Ultrasonic-Assisted Extraction of Polysaccharides from Brassica rapa L. and Its Effects on Gut Microbiota in Humanized Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Extraction Process
2.3. Determination of Chemical Composition
2.4. Determination of Monosaccharide and Molecular Weight
2.5. FT-IR and SEM Analysis
2.6. Animal Experiments
Construction of a Humanized Microbiota Mice Model
2.7. Measurement of Short-Chain Fatty Acids (SCFAs)
2.8. 16S rDNA Gene and Bioinformatics Analysis
2.9. Statistical Analysis
3. Results
3.1. Preparation Process of BRAP
3.2. Structural Analysis and Chemical Composition of BRAP1-1
3.2.1. Molecular Weight Determination and Monosaccharide Composition
3.2.2. FT-IR Analysis
3.2.3. SEM Analysis
3.2.4. Chemical Composition
3.3. Gut Microbiota Analysis
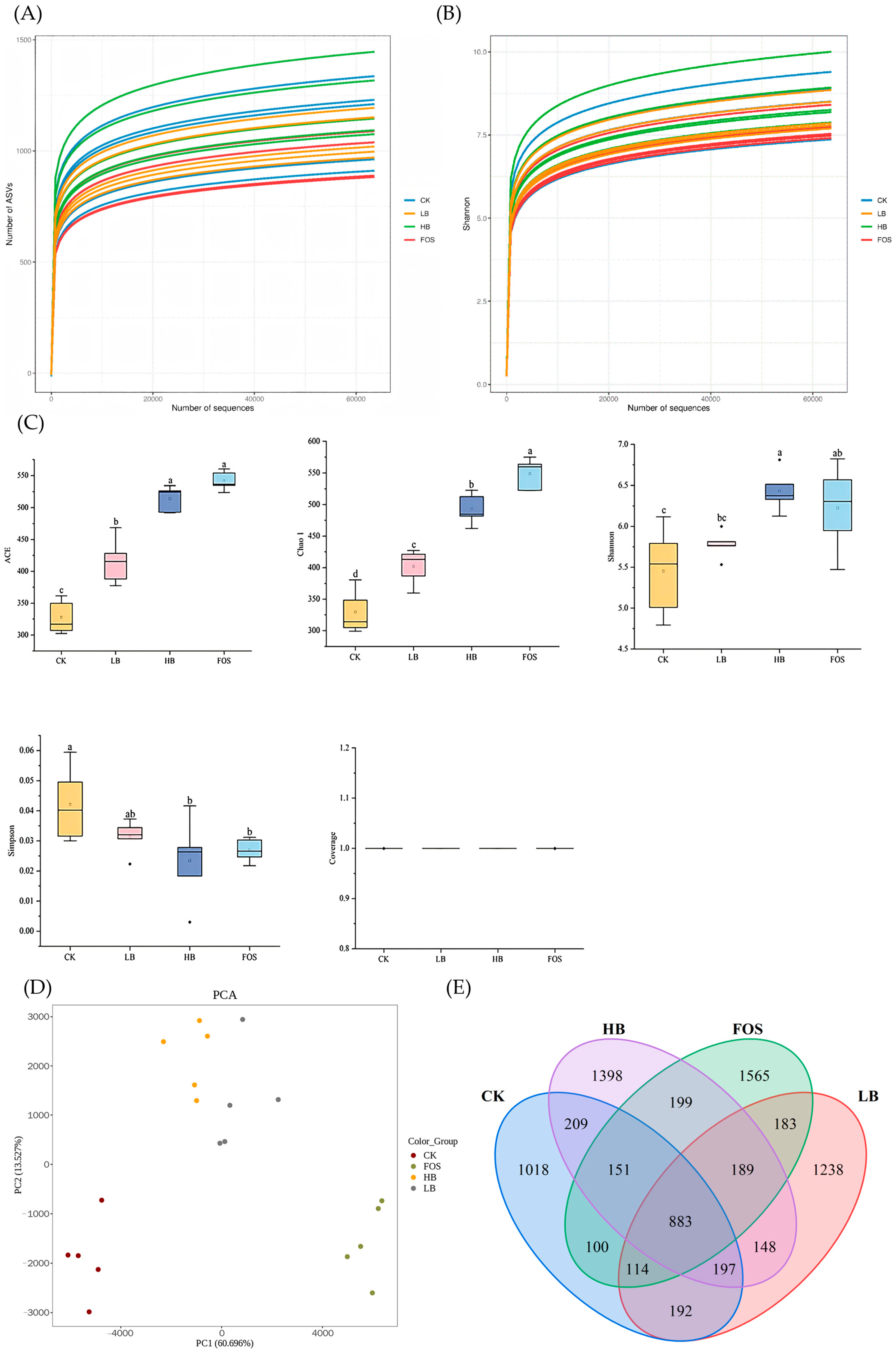
3.3.1. Dilution Curves and Shannon Index Curves
3.3.2. Microbial Alpha Diversity
3.3.3. Microbial Beta Diversity
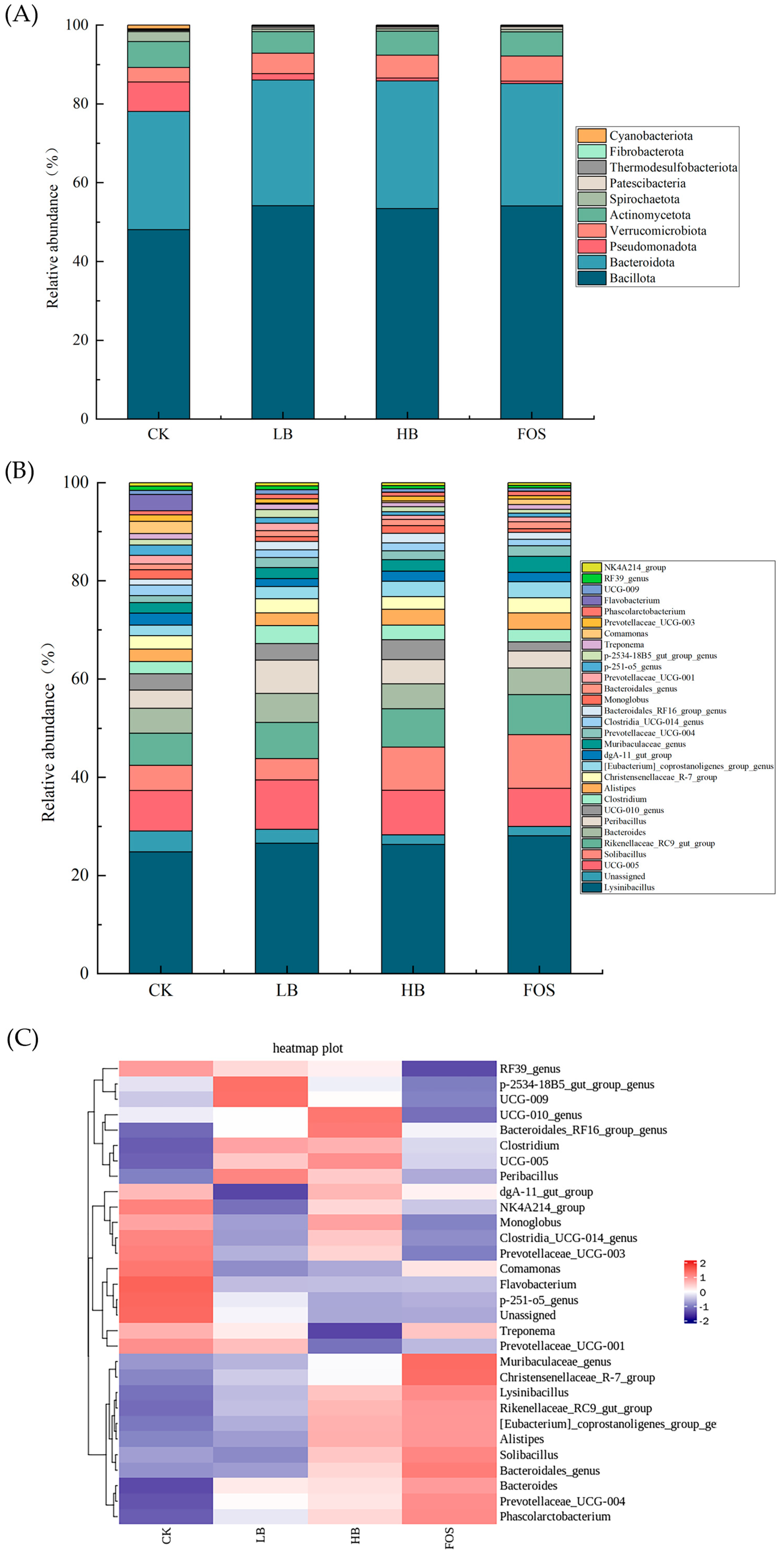
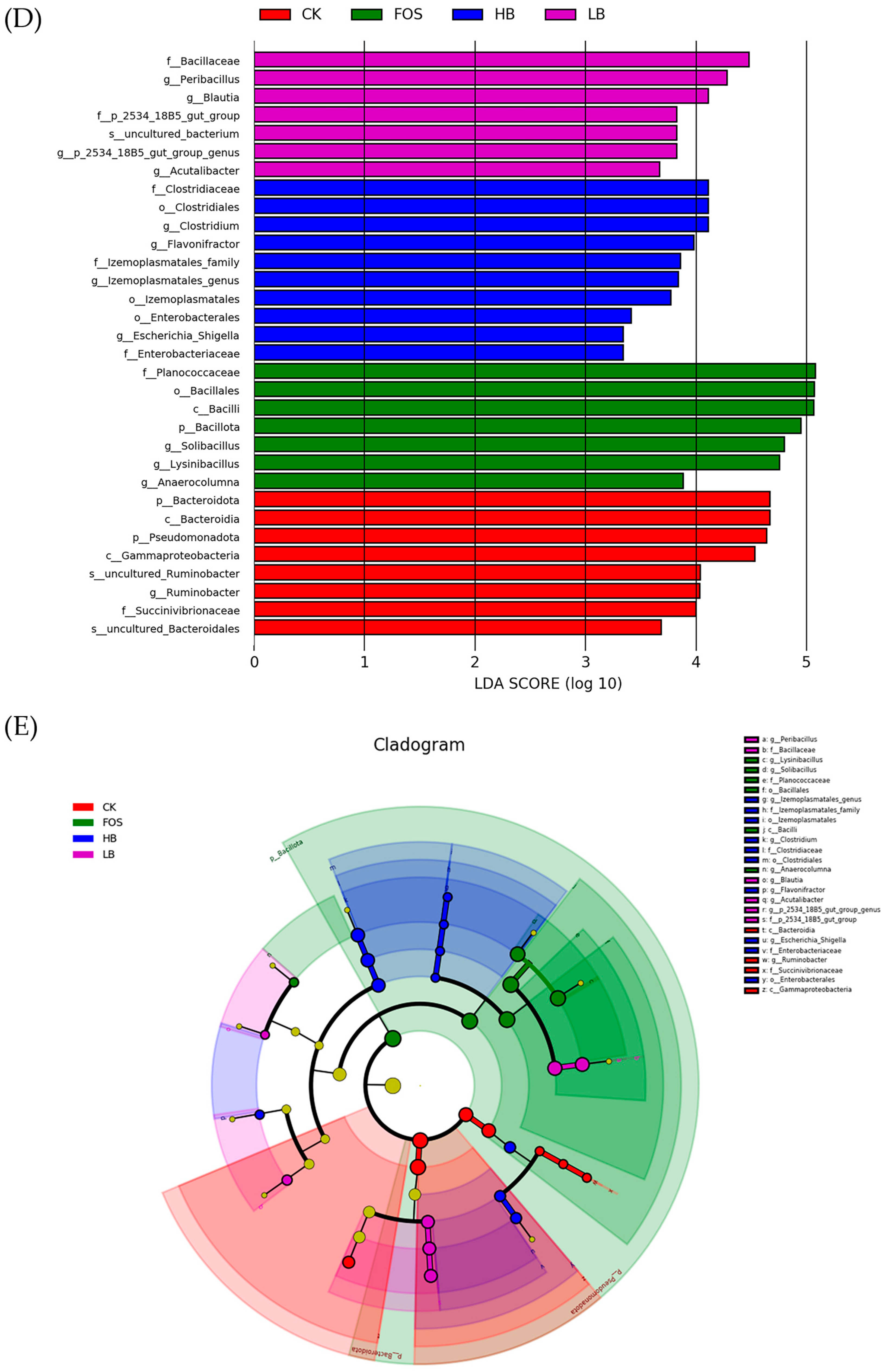
3.3.4. Modulation of the Gut Microbiota Structure by BRAP1-1
3.4. Effect of BRAP1-1 on SCFAs in Mice Feces
3.5. Relationship Between SCFAs and Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, W.Y.; Wang, C.X.; Mayhesumu, X.; Pan, L.; Dang, Y.; Lili, A.; Abuduwaili, A.; Mansur, S. Isolation, Structural elucidation, antioxidant and hypoglycemic activity of polysaccharides of Brassica rapa L. Molecules 2022, 27, 3002. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.Y.; Zhu, H.K.; Liu, C.; Zhang, W.Y.; Li, J.Y.; Hu, B.; Guo, Y.H.; Cheng, Y.L.; Pi, F.W.; Xie, Y.F.; et al. Bioactive compound from the Tibetan turnip (Brassica rapa L.) elicited anti-hypoxia effects in OGD/R-injured HT22 cells by activating the PI3K/AKT pathway. Food Funct. 2021, 12, 2901–2913. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.J.; Zhang, W.Y.; Liu, L.; Cheng, Y.L.; Guo, Y.H.; Yao, W.R.; Qian, H. Fractionation, characterization and anti-fatigue activity of polysaccharides from Brassica Rapa L. Process Biochem. 2021, 106, 163–175. [Google Scholar] [CrossRef]
- Hamed, Y.S.; Ahsan, H.M.; Hussain, M.; Ahmad, I.; Tian, B.; Wang, J.; Zou, X.G.; Bu, T.; Ming, C.; Rayan, A.M.; et al. Polysaccharides from Brassica rapa root: Extraction, purification, structural features, and biological activities. A review. Int. J. Biol. Macromol. 2024, 254, 128023. [Google Scholar] [CrossRef] [PubMed]
- Wufuer, R.; Bai, J.; Liu, Z.; Zhou, K.; Taoerdahong, H.Q. Biological activity of Brassica rapa L. polysaccharides on RAW264.7 macrophages and on tumor cells. Bioorg Med. Chem. 2020, 28, 115330. [Google Scholar] [CrossRef]
- Xu, S.Y.; Chen, X.Q.; Liu, Y.; Cheong, K.L. Ultrasonic/microwave-assisted extraction, simulated digestion, and fermentation in vitro by human intestinal flora of polysaccharides from Porphyra haitanensis. Int. J. Biol. Macromol. 2020, 152, 748–756. [Google Scholar] [CrossRef]
- Wang, N.; Qin, J.Y.; Chen, Z.S.; Wu, J.Y.; Xiang, W.Z. Optimization of ultrasonic-assisted extraction, characterization and antioxidant and immunoregulatory activities of Arthrospira Platensis polysaccharides. Molecules 2024, 29, 4645. [Google Scholar] [CrossRef]
- Yusoff, I.M.; Taher, Z.M.; Rahmat, Z.; Chua, L.S. A review of ultrasound-assisted extraction for plant bioactive compounds: Phenolics, flavonoids, thymols, saponins and proteins. Food Res. Int. 2022, 157, 111268. [Google Scholar] [CrossRef]
- Hu, X.T.; Xu, F.R.; Li, J.L.; Li, J.; Mo, C.; Zhao, M.; Wang, L.F. Ultrasonic-assisted extraction of polysaccharides from coix seeds: Optimization, purification, and in vitro digestibility. Food Chem. 2022, 374, 131636. [Google Scholar] [CrossRef]
- Lin, T.T.; Liu, Y.; Lai, C.J.S.; Yang, T.T.; Xie, J.B.; Zhang, Y.Q. The effect of ultrasound assisted extraction on structural composition, antioxidant activity and immunoregulation of polysaccharides from Ziziphus jujuba Mill var. Spinosa seeds. Ind. Crops Prod. 2018, 125, 150–159. [Google Scholar] [CrossRef]
- Shirsath, S.R.; Sonawane, S.H.; Gogate, P.R. Intensification of extraction of natural products using ultrasonic irradiations—A review of current status. Chem. Eng. Process. Process Intensif. 2012, 53, 10–23. [Google Scholar] [CrossRef]
- Raza, A.; LI, F.; XU, X.Q.; Tang, J. Optimization of ultrasonicassisted extraction of antioxidant polysaccharides from the stem of Trapa quadrispinosa using response surface methodology. Int. J. Biol. Macromol. 2017, 94, 335–344. [Google Scholar] [CrossRef]
- Li, J.; Li, B.; Geng, P.; Song, A.X.; Wu, J.Y. Ultrasonic degradation kinetics and rheological profiles of a food polysaccharide (Konjac glucomannan) in water. Food Hydrocoll. 2017, 70, 14–19. [Google Scholar] [CrossRef]
- Yan, J.K.; Wang, Y.Y.; Ma, H.L.; Wang, Z.B. Ultrasonic effects on the degradation kinetics, preliminary characterization and antioxidant activities of polysaccharides from Phellinus linteus mycelia. Ultrason. Sonochem. 2016, 29, 251–257. [Google Scholar] [CrossRef]
- Tang, W.; Lin, L.H.; Xie, J.H.; Wang, Z.J.; Wang, H.; Dong, Y.J.; Shen, M.Y.; Xie, M.Y. Effect of ultrasonic treatment on the physicochemical properties and antioxidant activities of polysaccharide from Cyclocarya paliurus. Carbohydr. Polym. 2016, 151, 305–312. [Google Scholar] [CrossRef]
- Yu, X.J.; Zhou, C.S.; Yang, H.; Huang, X.Y.; Ma, H.L.; Qin, X.P.; Hu, J.L. Effect of ultrasonic treatment on the degradation and inhibition cancer cell lines of polysaccharides from Porphyra yezoensis. Carbohydr. Polym. 2015, 117, 650–656. [Google Scholar] [CrossRef]
- Yao, Y.; Zhu, Y.Y.; Gao, Y.; Ren, G.X. Effect of ultrasonic treatment on immunological activities of polysaccharides from adlay. Int. J. Biol. Macromol. 2015, 80, 246–252. [Google Scholar] [CrossRef]
- Lee, S.; Goodson, M.; Vang, W.; Kalanetra, K.; Barile, D.; Raybould, H. 2′-fucosyllactose supplementation improves gut-brain signaling and diet-induced obese phenotype and changes the gut microbiota in high fat-fed mice. Nutrients 2020, 12, 1003. [Google Scholar] [CrossRef]
- Li, X.J.; Guo, R.; Wu, X.J.; Liu, X.; Ai, L.Z.; Sheng, Y.; Song, Z.B.; Wu, Y. Dynamic digestion of tamarind seed polysaccharide: Indigestibility in gastrointestinal simulations and gut microbiota changes in vitro. Carbohydr. Polym. 2020, 239, 116194. [Google Scholar] [CrossRef]
- Akhtar, H.M.S.; Abdin, M.; Ahmed, S.; Aslam, F. Digestion by saliva, simulated gastric and small intestinal juices and in vitro fermentation by human gut microbiota of polysaccharides from cicer arietinum l. hulls. J. Microbiol. Biotechnol. Food Sci. 2021, 11, e3966. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, N.; Du, M.X.; Sun, Y.T.; Wang, K.; Wang, Y.J.; Li, D.H.; Yu, H.; Song, Y.; Bai, B. The mouse gut microbial biobank expands the coverage of cultured bacteria. Nat. Commun. 2020, 11, 79. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.Q.; Ye, H.; Hu, B.; Zhou, L.; Jabbar, S.; Zeng, X.X.; Shen, W.B. Optimization of extraction, characterization and antioxidant activity of polysaccharides from Brassica rapa L. Int. J. Biol. Macromol. 2016, 82, 979–988. [Google Scholar] [CrossRef]
- Campestrini, L.H.; Silveira, J.L.; Duarte, M.E.; Koop, H.S.; Noseda, M.D. NMR and rheological study of Aloe barbadensis partially acetylated glucomannan. Carbohydr. Polym. 2013, 94, 511–519. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Rifi, M.; Radwan, Z.; Sari-Chmayssem, N.; Kassir, R.; Fajloun, Z.; Abdel Rahman, A.; El-Sabban, M.; Prévostel, C.; Dassouki, Z.; Mawlawi, H. Exploring the antineoplastic properties of the lebanese Jania Rubens against colorectal cancer. Metabolites 2025, 15, 90. [Google Scholar] [CrossRef]
- Salvador, L.D.; Suganuma, T.; Kitahara, K.; Tanoue, H.; Ichiki, M. Monosaccharide composition of sweetpotato fiber and cell wall polysaccharides from sweetpotato, cassava, and potato analyzed by the high-performance anion exchange chromatography with pulsed amperometric detection method. J. Agric. Food Chem. 2000, 48, 3448–3454. [Google Scholar] [CrossRef]
- Zheng, Z.M.; Huang, Q.L.; Ling, C.Q. Water-soluble yeast β-glucan fractions with different molecular weights: Extraction and separation by acidolysis assisted-size exclusion chromatography and their association with proliferative activity. Int. J. Biol. Macromol. 2018, 123, 269–279. [Google Scholar] [CrossRef]
- Gong, X.B.; Zhang, Z.P.; Shi, X.; Zhu, Y.R.; Ali, F.; Dong, Y.; Zhang, F.; Zhang, B.S. Structural elucidation and anti-psoriasis activity of a novel polysaccharide from Saussurea costus. Carbohydr. Polym. 2024, 333, 121963. [Google Scholar] [CrossRef]
- Basu, S.; Ghosh, M.; Bhunia, R.K.; Ganguly, J.; Banik, B.K. Polysaccharides from Dolichos biflorus linn and Trachyspermum ammi linn seeds: Isolation, characterization and remarkable antimicrobial activity. Chem. Cent. J. 2017, 11, 118. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Yan, C.H.; Lin, X.P.; Ai, C.Q.; Dong, X.P.; Shao, L.; Wang, S.T.; Song, S.; Zhu, B.W. Responses of the gut microbiota and metabolite profiles to sulfated polysaccharides from sea cucumber in humanized microbiota mice. Food Funct. 2022, 13, 4171–4183. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Lundberg, R.; KrychA, L.; Metzdorff, S.B. A humanized diet profile may facilitate colonization and immune stimulation in human microbiota-colonized mice. Front. Microbiol. 2020, 11, 1336. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Ping, Y.; Li, C.X.; Wang, L.H.; Zhao, H. Effects of Atractylodes Macrocephala Rhizoma polysaccharide on intestinal microbiota composition in rats with mammary gland hyperplasia. Front. Endocrinol. 2023, 13, 1102605. [Google Scholar] [CrossRef]
- Leong, Y.K.; Yang, F.C.; Chang, J.S. Extraction of polysaccharides from edible mushrooms: Emerging technologies and recent advances. Carbohydr. Polym. 2021, 251, 117006. [Google Scholar] [CrossRef]
- Zhang, H.j.; Li, H.Z.; Zhang, Z.J.; Hou, T.Y. Optimization of ultrasound-assisted extraction of polysaccharides from perilla seed meal by response surface methodology: Characterization and in vitro antioxidant activities. J. Food Sci. 2021, 86, 306–318. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, J.; Kan, Q.X.; Song, M.Y.; Hou, T.; An, S.Y.; Lin, H.Y.; Chen, H.Z. Extraction, structural characterization, and immunomodulatory activity of a high molecular weight polysaccharide from Ganoderma lucidum. Front. Nutr. 2022, 9, 846080. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.; Deng, Y.F.; Chen, G.H. Effect of extraction method on the structure and bioactivity of polysaccharides from activated sludge. Water Res. 2024, 253, 121196. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, F.M.; Li, Q.; Chen, H.; Zhang, W.J.; Yu, P.; Wu, X.Y. Structure characterization of one polysaccharide from Lepidium meyenii Walp. and its antioxidant activity and protective effect against H2O2-induced induced injury RAW264.7 cells. Int. J. Biol. Macromol. 2018, 118, 816–833. [Google Scholar] [CrossRef]
- Zhou, C.X.; Yu, X.J.; Zhang, Y.Z.; He, R.H.; Ma, H.L. Ultrasonic degradation, purification and analysis of structure and antioxidant activity of polysaccharide from Porphyra yezoensis udea. Carbohydr. Polym. 2012, 87, 2046–2051. [Google Scholar] [CrossRef]
- Ji, X.L.; Guo, J.H.; Ding, D.Q.; Gao, J.; Hao, L.R.; Guo, X.D.; Liu, Y.Q. Structural characterization and antioxidant activity of a novel high-molecular-weight polysaccharide from Ziziphus Jujuba cv. Muzao. J. Food Meas. Charact. 2022, 16, 2191–2200. [Google Scholar] [CrossRef]
- Wang, W.; Zou, Y.; Li, Q.; Mao, R.; Shao, X.; Jin, D.; Zheng, D.; Zhao, T.; Zhu, H.; Zhang, L.; et al. Immunomodulatory effects of a polysaccharide purified from Lepidium meyenii Walp. on macrophages. Process Biochem. 2016, 51, 542–553. [Google Scholar] [CrossRef]
- Huang, N.; Yang, Y.N.; Huang, J.; Shao, H.Y.; Li, Y.L.; Qin, S.H.; Li, H.F.; Shen, X.J.; Yang, L.; Hu, J.M. Structure characterization and immunoactivity on dendritic cells of two neutral polysaccharides from Dictyophora rubrovalvata. Nat. Prod. Bioprospect. 2024, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.H.; Liu, J.; Xia, X.W.; Wong, I.N.; Chung, S.K.; Xu, B.J.; El-Seedi, H.R.; Wang, B.; Huang, R.M. Sulfated heteropolysaccharides from Undaria pinnatifida: Structural characterization and transcript-metabolite profiling of immunostimulatory effects on RAW264.7 cells. Food Chem. X 2022, 13, 100251. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.M.; Jiang, L.; Wang, W.J.; Xiao, Y.H.; Liu, S.C.; Luo, Y.; Shen, M.Y.; Xie, J.H. Effects of Mesona chinensis Benth polysaccharide on physicochemical and rheological properties of sweet potato starch and its interactions. Food Hydrocoll. 2020, 99, 105371. [Google Scholar] [CrossRef]
- Kong, Q.H.; Zhang, R.F.; You, L.J.; Ma, Y.X.; Liao, L.; Pedisi’c, S. In vitro fermentation characteristics of polysaccharide from Sargassum fusiforme and its modulation effects on gut microbiota. Food Chem. Toxicol. 2021, 151, 112145. [Google Scholar] [CrossRef]
- Zheng, Y.J.; Tian, H.L.; Li, Y.; Wang, X.; Shi, P.Q. Effects of carboxymethylation, hydroxypropylation and dual enzyme hydrolysis combination with heating on physicochemical and functional properties and antioxidant activity of coconut cake dietary fibre. Food Chem. 2021, 336, 127688. [Google Scholar] [CrossRef]
- Fărcas, A.C.; Socaci, S.A.; Nemes, S.A.; Salantă, L.C.; Chis, M.S.; Pop, C.R.; Borsa, A.; Diaconeasa, Z.; Vodnar, D.C. Cereal waste valorization through conventional and current extraction techniques-an up-to-date overview. Foods 2022, 11, 2454. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, H.H.; Teng, Y.; Zhang, S.T.; Zhu, B.W.; Xia, X.D. Gut microbiota mediates the anti-colitis effects of polysaccharides derived from Rhopilema esculentum Kishinouye in mice. Food Funct. 2023, 14, 1989–2007. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, W.G.; Wang, H.T.; Yao, L.Y.; He, Z.Y.; Sun, M.; Feng, T.; Yu, C.; Yue, H. Flash extraction of ulvan polysaccharides from marine green macroalga Ulva linza and evaluation of its antioxidant and gut microbiota modulation activities. Int. J. Biol. Macromol. 2024, 262, 130174. [Google Scholar] [CrossRef]
- Ying, M.X.; Yu, Q.; Zheng, B.; Wang, H.; Wang, J.Q.; Chen, S.P.; Nie, S.P.; Xie, M.Y. Cultured Cordyceps sinensis polysaccharides modulate intestinal mucosal immunity and gut microbiota in cyclophosphamide-treated mice. Carbohydr. Polym. 2020, 235, 115957. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef]
- Fouts, D.E.; Szpakowski, S.; Purushe, J.; Torralba, M.; Waterman, R.C.; MacNeil, M.D.; Alexander, L.J.; Nelson, K.E. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS ONE 2012, 7, e48289. [Google Scholar] [CrossRef]
- Hardalo, C.; Edberg, S.C. Pseudomonas aeruginosa: Assessment of risk from drinking water. Crit. Rev. Microbiol. 1997, 23, 47–75. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Liu, X.L.; Chang, S.H.; Zhang, C.; Du, W.C.; Hou, F.J. Effect of Cistanche deserticola on rumen microbiota and rumen function in grazing sheep. Front. Microbiol. 2022, 13, 840725. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS ONE 2016, 11, e0146162. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Karami, A.; Kania, P.; Jubury, A.A. Gut microbiota in rainbow trout Oncorhynchus mykiss with different susceptibility to Flavobacterium psychrophilum infection. Aquaculture 2024, 596, 741841. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, X.; Kong, X. Research progress on prebiotic effects of polysaccharides from edible and medicinal fungi and their effects on gut microecosystem. Acta Edulis Fungi. 2022, 29, 107–116. [Google Scholar]
- Bielawska, K.; Dziakowska, I.; Roszkowska-Jakimiec, W. Chromatographic determination of fatty acids in biological material. Toxicol. Mech. Methods 2010, 20, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Song, W.S.; Park, H.G.; Kim, S.M.; Jo, S.H.; Kim, B.J.; Theberge, A.B.; Kim, Y.J. Chemical derivatization-based LC–MS/MS method for quantitation of gut microbial short-chain fatty acids. J. Ind. Eng. Chem. 2020, 83, 297–302. [Google Scholar] [CrossRef]
- Malairaj, S.; Veeraperumal, S.; Yao, W.Z.; Subramanian, M.; Tan, K.; Zhong, S.Y.; Cheong, K.L. Porphyran from Porphyra haitanensis enhances intestinal barrier function and regulates gut microbiota composition. Mar. Drugs 2023, 21, 265. [Google Scholar] [CrossRef]
- Chang, S.H.; Ko, Y.F.; Liau, J.C.; Wu, C.Y.; Hwang, T.L.; Ojcius, D.M.; Young, J.D.; Martel, J. Hirsutella sinensis polysaccharides and Parabacteroides goldsteinii reduce lupus severity in imiquimod-treated mice. Biomed. J. 2024, 18, 100754. [Google Scholar] [CrossRef]
- Ji, R.; Wang, Z.B.; Kuang, H.X. Extraction, purification, structural characterization, and biological activity of polysaccharides from Schisandra chinensis: A review. Int. J. Biol. Macromol. 2024, 271, 132590. [Google Scholar] [CrossRef]
- He, L.; Yan, X.T.; Liang, J.; Li, S.J.; He, H.R.; Xiong, Q.P.; Lai, X.P.; Hou, S.Z.; Huang, S. Comparison of different extraction methods for polysaccharides from Dendrobium officinale stem. Carbohydr. Polym. 2018, 198, 101–108. [Google Scholar] [CrossRef]
- He, J.L.; Guo, H.; Wei, S.Y.; Zhou, J.; Wu, D.T. Effects of different extraction methods on the structural properties and bioactivities of polysaccharides extracted from Qingke (Tibetan hulless barley). J. Cereal Sci. 2020, 92, 102906. [Google Scholar] [CrossRef]
- Borjigin, G.; Wei, F.S.; Jiang, S.; Li, Q.; Yang, C.J. Extraction, purification, structural characterization and biological activity of polysaccharides from Fritillaria: A review. Int. J. Biol. Macromol. 2023, 242, 124817. [Google Scholar] [CrossRef]
- Ji, X.L.; Cheng, Y.Q.; Tian, J.Y.; Zhang, S.Q.; Jing, Y.S.; Shi, M.M.; Jing, Y.S. Structural characterization of polysaccharide from jujube (Ziziphus jujuba Mill.) fruit. Chem. Biol. Technol. Agric. 2021, 8, 54. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, Q.; Liu, B.; Chen, F.; Wang, M.F. Holothurian fucosylated chondroitin sulfates and their potential benefits for human health: Structures and biological activities. Carbohydr. Polym. 2022, 275, 118691. [Google Scholar] [CrossRef]
- Shao, S.; Wang, D.D.; Zheng, W.; Li, X.Y.; Zhang, H.; Zhao, D.Q.; Wang, M.X. A unique polysac- charide from Hericium erinaceus mycelium ameliorates acetic acid-induced ulcerative colitis rats by modulating the composition of the gut microbiota, short chain fatty acids levels and GPR41/43 respectors. Int. Immunopharmacol. 2019, 71, 411–422. [Google Scholar] [CrossRef]
- Aldars-García, L.; Marin, A.C.; Chaparro, M.; Gisbert, J.P. The interplay between immune system and microbiota in inflammatory bowel disease: A Narrative Review. Int. J. Mol. Sci. 2021, 22, 3076. [Google Scholar] [CrossRef]
- Mills, R.H.; Dulai, P.S.; Vázquez-Baeza, Y.; Sauceda, C.; Daniel, N.; Gerner, R.R.; Batachari, L.E.; Malfavon, M.; Zhu, Q. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat. Microbiol. 2022, 7, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.T.; Howell, K.; Suleria, H.; Zhang, P.Z.; Gu, C.H. Sugarcane polyphenol and fiber to affect production of short-chain fatty acids and microbiota composition using in vitro digestion and pig faecal fermentation model. Food Chem. 2022, 385, 132665. [Google Scholar] [CrossRef] [PubMed]





| Item | Carbohydrate (%) | Protein (%) | Uronic Acid (%) | Sulfuric Radical (%) |
|---|---|---|---|---|
| BRAP | 38.17 ± 1.67 b | 1.86 ± 0.23 a | 11.32 ± 1.38 b | 1.25 ± 0.11 a |
| BRAP1-1 | 73.68 ± 1.81 a | 0.48 ± 0.16 b | 26.11 ± 1.25 a | 0.56 ± 0.12 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Wang, W.; Li, W.; Wang, Z.; Bi, K.; Li, Y.; Wu, Y.; Zhao, Y.; Yang, R.; Du, Q. Ultrasonic-Assisted Extraction of Polysaccharides from Brassica rapa L. and Its Effects on Gut Microbiota in Humanized Mice. Foods 2025, 14, 1994. https://doi.org/10.3390/foods14111994
Zhang M, Wang W, Li W, Wang Z, Bi K, Li Y, Wu Y, Zhao Y, Yang R, Du Q. Ultrasonic-Assisted Extraction of Polysaccharides from Brassica rapa L. and Its Effects on Gut Microbiota in Humanized Mice. Foods. 2025; 14(11):1994. https://doi.org/10.3390/foods14111994
Chicago/Turabian StyleZhang, Mengying, Wei Wang, Wei Li, Zhipeng Wang, Kaiyue Bi, Yanbo Li, Yuhan Wu, Yu Zhao, Rui Yang, and Qingping Du. 2025. "Ultrasonic-Assisted Extraction of Polysaccharides from Brassica rapa L. and Its Effects on Gut Microbiota in Humanized Mice" Foods 14, no. 11: 1994. https://doi.org/10.3390/foods14111994
APA StyleZhang, M., Wang, W., Li, W., Wang, Z., Bi, K., Li, Y., Wu, Y., Zhao, Y., Yang, R., & Du, Q. (2025). Ultrasonic-Assisted Extraction of Polysaccharides from Brassica rapa L. and Its Effects on Gut Microbiota in Humanized Mice. Foods, 14(11), 1994. https://doi.org/10.3390/foods14111994






