Food allergy. Nat. Rev. Dis. Prim. 2018, 4, 17098. [Google Scholar] [CrossRef]
- Berin, M.C.; Lozano-Ojalvo, D.; Agashe, C.; Baker, M.G.; Bird, J.A.; Nowak-Wegrzyn, A. Acute FPIES reactions are associated with an IL-17 inflammatory signature. J. Allergy Clin. Immunol. 2021, 148, 895–901.e6. [Google Scholar] [CrossRef]
- Morita, H.; Nomura, I.; Orihara, K.; Yoshida, K.; Akasawa, A.; Tachimoto, H.; Ohtsuka, Y.; Namai, Y.; Futamura, M.; Shoda, T. Antigen-specific T-cell responses in patients with non–IgE-mediated gastrointestinal food allergy are predominantly skewed to TH2. J. Allergy Clin. Immunol. 2013, 131, 590–592.e6. [Google Scholar] [CrossRef]
- Bol-Schoenmakers, M.; Rezende, M.M.; Bleumink, R.; Boon, L.; Man, S.; Hassing, I.; Fiechter, D.; Pieters, R.; Smit, J. Regulation by intestinal γδ T cells during establishment of food allergic sensitization in mice. Allergy 2011, 66, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.; Cianferoni, A.; Spergel, J.; Aceves, S.; Holbreich, M.; Venter, C.; Rothenberg, M.; Terreehorst, I.; Muraro, A.; Lucendo, A. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy 2016, 71, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Pecora, V.; Prencipe, G.; Valluzzi, R.; Dahdah, L.; Insalaco, A.; Cianferoni, A.; De Benedetti, F.; Fiocchi, A. Inflammatory events during food protein-induced enterocolitis syndrome reactions. Pediatr. Allergy Immunol. 2017, 28, 464–470. [Google Scholar] [CrossRef]
- Chinthrajah, R.S.; Hernandez, J.D.; Boyd, S.D.; Galli, S.J.; Nadeau, K.C. Molecular and cellular mechanisms of food allergy and food tolerance. J. Allergy Clin. Immunol. 2016, 137, 984–997. [Google Scholar] [CrossRef]
- EFSA Panel on Genetically Modified Organisms (GMO Panel). Scientific Opinion on the assessment of allergenicity of GM plants and microorganisms and derived food and feed. EFSA J. 2010, 8, 1700. [Google Scholar] [CrossRef]
- Dramburg, S.; Hilger, C.; Santos, A.F.; de Las Vecillas, L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; Altmann, F.; Arruda, K.L.; Asero, R. EAACI molecular allergology user’s guide 2.0. Pediatr. Allergy Immunol. 2023, 34, e13854. [Google Scholar] [CrossRef]
- Zhou, F.; He, S.; Sun, H.; Wang, Y.; Zhang, Y. Advances in epitope mapping technologies for food protein allergens: A review. Trends Food Sci. Technol. 2021, 107, 226–239. [Google Scholar] [CrossRef]
- Kamath, S.D.; Bublin, M.; Kitamura, K.; Matsui, T.; Ito, K.; Lopata, A.L. Cross-reactive epitopes and their role in food allergy. J. Allergy Clin. Immunol. 2023, 151, 1178–1190. [Google Scholar] [CrossRef]
- Liu, C.; Sathe, S.K. Food allergen epitope mapping. J. Agric. Food Chem. 2018, 66, 7238–7248. [Google Scholar] [CrossRef]
- Lee, A.S.E.; Suprun, M.; Sampson, H. Epitope-based IgE assays and their role in providing diagnosis and prognosis of food allergy. J. Allergy Clin. Immunol. Pract. 2023, 11, 2983–2988. [Google Scholar] [CrossRef]
- Sun, N.; Liu, Y.; Liu, K.; Wang, S.; Liu, Q.; Lin, S. Gastrointestinal fate of food allergens and its relationship with allergenicity. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3376–3404. [Google Scholar] [CrossRef] [PubMed]
- Bøgh, K.L.; Madsen, C.B. Food allergens: Is there a correlation between stability to digestion and allergenicity? Crit. Rev. Food Sci. Nutr. 2016, 56, 1545–1567. [Google Scholar] [CrossRef] [PubMed]
- Vanga, S.K.; Singh, A.; Raghavan, V. Review of conventional and novel food processing methods on food allergens. Crit. Rev. Food Sci. Nutr. 2017, 57, 2077–2094. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Lin, S.; Sun, N. How does food matrix components affect food allergies, food allergens and the detection of food allergens? A systematic review. Trends Food Sci. Technol. 2022, 127, 280–290. [Google Scholar] [CrossRef]
- Yang, H.; Qu, Y.; Gao, Y.; Sun, S.; Ding, R.; Cang, W.; Wu, R.; Wu, J. Role of the dietary components in food allergy: A comprehensive review. Food Chem. 2022, 386, 132762. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Wang, Z.; Meng, X.; Hoffmann-Sommergruber, K.; Cavallari, N.; Wu, Y.; Gao, J.; Li, X.; Chen, H. Goblet cell-associated antigen passage: A gatekeeper of the intestinal immune system. Immunology 2023, 170, 1–12. [Google Scholar] [CrossRef]
- Newberry, R.D.; Hogan, S.P. Intestinal epithelial cells in tolerance and allergy to dietary antigens. J. Allergy Clin. Immunol. 2021, 147, 45–48. [Google Scholar] [CrossRef]
- Perrier, C.; Corthesy, B. Gut permeability and food allergies. Clin. Exp. Allergy 2011, 41, 20–28. [Google Scholar] [CrossRef]
- Noah, T.K.; Knoop, K.A.; McDonald, K.G.; Gustafsson, J.K.; Waggoner, L.; Vanoni, S.; Batie, M.; Arora, K.; Naren, A.P.; Wang, Y.-H. IL-13–induced intestinal secretory epithelial cell antigen passages are required for IgE-mediated food-induced anaphylaxis. J. Allergy Clin. Immunol. 2019, 144, 1058–1073.e1053. [Google Scholar] [CrossRef]
- Knoop, K.A.; Kulkarni, D.H.; McDonald, K.G.; Gustafsson, J.K.; Davis, J.E.; Floyd, A.N.; Newberry, R.D. In vivo labeling of epithelial cell–associated antigen passages in the murine intestine. Lab Anim. 2020, 49, 79–88. [Google Scholar] [CrossRef]
- Niess, J.H.; Brand, S.; Gu, X.; Landsman, L.; Jung, S.; McCormick, B.A.; Vyas, J.M.; Boes, M.; Ploegh, H.L.; Fox, J.G. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 2005, 307, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Parrish, A.; Boudaud, M.; Kuehn, A.; Ollert, M.; Desai, M.S. Intestinal mucus barrier: A missing piece of the puzzle in food allergy. Trends Mol. Med. 2022, 28, 36–50. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, B.; Zhang, Y.; Zeng, H. Food allergy and gut microbiota. Trends Food Sci. Technol. 2023, 140, 104141. [Google Scholar] [CrossRef]
- Breiteneder, H.; Mills, E.C. Molecular properties of food allergens. J. Allergy Clin. Immunol. 2005, 115, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Wegrzyn, A.; Szajewska, H.; Lack, G. Food allergy and the gut. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 241–257. [Google Scholar] [CrossRef]
- Lee, K.H.; Song, Y.; Wu, W.; Yu, K.; Zhang, G. The gut microbiota, environmental factors, and links to the development of food allergy. Clin. Mol. Allergy 2020, 18, 5. [Google Scholar] [CrossRef]
- Poto, R.; Fusco, W.; Rinninella, E.; Cintoni, M.; Kaitsas, F.; Raoul, P.; Caruso, C.; Mele, M.C.; Varricchi, G.; Gasbarrini, A. The role of gut microbiota and leaky gut in the pathogenesis of food allergy. Nutrients 2023, 16, 92. [Google Scholar] [CrossRef]
- Santos, A.F.; Riggioni, C.; Agache, I.; Akdis, C.A.; Akdis, M.; Alvarez-Perea, A.; Alvaro-Lozano, M.; Ballmer-Weber, B.; Barni, S.; Beyer, K. EAACI guidelines on the diagnosis of IgE-mediated food allergy. Allergy 2023, 78, 3057–3076. [Google Scholar] [CrossRef]
- Tedner, S.G.; Asarnoj, A.; Thulin, H.; Westman, M.; Konradsen, J.R.; Nilsson, C. Food allergy and hypersensitivity reactions in children and adults—A review. J. Intern. Med. 2022, 291, 283–302. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Meng, X.; Wu, Y.; Gao, J.; Chen, H.; Li, X. Extracellular adenosine triphosphate: A new gateway for food allergy mechanism research? Food Chem. 2024, 141821. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, X.; Shen, Y.; Meng, X.; Wu, Y.; Tong, P.; Li, X.; Chen, H.; Gao, J. Quantifying allergenic proteins using antibody-based methods or liquid chromatography–mass spectrometry/mass spectrometry: A review about the influence of food matrix, extraction, and sample preparation. Compr. Rev. Food Sci. Food Saf. 2024, 23, e70029. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Abril, A.G.; Pazos, M.; Calo-Mata, P.; Villa, T.G.; Barros-Velázquez, J. Proteins and peptides: Proteomics approaches for food authentication and allergen profiling. Curr. Opin. Food Sci. 2024, 101172. [Google Scholar] [CrossRef]
- Birse, N.; Burns, D.T.; Walker, M.J.; Quaglia, M.; Elliott, C.T. Food allergen analysis: A review of current gaps and the potential to fill them by matrix-assisted laser desorption/ionization. Compr. Rev. Food Sci. Food Saf. 2023, 22, 3984–4003. [Google Scholar] [CrossRef] [PubMed]
- Oeo-Santos, C.; Mas, S.; Benedé, S.; López-Lucendo, M.; Quiralte, J.; Blanca, M.; Mayorga, C.; Villalba, M.; Barderas, R. A recombinant isoform of the Ole e 7 olive pollen allergen assembled by de novo mass spectrometry retains the allergenic ability of the natural allergen. J. Proteom. 2018, 187, 39–46. [Google Scholar] [CrossRef]
- Bogdanov, B.; Smith, R.D. Proteomics by FTICR mass spectrometry: Top down and bottom up. Mass Spectrom. Rev. 2005, 24, 168–200. [Google Scholar] [CrossRef]
- Bianco, M.; Ventura, G.; Calvano, C.D.; Losito, I.; Cataldi, T.R. Food allergen detection by mass spectrometry: From common to novel protein ingredients. Proteomics 2023, 23, 2200427. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Liu, G.; Fu, L. Food allergomics based on high-throughput and bioinformatics technologies. Food Res. Int. 2020, 130, 108942. [Google Scholar] [CrossRef]
- Barre, A.; Pichereaux, C.; Simplicien, M.; Burlet-Schiltz, O.; Benoist, H.; Rougé, P. A proteomic-and bioinformatic-based identification of specific allergens from edible insects: Probes for future detection as food ingredients. Foods 2021, 10, 280. [Google Scholar] [CrossRef]
- Yang, Y.; He, X.; Li, F.; He, S.; Liu, M.; Li, M.; Xia, F.; Su, W.; Liu, G. Animal-derived food allergen: A review on the available crystal structure and new insights into structural epitope. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13340. [Google Scholar] [CrossRef]
- Sharma, N.; Patiyal, S.; Dhall, A.; Pande, A.; Arora, C.; Raghava, G.P. AlgPred 2.0: An improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief. Bioinform. 2021, 22, bbaa294. [Google Scholar] [CrossRef]
- Sheng, K.; Jiang, H.; Fang, Y.; Wang, L.; Jiang, D. Emerging electrochemical biosensing approaches for detection of allergen in food samples: A review. Trends Food Sci. Technol. 2022, 121, 93–104. [Google Scholar] [CrossRef]
- Morais, S.; Tortajada-Genaro, L.A.; Maquieira, Á.; Martinez, M.-Á.G. Biosensors for food allergy detection according to specific IgE levels in serum. TrAC Trends Anal. Chem. 2020, 127, 115904. [Google Scholar] [CrossRef]
- Peng, P.; Liu, C.; Li, Z.; Xue, Z.; Mao, P.; Hu, J.; Xu, F.; Yao, C.; You, M. Emerging ELISA derived technologies for in vitro diagnostics. TrAC Trends Anal. Chem. 2022, 152, 116605. [Google Scholar] [CrossRef]
- Huang, J.; Liu, C.; Wang, Y.; Wang, C.; Xie, M.; Qian, Y.; Fu, L. Application of in vitro and in vivo models in the study of food allergy. Food Sci. Hum. Wellness 2018, 7, 235–243. [Google Scholar] [CrossRef]
- Oettgen, H.C. Mast cells in food allergy: Inducing immediate reactions and shaping long-term immunity. J. Allergy Clin. Immunol. 2023, 151, 21–25. [Google Scholar] [CrossRef]
- Castan, L.; Bøgh, K.L.; Maryniak, N.Z.; Epstein, M.M.; Kazemi, S.; O’Mahony, L.; Bodinier, M.; Smit, J.J.; Van Bilsen, J.H.; Blanchard, C. Overview of in vivo and ex vivo endpoints in murine food allergy models: Suitable for evaluation of the sensitizing capacity of novel proteins? Allergy 2020, 75, 289–301. [Google Scholar] [CrossRef]
- Liu, T.; Navarro, S.; Lopata, A.L. Current advances of murine models for food allergy. Mol. Immunol. 2016, 70, 104–117. [Google Scholar] [CrossRef]
- Larsen, J.M.; Bøgh, K.L. Animal models of allergen-specific immunotherapy in food allergy: Overview and opportunities. Clin. Exp. Allergy 2018, 48, 1255–1274. [Google Scholar] [CrossRef]
- Monaci, L.; Nørgaard, J.V.; van Hengel, A.J. Feasibility of a capillary LC/ESI-Q-TOF MS method for the detection of milk allergens in an incurred model food matrix. Anal. Methods 2010, 2, 967–972. [Google Scholar] [CrossRef]
- Di Stasio, L.; Picariello, G.; Mongiello, M.; Nocerino, R.; Canani, R.B.; Bavaro, S.; Monaci, L.; Ferranti, P.; Mamone, G. Peanut digestome: Identification of digestion resistant IgE binding peptides. Food Chem. Toxicol. 2017, 107, 88–98. [Google Scholar] [CrossRef]
- Pilolli, R.; De Angelis, E.; Monaci, L. Streamlining the analytical workflow for multiplex MS/MS allergen detection in processed foods. Food Chem. 2017, 221, 1747–1753. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Blundell, T.L.; Mizuguchi, K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001, 310, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Fiers, M.W.; Kleter, G.A.; Nijland, H.; Peijnenburg, A.A.; Nap, J.P.; Van Ham, R.C. Allermatch™, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. BMC Bioinform. 2004, 5, 133. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006, 34, W202–W209. [Google Scholar] [CrossRef]
- Hu, W.; Xie, F.; Wu, Y.; Meng, X.; Yang, A.; Wu, Z.; Gao, J.; Li, X.; Chen, H. Identification and Validation of Key Amino Acids in IgE Linear Epitopes of β-Lactoglobulin: Comparison of Recognition Patterns of Chinese Bovine Milk-Allergic Sera with Different Symptoms. J. Agric. Food Chem. 2025, 73, 5537–5547. [Google Scholar] [CrossRef]
- Garino, C.; Coïsson, J.D.; Arlorio, M. In silico allergenicity prediction of several lipid transfer proteins. Comput. Biol. Chem. 2016, 60, 32–42. [Google Scholar] [CrossRef]
- Wang, L.; Niu, D.; Zhao, X.; Wang, X.; Hao, M.; Che, H. A comparative analysis of novel deep learning and ensemble learning models to predict the allergenicity of food proteins. Foods 2021, 10, 809. [Google Scholar] [CrossRef]
- Chen, W.-M.; Shao, Y.-H.; Wang, Z.; Liu, J.; Tu, Z.-C. Simulated in vitro digestion of α-lactalbumin modified by phosphorylation: Detection of digestive products and allergenicity. Food Chem. 2022, 372, 131308. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Xavier, A.A.; Mariutti, L.R. Static and semi-dynamic in vitro digestion methods: State of the art and recent achievements towards standardization. Curr. Opin. Food Sci. 2021, 41, 260–273. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Prodić, I.; Krstić Ristivojević, M.; Smiljanić, K. Antioxidant properties of protein-rich plant foods in gastrointestinal digestion—Peanuts as our antioxidant friend or foe in allergies. Antioxidants 2023, 12, 886. [Google Scholar] [CrossRef] [PubMed]
- Prodić, I.; Smiljanić, K.; Simović, A.; Radosavljević, J.; Ćirković Veličković, T. Thermal processing of peanut grains impairs their mimicked gastrointestinal digestion while downstream defatting treatments affect digestomic profiles. Foods 2019, 8, 463. [Google Scholar] [CrossRef] [PubMed]
- Ménard, O.; Lesmes, U.; Shani-Levi, C.; Calahorra, A.A.; Lavoisier, A.; Morzel, M.; Rieder, A.; Feron, G.; Nebbia, S.; Mashiah, L. Static in vitro digestion model adapted to the general older adult population: An INFOGEST international consensus. Food Funct. 2023, 14, 4569–4582. [Google Scholar] [CrossRef]
- Prodic, I.; Stanic-Vucinic, D.; Apostolovic, D.; Mihailovic, J.; Radibratovic, M.; Radosavljevic, J.; Burazer, L.; Milcic, M.; Smiljanic, K.; van Hage, M. Influence of peanut matrix on stability of allergens in gastric-simulated digesta: 2S albumins are main contributors to the IgE reactivity of short digestion-resistant peptides. Clin. Exp. Allergy 2018, 48, 731–740. [Google Scholar] [CrossRef]
- Karnaneedi, S.; Johnston, E.B.; Bose, U.; Juhász, A.; Broadbent, J.A.; Ruethers, T.; Jerry, E.M.; Kamath, S.D.; Limviphuvadh, V.; Stockwell, S. The allergen profile of two edible insect species—Acheta domesticus and Hermetia illucens. Mol. Nutr. Food Res. 2024, 68, 2300811. [Google Scholar] [CrossRef]
- Bianco, M.; Calvano, C.D.; Ventura, G.; Losito, I.; Cataldi, T.R. Determination of hidden milk allergens in meat-based foodstuffs by liquid chromatography coupled to electrospray ionization and high-resolution tandem mass spectrometry. Food Control 2022, 131, 108443. [Google Scholar] [CrossRef]
- Ho, C.-W.; Hsu, J.-L.; Chen, S.-H.; Liaw, E.-T.; Liu, S.-S.; Huang, E.S.; Chen, Y.-K.; Huang, C.-C.J.; Yu, H.-S. Development and validation of mass spectrometry-based method for detecting shrimp allergen tropomyosin. LWT 2021, 152, 112367. [Google Scholar] [CrossRef]
- Fan, S.; Ma, J.; Liu, Z.; Ning, Y.; Cao, M.; Li, Q.; Zhang, Y. Determination of egg and milk allergen in food products by liquid chromatography-tandem mass spectrometry based on signature peptides and isotope-labeled internal standard. Food Sci. Hum. Wellness 2023, 12, 728–736. [Google Scholar] [CrossRef]
- Hamzelou, S.; Belobrajdic, D.; Juhász, A.; Brook, H.; Bose, U.; Colgrave, M.L.; Broadbent, J.A. Nutrition, allergenicity and physicochemical qualities of food-grade protein extracts from Nannochloropsis oculate. Food Chem. 2023, 424, 136459. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, L.; Raghavan, V.; Liu, Y.; Wang, J. Analysis of epitopes and structural responses in egg allergen Gal d 1 using bioinformatic tools and molecular dynamics simulation. Curr. Res. Food Sci. 2025, 101048. [Google Scholar] [CrossRef]
- He, S.; Gao, K.; Pan, T.; Wu, Y.; Di, D.; Li, X.; Sun, H.; Zhang, Y. Exploring the allergenic potential of sesame oleosins: Isolation and bioinformatics analysis. Int. J. Biol. Macromol. 2024, 280, 135997. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.S.; Schein, C.H.; Braun, W. The updated Structural Database of Allergenic Proteins (SDAP 2.0) provides 3D models for allergens and incorporated bioinformatics tools. J. Allergy Clin. Immunol. Glob. 2023, 2, 100162. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Luo, C.; Xu, L.; Lin, H.; Li, Z.; Pavase, T.R.; Wu, Y.; Chen, Y. Comparison of digestibility and potential allergenicity of raw shrimp (Litopenaeus vannamei) extracts in static and dynamic digestion systems. Food Chem. 2021, 345, 128831. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, A.; Benedé, S.; Tedeschi, T.; Sforza, S.; Recio, I.; Miralles, B. In vitro simulated semi-dynamic gastrointestinal digestion: Evaluation of the effects of processing on whey proteins digestibility and allergenicity. Food Funct. 2022, 13, 1593–1602. [Google Scholar] [CrossRef]
- Kamemura, N.; Sugimoto, M.; Tamehiro, N.; Adachi, R.; Tomonari, S.; Watanabe, T.; Mito, T. Cross-allergenicity of crustacean and the edible insect Gryllus bimaculatus in patients with shrimp allergy. Mol. Immunol. 2019, 106, 127–134. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Li, R.; Timira, V.; Dasanayaka, B.P.; Zhang, Z.; Zhang, J.; Lin, H.; Li, Z. Evaluation of poly-and monoclonal antibody-based sandwich enzyme-linked immunosorbent assay (ELISA) for their performance to detect crustacean residues in processed foods. Food Control 2022, 138, 108983. [Google Scholar] [CrossRef]
- Liang, J.; Taylor, S.L.; Baumert, J.; Lee, N.A. Development of a sensitive sandwich ELISA with broad species specificity for improved fish allergen detection. Food Chem. 2022, 396, 133656. [Google Scholar] [CrossRef]
- Civera, A.; Galan-Malo, P.; Segura-Gil, I.; Mata, L.; Tobajas, A.P.; Sánchez, L.; Pérez, M.D. Development of sandwich ELISA and lateral flow immunoassay to detect almond in processed food. Food Chem. 2022, 371, 131338. [Google Scholar] [CrossRef]
- Schwager, C.; Kull, S.; Behrends, J.; Röckendorf, N.; Schocker, F.; Frey, A.; Homann, A.; Becker, W.-M.; Jappe, U. Peanut oleosins associated with severe peanut allergy—Importance of lipophilic allergens for comprehensive allergy diagnostics. J. Allergy Clin. Immunol. 2017, 140, 1331–1338.e1338. [Google Scholar] [CrossRef]
- Lu, Q.; Zuo, L.; Wu, Z.; Li, X.; Tong, P.; Wu, Y.; Fan, Q.; Chen, H.; Yang, A. Characterization of the protein structure of soymilk fermented by Lactobacillus and evaluation of its potential allergenicity based on the sensitized-cell model. Food Chem. 2022, 366, 130569. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wang, J.; Ni, S.; Wang, C.; Wang, Y. Identification of allergenic epitopes and critical amino acids of major allergens in Chinese shrimp (Penaeus chinensis) by immunoinformatics coupled with competitive-binding strategy. J. Agric. Food Chem. 2018, 66, 2944–2953. [Google Scholar] [CrossRef] [PubMed]
- Klueber, J.; Costa, J.; Randow, S.; Codreanu-Morel, F.; Verhoeckx, K.; Bindslev-Jensen, C.; Ollert, M.; Hoffmann-Sommergruber, K.; Morisset, M.; Holzhauser, T. Homologous tropomyosins from vertebrate and invertebrate: Recombinant calibrator proteins in functional biological assays for tropomyosin allergenicity assessment of novel animal foods. Clin. Exp. Allergy 2020, 50, 105–116. [Google Scholar] [CrossRef]
- Yang, A.; Zuo, L.; Cheng, Y.; Wu, Z.; Li, X.; Tong, P.; Chen, H. Degradation of major allergens and allergenicity reduction of soybean meal through solid-state fermentation with microorganisms. Food Funct. 2018, 9, 1899–1909. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Shao, H.; Zhang, X.; Yang, F.; Wang, J.; Tan, S.; Chen, H.; Li, X. Comparison of cow’s milk allergy models highlighted higher humoral and Th2 immune responses in BALB/c than C3H/HeNCrl mice. Food Chem. Toxicol. 2024, 184, 114315. [Google Scholar] [CrossRef]
- Jia, X.-D.; Li, N.; Wu, Y.-N.; Yang, X.-G. Studies on BN rats model to determine the potential allergenicity of proteins from genetically modified foods. World J. Gastroenterol. 2005, 11, 5381. [Google Scholar] [CrossRef]
- Fu, C.J.; Jez, J.M.; Kerley, M.S.; Allee, G.L.; Krishnan, H.B. Identification, characterization, epitope mapping, and three-dimensional modeling of the α-subunit of β-conglycinin of soybean, a potential allergen for young pigs. J. Agric. Food Chem. 2007, 55, 4014–4020. [Google Scholar] [CrossRef]
- Domon, E.; Takagi, H.; Hirose, S.; Sugita, K.; Kasahara, S.; Ebinuma, H.; Takaiwa, F. 26-Week oral safety study in macaques for transgenic rice containing major human T-cell epitope peptides from Japanese cedar pollen allergens. J. Agric. Food Chem. 2009, 57, 5633–5638. [Google Scholar] [CrossRef]
- Wu, L.; Li, G.; Xu, X.; Zhu, L.; Huang, R.; Chen, X. Application of nano-ELISA in food analysis: Recent advances and challenges. TrAC Trends Anal. Chem. 2019, 113, 140–156. [Google Scholar] [CrossRef]
- Ahmed, S.; Ning, J.; Peng, D.; Chen, T.; Ahmad, I.; Ali, A.; Lei, Z.; Abu bakr Shabbir, M.; Cheng, G.; Yuan, Z. Current advances in immunoassays for the detection of antibiotics residues: A review. Food Agric. Immunol. 2020, 31, 268–290. [Google Scholar] [CrossRef]
- He, S.; Xiong, M.; Li, L.; Yan, Y.; Li, J.; Feng, Z.; Li, Y.; Zhao, J.; Dong, Y.; Li, X. One-step purification of IgE epitope-specific antibody using immunomagnetic beads and highly sensitive detection of bovine β-lactoglobulin for the prediction of milk allergenicity in foods. J. Agric. Food Chem. 2023, 71, 14068–14078. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Li, X.; Wu, Y.; Wu, S.; Wu, Z.; Yang, A.; Tong, P.; Yuan, J.; Gao, J.; Chen, H. Highly sensitive detection of bovine β-lactoglobulin with wide linear dynamic range based on platinum nanoparticles probe. J. Agric. Food Chem. 2018, 66, 11830–11838. [Google Scholar] [CrossRef]
- Pandey, A.K.; Varshney, R.K.; Sudini, H.K.; Pandey, M.K. An improved enzyme-linked immunosorbent assay (ELISA) based protocol using seeds for detection of five major peanut allergens Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8. Front. Nutr. 2019, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Aquino, A.; Conte-Junior, C.A. A systematic review of food allergy: Nanobiosensor and food allergen detection. Biosensors 2020, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Gaudenzio, N.; Tsai, M. Mast cells in inflammation and disease: Recent progress and ongoing concerns. Annu. Rev. Immunol. 2020, 38, 49–77. [Google Scholar] [CrossRef]
- Passante, E.; Frankish, N. The RBL-2H3 cell line: Its provenance and suitability as a model for the mast cell. Inflamm. Res. 2009, 58, 737–745. [Google Scholar] [CrossRef]
- Akula, S.; Paivandy, A.; Fu, Z.; Thorpe, M.; Pejler, G.; Hellman, L. How relevant are bone marrow-derived mast cells (BMMCs) as models for tissue mast cells? A comparative transcriptome analysis of BMMCs and peritoneal mast cells. Cells 2020, 9, 2118. [Google Scholar] [CrossRef]
- Jiang, D.; Jiang, H.; Ji, J.; Sun, X.; Qian, H.; Zhang, G.; Tang, L. Mast-cell-based fluorescence biosensor for rapid detection of major fish allergen parvalbumin. J. Agric. Food Chem. 2014, 62, 6473–6480. [Google Scholar] [CrossRef]
- Vogel, L.; Lüttkopf, D.; Hatahet, L.; Haustein, D.; Vieths, S. Development of a functional in vitro assay as a novel tool for the standardization of allergen extracts in the human system. Allergy 2005, 60, 1021–1028. [Google Scholar] [CrossRef]
- Santos, A.F.; Alpan, O.; Hoffmann, H.J. Basophil activation test: Mechanisms and considerations for use in clinical trials and clinical practice. Allergy 2021, 76, 2420–2432. [Google Scholar] [CrossRef]
- Hemmings, O.; Kwok, M.; McKendry, R.; Santos, A.F. Basophil activation test: Old and new applications in allergy. Curr. Allergy Asthma Rep. 2018, 18, 77. [Google Scholar] [CrossRef] [PubMed]
- Sabato, V.; Van Hengel, A.; De Knop, K.; Verweij, M.; Hagendorens, M.; Bridts, C.; De Clerck, L.; Schiavino, D.; Stevens, W.; Ebo, D. Human basophils: A unique biological instrument to detect the allergenicity of food. J. Investig. Allergol. Clin. Immunol. Off. Organ Int. Assoc. Asthmology (INTERASMA) Soc. Latinoam. Alerg. Inmunol./Int. Assoc. Asthmology-Barc. 2011, 21, 179–184. [Google Scholar]
- Jiménez-Saiz, R.; Chu, D.K.; Mandur, T.S.; Walker, T.D.; Gordon, M.E.; Chaudhary, R.; Koenig, J.; Saliba, S.; Galipeau, H.J.; Utley, A. Lifelong memory responses perpetuate humoral TH2 immunity and anaphylaxis in food allergy. J. Allergy Clin. Immunol. 2017, 140, 1604–1615.e1605. [Google Scholar] [CrossRef] [PubMed]
- Justice, M.J.; Siracusa, L.D.; Stewart, A.F. Technical approaches for mouse models of human disease. Dis. Models Mech. 2011, 4, 305–310. [Google Scholar] [CrossRef]
- Burton, O.T.; Stranks, A.J.; Tamayo, J.M.; Koleoglou, K.J.; Schwartz, L.B.; Oettgen, H.C. A humanized mouse model of anaphylactic peanut allergy. J. Allergy Clin. Immunol. 2017, 139, 314–322.e9. [Google Scholar] [CrossRef]
- Fotschki, J.; Szyc, A.; Laparra, J.; Markiewicz, L.; Wróblewska, B. Immune-modulating properties of horse milk administered to mice sensitized to cow milk. J. Dairy Sci. 2016, 99, 9395–9404. [Google Scholar] [CrossRef]
- Wavrin, S.; Bernard, H.; Wal, J.-M.; Adel-Patient, K. Cutaneous or respiratory exposures to peanut allergens in mice and their impacts on subsequent oral exposure. Int. Arch. Allergy Immunol. 2014, 164, 189–199. [Google Scholar] [CrossRef]
- Li, Q.; Hu, W.; Zhao, J.; Wang, J.; Dai, Y.; Zhao, Y.; Meng, Q.; Li, N. Supplementation transgenic cow’s milk containing recombinant human lactoferrin enhances systematic and intestinal immune responses in piglets. Mol. Biol. Rep. 2014, 41, 2119–2128. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Chen, X.; Li, X. Food Allergenicity Evaluation Methods: Classification, Principle, and Applications. Foods 2025, 14, 2005. https://doi.org/10.3390/foods14122005
Zhou H, Chen X, Li X. Food Allergenicity Evaluation Methods: Classification, Principle, and Applications. Foods. 2025; 14(12):2005. https://doi.org/10.3390/foods14122005
Chicago/Turabian StyleZhou, Huiqiao, Xiao Chen, and Xin Li. 2025. "Food Allergenicity Evaluation Methods: Classification, Principle, and Applications" Foods 14, no. 12: 2005. https://doi.org/10.3390/foods14122005
APA StyleZhou, H., Chen, X., & Li, X. (2025). Food Allergenicity Evaluation Methods: Classification, Principle, and Applications. Foods, 14(12), 2005. https://doi.org/10.3390/foods14122005






